Tacrolimus Modulates TGF-β Signaling–Related Genes and MicroRNAs in Human Retinal Pigment Epithelial Cells Activated by Lipopolysaccharide
Abstract
1. Introduction
2. Results
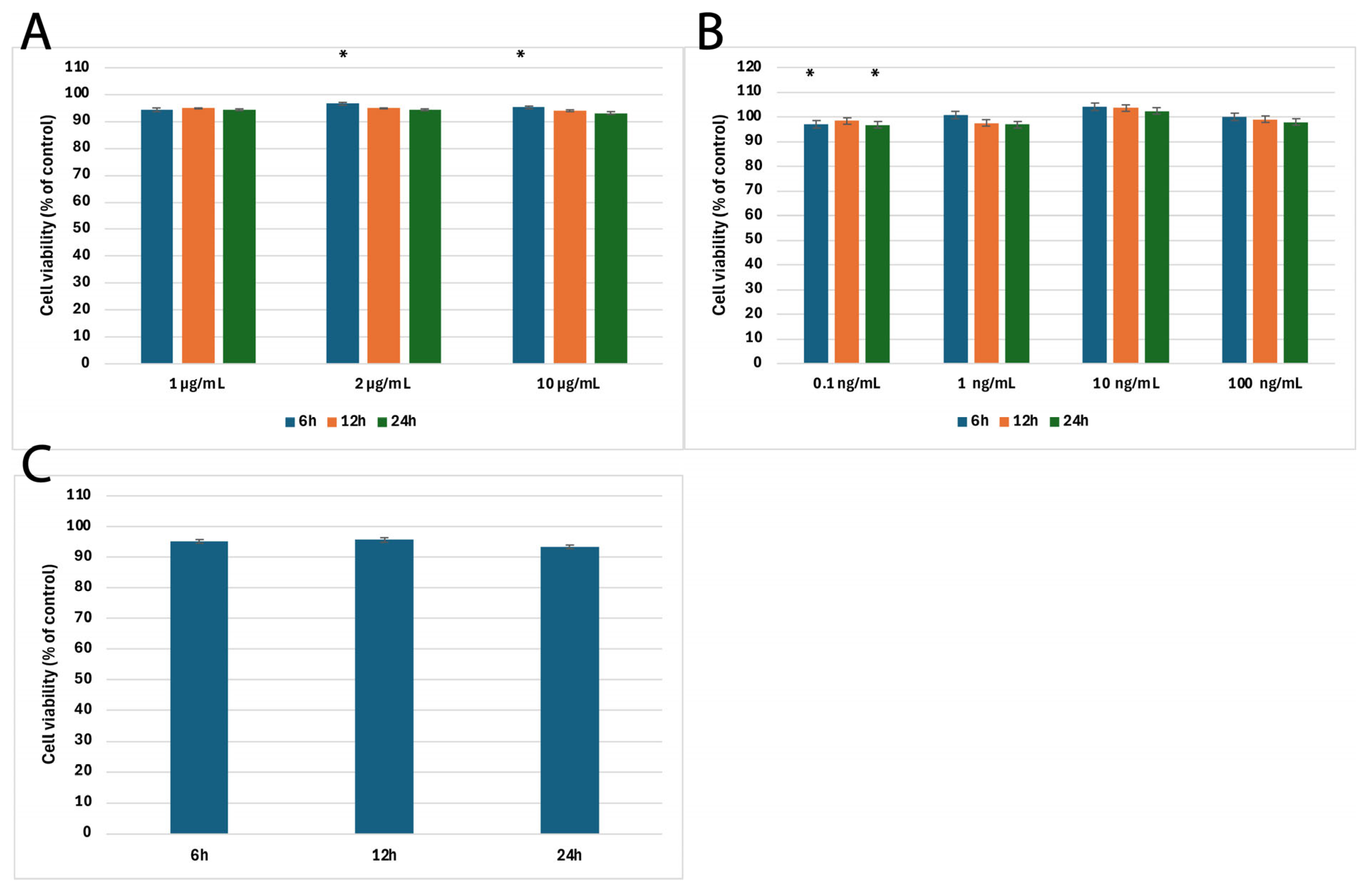
2.1. Results of MTT Cytotoxicity Assay
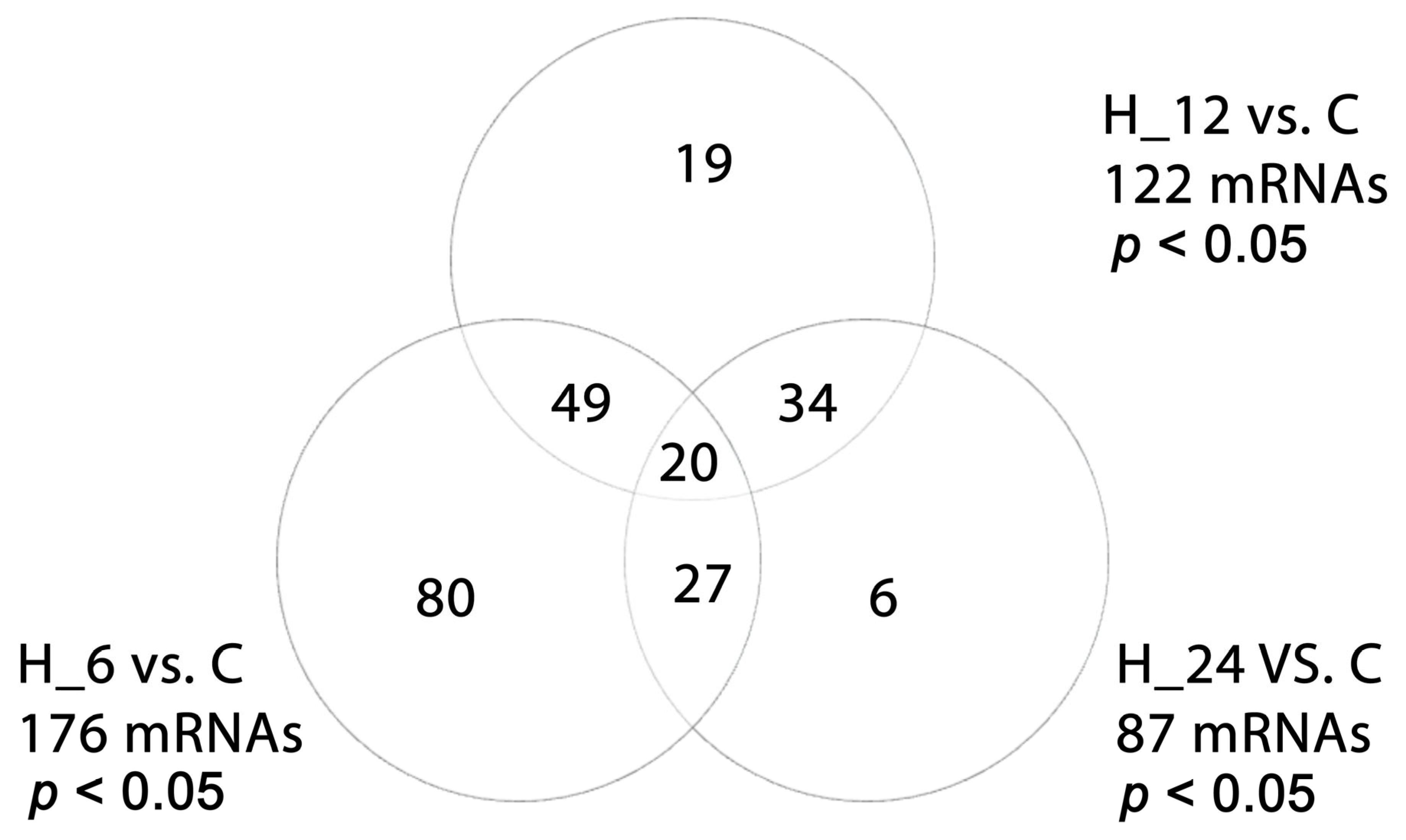
2.2. Differential Microarray Expression of TGF-β-Related Genes in H-RPE Cells Treated with LPS, Tacrolimus, and Their Combination
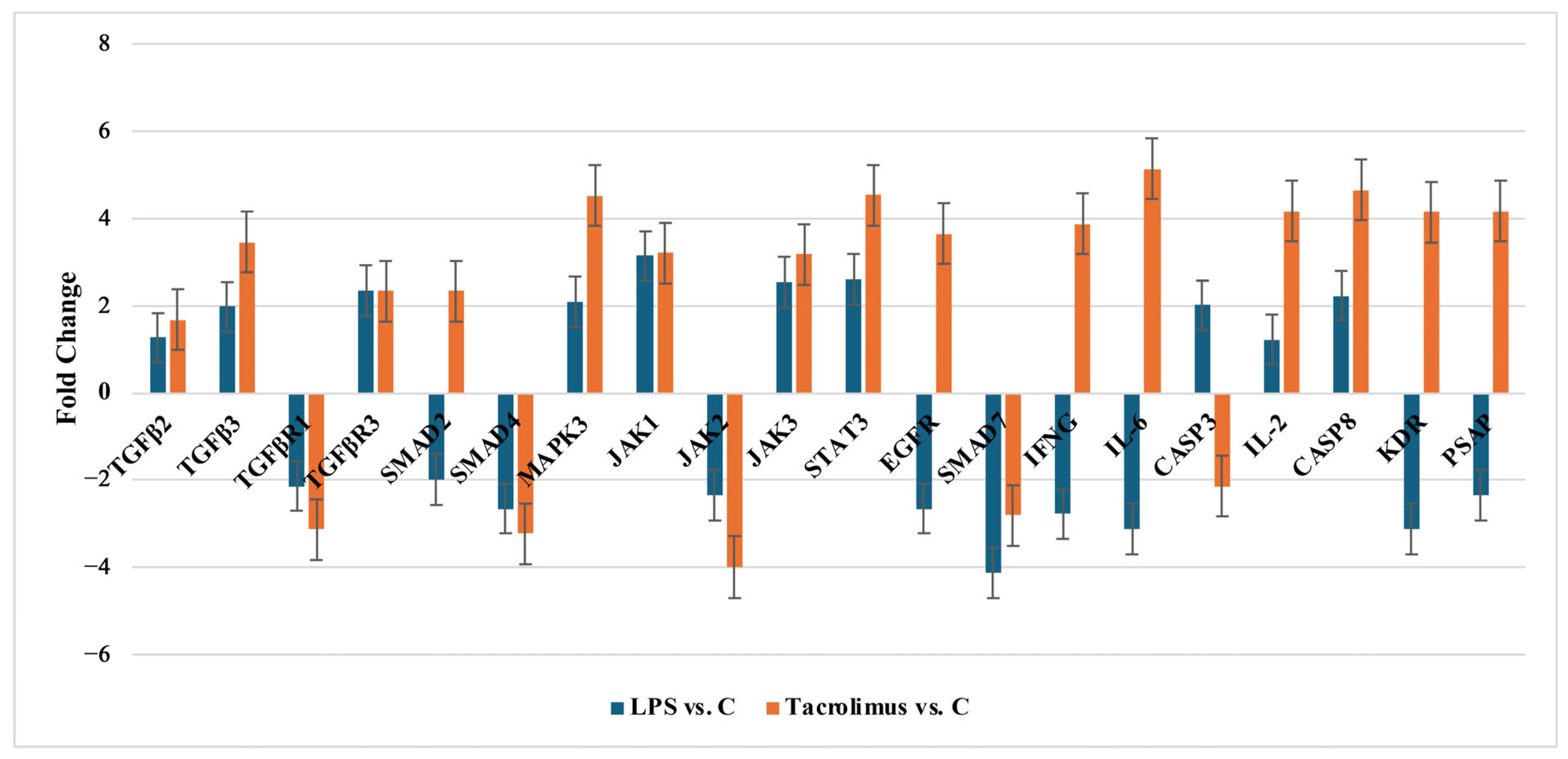
2.2.1. Changes of Selected Genes Related to TGF-β-Family in H-RPE Cells Treated with LPS or Tacrolimus via RT-qPCR
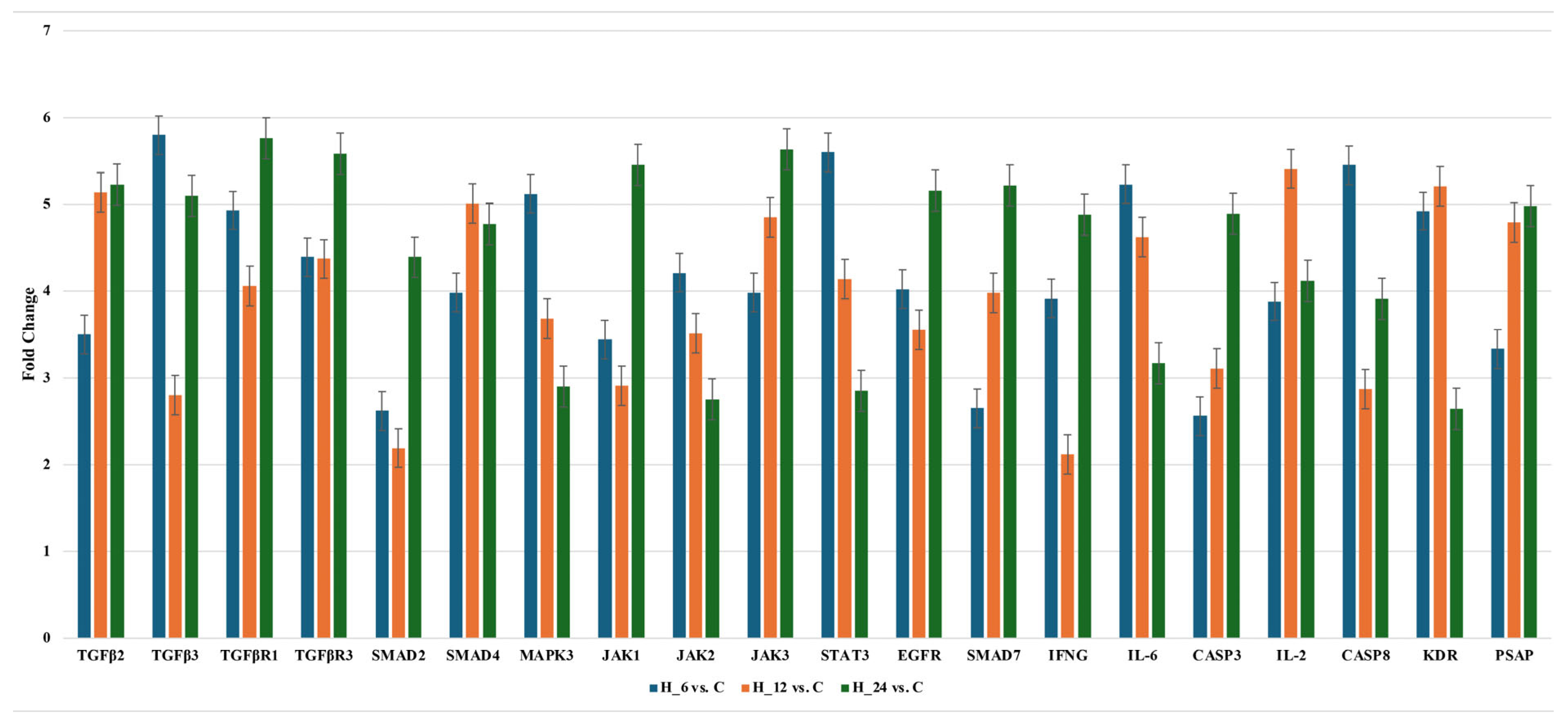
2.2.2. Changes of Selected Genes Related to TGF-β-Family in H-RPE Cells Treated with LPS and Tacrolimus via RT-qPCR
2.3. Differential miRNA Expression in H-RPE Cells Treated with LPS, Tacrolimus, or Their Combination
2.4. Results of Concentration of Selected Proteins Enzyme-Linked Immunosorbent Assay (ELISA Assay)
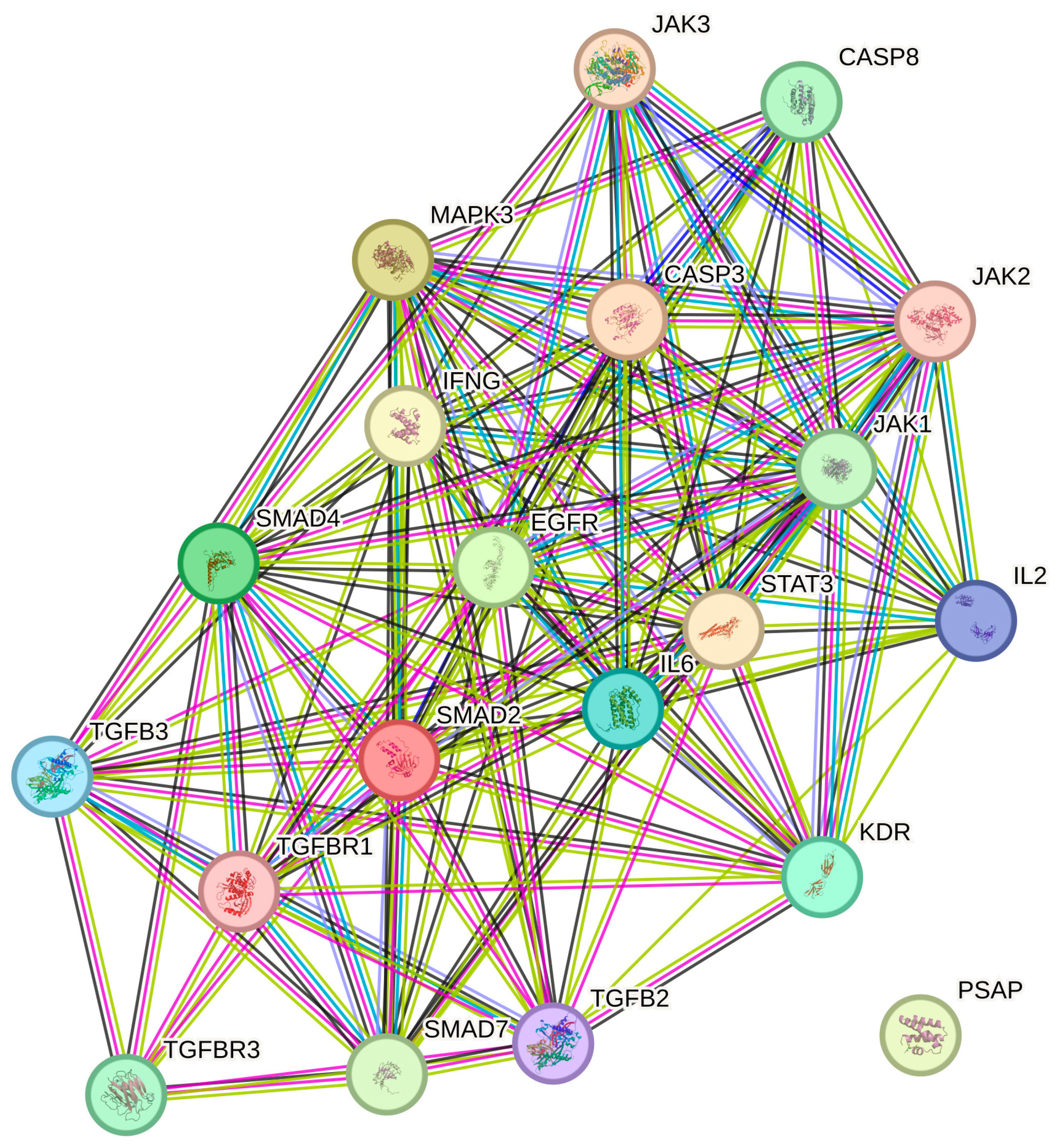
2.5. STRING Analysis
3. Discussion
4. Materials and Methods
4.1. Study Design
4.2. Cell Culture and Cytotoxicity Assay
4.3. Isolation of the Total Ribonucleic Acid (RNA)
4.4. Messenger RNA and miRNAs Microarray Analysis
4.5. RT-qPCR
4.6. ELISA Assay
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACTB | Beta-Actin |
| AMD | Age-Related Macular Degeneration |
| ANOVA | Analysis of Variance |
| C | Control Culture |
| CASP3/8 | Caspase 3/Caspase 8 |
| DR | Diabetic Retinopathy |
| EGFR | Epidermal Growth Factor Receptor |
| ELISA | Enzyme-Linked Immunosorbent Assay |
| EMT | Epithelial–Mesenchymal Transition |
| FBS | Fetal Bovine Serum |
| FDR | False Discovery Rate |
| FGF-B | Fibroblast Growth Factor-Basic |
| GAPDH | Glyceraldehyde-3-Phosphate Dehydrogenase |
| H-RPE | Human Retinal Pigment Epithelial (cells) |
| IL2/6 | Interleukin 2/Interleukin 6 |
| IFNG | Interferon Gamma |
| JAK1/2/3 | Janus Kinase 1/2/3 |
| KDR (VEGFR-2) | Kinase Insert Domain Receptor (Vascular Endothelial Growth Factor Receptor 2) |
| LPS | Lipopolysaccharide |
| MAPK3 | Mitogen-Activated Protein Kinase 3 (ERK1) |
| miRNA | microRNA |
| MTT | 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide |
| NFAT | Nuclear Factor of Activated T Cells |
| PCR | Polymerase Chain Reaction |
| PPI | Protein–Protein Interaction |
| PVR | Proliferative Vitreoretinopathy |
| PSAP | Prosaposin |
| RNA | Ribonucleic Acid |
| RT-qPCR | Quantitative Reverse-Transcription Polymerase Chain Reaction |
| SMAD2/4/7 | Mothers Against Decapentaplegic Homolog 2/4/7 |
| STAT3 | Signal Transducer and Activator of Transcription 3 |
| STRING | Search Tool for the Retrieval of Interacting Genes/Proteins |
| TGF-β1/2/3 | Transforming Growth Factor Beta 1/2/3 |
| TGF-βR1/3 | Transforming Growth Factor Beta Receptor 1/3 |
| VEGFR-2 | Vascular Endothelial Growth Factor Receptor 2 |
Appendix A
| mRNA | LPS vc. C | Tacrolimus vs. C |
|---|---|---|
| TGF-β2 | NS | p = 0.043 |
| TGF-β3 | p = 0.021 | p = 0.012 |
| TGF-βR1 | p = 0.041 | p = 0.013 |
| TGF-βR3 | p = 0.003 | p = 0.011 |
| SMAD2 | NS | p = 0.025 |
| SMAD4 | p = 0.009 | p = 0.028 |
| SMAD7 | p = 0.001 | p = 0.029 |
| JAK1 | p = 0.019 | p = 0.013 |
| JAK2 | p = 0.041 | p = 0.027 |
| JAK3 | p = 0.039 | p = 0.021 |
| STAT3 | p = 0.047 | p = 0.001 |
| EGFR | p = 0.023 | p = 0.017 |
| MAPK3 | p = 0.041 | p = 0.001 |
| IFNG | p = 0.034 | p = 0.031 |
| IL-6 | p = 0.002 | p = 0.012 |
| CASP3 | p = 0.049 | p = 0.043 |
| CASP8 | p = 0.047 | p = 0.013 |
| IL-2 | NS | p = 0.014 |
| KDR | p = 0.022 | p = 0.019 |
| PSAP | p = 0.039 | p = 0.038 |
| mRNA | ANOVA p-Value | Scheffé’s Post Hoc Test Results | ||
|---|---|---|---|---|
| H_6 vs. C | H_12 vs. C | H_24 vs. C | ||
| TGF-β2 | 0.002 | p = 0.021 | p = 0.008 | p = 0.016 |
| TGF-β3 | 0.005 | NS | p = 0.033 | p = 0.009 |
| TGF-βR1 | 0.001 | p = 0.006 | p = 0.018 | p = 0.041 |
| TGF-βR3 | 0.010 | p = 0.028 | p = 0.019 | NS |
| SMAD2 | 0.012 | p = 0.036 | p = 0.031 | p = 0.007 |
| SMAD4 | 0.020 | NS | p = 0.044 | p = 0.009 |
| SMAD7 | 0.003 | p = 0.022 | p = 0.005 | p = 0.019 |
| JAK1 | 0.008 | p = 0.017 | p = 0.011 | p = 0.038 |
| JAK2 | 0.018 | p = 0.031 | p = 0.006 | p = 0.045 |
| JAK3 | 0.035 | NS | p = 0.027 | p = 0.033 |
| STAT3 | 0.002 | p = 0.015 | p = 0.009 | p = 0.007 |
| EGFR | 0.009 | p = 0.023 | p = 0.032 | p = 0.041 |
| MAPK3 | 0.006 | p = 0.037 | p = 0.004 | p = 0.033 |
| IFNG | 0.004 | p = 0.029 | p = 0.007 | p = 0.015 |
| IL6 | 0.005 | p = 0.034 | p = 0.016 | p = 0.008 |
| CASP3 | 0.001 | p = 0.022 | p = 0.009 | p = 0.005 |
| CASP8 | 0.004 | p = 0.033 | p = 0.021 | p = 0.008 |
| IL-2 | 0.031 | NS | p = 0.044 | NS |
| KDR | 0.014 | p = 0.008 | NS | NS |
| PSAP | 0.003 | p = 0.025 | p = 0.038 | p = 0.012 |
References
- Markitantova, Y.; Simirskii, V. Endogenous and Exogenous Regulation of Redox Homeostasis in Retinal Pigment Epithelium Cells: An Updated Antioxidant Perspective. Int. J. Mol. Sci. 2023, 24, 10776. [Google Scholar] [CrossRef] [PubMed]
- Taylor, A.W.; Hsu, S.; Ng, T.F. The Role of Retinal Pigment Epithelial Cells in Regulation of Macrophages/Microglial Cells in Retinal Immunobiology. Front. Immunol. 2021, 12, 724601. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Zhou, J.; Li, D. Functions and Diseases of the Retinal Pigment Epithelium. Front. Pharmacol. 2021, 12, 727870. [Google Scholar] [CrossRef] [PubMed]
- Pastor, J.C.; Rojas, J.; Pastor-Idoate, S.; Di Lauro, S.; Gonzalez-Buendia, L.; Delgado-Tirado, S. Proliferative Vitreoretinopathy: A New Concept of Disease Pathogenesis and Practical Consequences. Prog. Retin. Eye Res. 2016, 51, 125–155. [Google Scholar] [CrossRef]
- Rizzo, S.; De Angelis, L.; Barca, F.; Bacherini, D.; Vannozzi, L.; Giansanti, F.; Faraldi, F.; Caporossi, T. Vitreoschisis and Retinal Detachment: New Insight in Proliferative Vitreoretinopathy. Eur. J. Ophthalmol. 2022, 32, 2833–2839. [Google Scholar] [CrossRef]
- Ananikas, K.; Stavrakas, P.; Kroupis, C.; Christou, E.E.; Brouzas, D.; Petrou, P.; Papakonstantinou, D. Molecular Biologic Milieu in Rhegmatogenous Retinal Detachment and Proliferative Vitreoretinopathy: A Literature Review. Ophthalmic Res. 2022, 65, 637–646. [Google Scholar] [CrossRef]
- Van Overdam, K.A.; Van Den Bosch, T.P.P.; Van Etten, P.G.; Uppal, G.S.; Veckeneer, M.; Verdijk, R.M. Novel Insights into the Pathophysiology of Proliferative Vitreoretinopathy: The Role of Vitreoschisis-induced Vitreous Cortex Remnants. Acta Ophthalmol. 2022, 100, E1749–E1759. [Google Scholar] [CrossRef]
- Yang, S.; Li, H.; Yao, H.; Zhang, Y.; Bao, H.; Wu, L.; Zhang, C.; Li, M.; Feng, L.; Zhang, J. Long Noncoding RNA ERLR Mediates Epithelial-Mesenchymal Transition of Retinal Pigment Epithelial Cells and Promotes Experimental Proliferative Vitreoretinopathy. Cell Death Differ. 2021, 28, 2351–2366. [Google Scholar] [CrossRef]
- Datlibagi, A.; Zein-El-Din, A.; Frohly, M.; Willermain, F.; Delporte, C.; Motulsky, E. Experimental Models to Study Epithelial-Mesenchymal Transition in Proliferative Vitreoretinopathy. Int. J. Mol. Sci. 2023, 24, 4509. [Google Scholar] [CrossRef]
- Li, X.; Zhao, M.; He, S. RPE Epithelial-Mesenchymal Transition Plays a Critical Role in the Pathogenesis of Proliferative Vitreoretinopathy. Ann. Transl. Med. 2020, 8, 263. [Google Scholar] [CrossRef]
- Mudhar, H.S. A Brief Review of the Histopathology of Proliferative Vitreoretinopathy (PVR). Eye 2020, 34, 246–250. [Google Scholar] [CrossRef] [PubMed]
- Zandi, S.; Pfister, I.B.; Traine, P.G.; Tappeiner, C.; Despont, A.; Rieben, R.; Skowronska, M.; Garweg, J.G. Biomarkers for PVR in Rhegmatogenous Retinal Detachment. PLoS ONE 2019, 14, e0214674. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Dai, C.; Sun, T. Inflammatory Mediators of Proliferative Vitreoretinopathy: Hypothesis and Review. Int. Ophthalmol. 2020, 40, 1587–1601. [Google Scholar] [CrossRef] [PubMed]
- Kimsa, M.; Strzalka-Mrozik, B.; Kimsa-Dudek, M.; Kruszniewska-Rajs, C.; Gola, J.; Adamska, J.; Mazurek, U. Transforming Growth Factor β-Related Genes in Human Retinal Pigment Epithelial Cells after Tacrolimus Treatment. Pharmacol. Rep. 2016, 68, 969–974. [Google Scholar] [CrossRef]
- Wu, F.; Eliott, D. Molecular Targets for Proliferative Vitreoretinopathy. Semin. Ophthalmol. 2021, 36, 218–223. [Google Scholar] [CrossRef]
- Kaneko, H.; Terasaki, H. Biological Involvement of MicroRNAs in Proliferative Vitreoretinopathy. Transl. Vis. Sci. Technol. 2017, 6, 5. [Google Scholar] [CrossRef]
- Peng, D.; Fu, M.; Wang, M.; Wei, Y.; Wei, X. Targeting TGF-β Signal Transduction for Fibrosis and Cancer Therapy. Mol. Cancer 2022, 21, 104. [Google Scholar] [CrossRef]
- Wilson, S.E. TGF Beta-1, -2 and -3 in the Modulation of Fibrosis in the Cornea and Other Organs. Exp. Eye Res. 2021, 207, 108594. [Google Scholar] [CrossRef]
- Zhou, M.; Geathers, J.S.; Grillo, S.L.; Weber, S.R.; Wang, W.; Zhao, Y.; Sundstrom, J.M. Role of Epithelial-Mesenchymal Transition in Retinal Pigment Epithelium Dysfunction. Front. Cell Dev. Biol. 2020, 8, 501. [Google Scholar] [CrossRef]
- Yeganeh, B.; Mukherjee, S.; Moir, L.M.; Kumawat, K.; Kashani, H.H.; Bagchi, R.A.; Baarsma, H.A.; Gosens, R.; Ghavami, S. Novel Non-Canonical TGF-β Signaling Networks: Emerging Roles in Airway Smooth Muscle Phenotype and Function. Pulm. Pharmacol. Ther. 2013, 26, 50–63. [Google Scholar] [CrossRef]
- Trelford, C.B.; Di Guglielmo, G.M. Canonical and Non-Canonical TGFβ Signaling Activate Autophagy in an ULK1-Dependent Manner. Front. Cell Dev. Biol. 2021, 9, 712124. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Myoung, H.; Kim, S.M. Review of Two Immunosuppressants: Tacrolimus and Cyclosporine. J. Korean Assoc. Oral Maxillofac. Surg. 2023, 49, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Daeschler, S.C.; Feinberg, K.; Harhaus, L.; Kneser, U.; Gordon, T.; Borschel, G.H. Advancing Nerve Regeneration: Translational Perspectives of Tacrolimus (FK506). Int. J. Mol. Sci. 2023, 24, 12771. [Google Scholar] [CrossRef] [PubMed]
- Satake, M.; Sakuraba, H.; Hiraga, H.; Yoshida, S.; Akemoto, Y.; Ota, S.; Murai, Y.; Hasui, K.; Kawaguchi, S.; Kikuchi, H. Tacrolimus Up-Regulates Expression of TGFβ Receptor Type II via ERK, Providing Protection against Intestinal Epithelial Injury. In Vivo 2022, 36, 1684–1693. [Google Scholar] [CrossRef]
- Di Lorenzo, F.; Duda, K.A.; Lanzetta, R.; Silipo, A.; De Castro, C.; Molinaro, A. A Journey from Structure to Function of Bacterial Lipopolysaccharides. Chem. Rev. 2022, 122, 15767–15821. [Google Scholar] [CrossRef]
- Page, M.J.; Kell, D.B.; Pretorius, E. The Role of Lipopolysaccharide-Induced Cell Signalling in Chronic Inflammation. Chronic Stress 2022, 6, 24705470221076390. [Google Scholar] [CrossRef]
- Ho, P.T.; Clark, I.M.; Le, L.T. MicroRNA-Based Diagnosis and Therapy. Int. J. Mol. Sci. 2022, 23, 7167. [Google Scholar] [CrossRef]
- Seyhan, A.A. Trials and Tribulations of MicroRNA Therapeutics. Int. J. Mol. Sci. 2024, 25, 1469. [Google Scholar] [CrossRef]
- Xu, X.; Hong, P.; Wang, Z.; Tang, Z.; Li, K. MicroRNAs in Transforming Growth Factor-Beta Signaling Pathway Associated with Fibrosis Involving Different Systems of the Human Body. Front. Mol. Biosci. 2021, 8, 707461. [Google Scholar] [CrossRef]
- Zou, X.-Z.; Liu, T.; Gong, Z.-C.; Hu, C.-P.; Zhang, Z. MicroRNAs-Mediated Epithelial-Mesenchymal Transition in Fibrotic Diseases. Eur. J. Pharmacol. 2017, 796, 190–206. [Google Scholar] [CrossRef]
- Zhang, H.; Zhou, Y.; Wen, D.; Wang, J. Noncoding RNAs: Master Regulator of Fibroblast to Myofibroblast Transition in Fibrosis. Int. J. Mol. Sci. 2023, 24, 1801. [Google Scholar] [CrossRef]
- Fujita, E.; Teramura, Y.; Shiraga, T.; Yoshioka, S.-I.; Iwatsubo, T.; Kawamura, A.; Kamimura, H. Pharmacokinetics and Tissue Distribution of Tacrolimus (FK506) after a Single or Repeated Ocular Instillation in Rabbits. J. Ocul. Pharmacol. Ther. 2008, 24, 309–319. [Google Scholar] [CrossRef]
- Bian, Z.-M.; Elner, S.G.; Khanna, H.; Murga-Zamalloa, C.A.; Patil, S.; Elner, V.M. Expression and Functional Roles of Caspase-5 in Inflammatory Responses of Human Retinal Pigment Epithelial Cells. Investig. Ophthalmol. Vis. Sci. 2011, 52, 8646–8656. Available online: https://iovs.arvojournals.org/article.aspx?articleid=2186809 (accessed on 13 March 2025). [CrossRef]
- Elner, S.G.; Petty, H.R.; Elner, V.M.; Yoshida, A.; Bian, Z.-M.; Yang, D.; Kindezelskii, A.L. TLR4 Mediates Human Retinal Pigment Epithelial Endotoxin Binding and Cytokine Expression. Trans. Am. Ophthalmol. Soc. 2005, 103, 126–137. [Google Scholar] [CrossRef]
- Nagano, J.; Iyonaga, K.; Kawamura, K.; Yamashita, A.; Ichiyasu, H.; Okamoto, T.; Suga, M.; Sasaki, Y.; Kohrogi, H. Use of Tacrolimus, a Potent Antifibrotic Agent, in Bleomycin-Induced Lung Fibrosis. Eur. Respir. J. 2006, 27, 460–469. [Google Scholar] [CrossRef]
- Ebihara, N.; Ohtomo, K.; Tokura, T.; Ushio, H.; Murakami, A. Effect of Tacrolimus on Chemokine Production by Corneal Myofibroblasts via Toll-like Receptors, Compared with Cyclosporine and Dexamethasone. Cornea 2011, 30, 702–708. [Google Scholar] [CrossRef]
- Patsenker, E.; Schneider, V.; Ledermann, M.; Saegesser, H.; Dorn, C.; Hellerbrand, C.; Stickel, F. Potent Antifibrotic Activity of mTOR Inhibitors Sirolimus and Everolimus but Not of Cyclosporine A and Tacrolimus in Experimental Liver Fibrosis. J. Hepatol. 2011, 55, 388–398. [Google Scholar] [CrossRef]
- Budi, E.H.; Schaub, J.R.; Decaris, M.; Turner, S.; Derynck, R. TGF-β as a Driver of Fibrosis: Physiological Roles and Therapeutic Opportunities. J. Pathol. 2021, 254, 358–373. [Google Scholar] [CrossRef]
- Rodari, M.M.; Cerf-Bensussan, N.; Parlato, M. Dysregulation of the Immune Response in TGF-β Signalopathies. Front. Immunol. 2022, 13, 1066375. [Google Scholar] [CrossRef]
- Harju, N.; Kauppinen, A.; Loukovaara, S. Fibrotic Changes in Rhegmatogenous Retinal Detachment. Int. J. Mol. Sci. 2025, 26, 1025. [Google Scholar] [CrossRef]
- Carpineto, P.; Licata, A.M.; Ciancaglini, M. Proliferative Vitreoretinopathy: A Reappraisal. J. Clin. Med. 2023, 12, 5287. [Google Scholar] [CrossRef] [PubMed]
- Rivelli, R.F.; Gonçalves, R.T.; Leite, M.; Santos, M.A.R.; Delgado, A.G.; Cardoso, L.R.; Takiya, C.M. Early Withdrawal of Calcineurin Inhibitor from a Sirolimus-Based Immunosuppression Stabilizes Fibrosis and the Transforming Growth Factor-β Signalling Pathway in Kidney Transplant. Nephrology 2015, 20, 168–176. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Guo, J.; Pang, X.-L.; Shang, W.-J.; Wang, Z.-G.; Wang, J.-X.; Yang, X.-L.; Feng, G.-W. Exploration of the Mechanism of NORAD Activation of TGF-Β1/Smad3 through miR-136-5p and Promotion of Tacrolimus-Induced Renal Fibrosis. Ren. Fail. 2023, 45, 2147083. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Wu, K.; Feng, F.; Wang, L.; Zhou, X.; Wang, W. Astragaloside IV Alleviates Silica-Induced Pulmonary Fibrosis via Inactivation of the TGF-Β1/Smad2/3 Signaling Pathway. Int. J. Mol. Med. 2021, 47, 16. [Google Scholar] [CrossRef]
- Ume, A. Friend or Foe? The Role of Transforming Growth Factor-β (TGFβ) Signaling in Calcineurin Inhibitor-Induced Renal Damage. Ph.D. Thesis, Wright State University, Dayton, OH, USA, 2023. [Google Scholar]
- Alqudah, A.; AbuDalo, R.; Qnais, E.; Wedyan, M.; Oqal, M.; McClements, L. The Emerging Importance of Immunophilins in Fibrosis Development. Mol. Cell. Biochem. 2023, 478, 1281–1291. [Google Scholar] [CrossRef]
- Ume, A.C.; Wenegieme, T.Y.; Shelby, J.N.; Paul-Onyia, C.D.B.; Waite, A.M.J.; Kamau, J.K.; Adams, D.N.; Susuki, K.; Bennett, E.S.; Ren, H.; et al. Tacrolimus Induces Fibroblast-to-Myofibroblast Transition via a TGF-β-Dependent Mechanism to Contribute to Renal Fibrosis. Am. J. Physiol.-Ren. Physiol. 2023, 324, F433–F445. [Google Scholar] [CrossRef]
- Liu, B.; Jiang, Q.; Chen, R.; Gao, S.; Xia, Q.; Zhu, J.; Zhang, F.; Shao, C.; Liu, X.; Li, X.; et al. Tacrolimus Ameliorates Bleomycin-Induced Pulmonary Fibrosis by Inhibiting M2 Macrophage Polarization via JAK2/STAT3 Signaling. Int. Immunopharmacol. 2022, 113, 109424. [Google Scholar] [CrossRef]
- Choe, J.-Y.; Park, K.-Y.; Park, S.-H.; Lee, S.-I.; Kim, S.-K. Regulatory Effect of Calcineurin Inhibitor, Tacrolimus, on IL-6/sIL-6R-Mediated RANKL Expression through JAK2-STAT3-SOCS3 Signaling Pathway in Fibroblast-like Synoviocytes. Arthritis Res. Ther. 2013, 15, R26. [Google Scholar] [CrossRef]
- Li, Y.; Wang, Y.; Li, J.; Ling, Z.; Chen, W.; Zhang, L.; Hu, Q.; Wu, T.; Cheng, B.; Wang, Y. Tacrolimus Inhibits Oral Carcinogenesis through Cell Cycle Control. Biomed. Pharmacother. 2021, 139, 111545. [Google Scholar] [CrossRef]
- Choi, Y.H. Tacrolimus Induces Apoptosis in Leukemia Jurkat Cells through Inactivation of the Reactive Oxygen Species-Dependent Phosphoinositide-3-Kinase/Akt Signaling Pathway. Biotechnol. Bioprocess Eng. 2022, 27, 183–192. [Google Scholar] [CrossRef]
- Tang, J.; Li, X.; Cheng, T.; Wu, J. miR-21-5p/SMAD7 Axis Promotes the Progress of Lung Cancer. Thorac. Cancer 2021, 12, 2307–2313. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Ye, M.; Wang, F.; Fang, J.; Wang, C.; Luo, J.; Liu, J.; Liu, J.; Liu, L.; Zhao, Q. MiR-21-3p Promotes Hepatocellular Carcinoma Progression via SMAD7/YAP1 Regulation. Front. Oncol. 2021, 11, 642030. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yang, N.; Cheng, Q.; Zhang, H.; Liu, F.; Shang, Y. MiR-21-5p in Macrophage-Derived Exosomes Targets Smad7 to Promote Epithelial Mesenchymal Transition of Airway Epithelial Cells. J. Asthma Allergy 2021, 14, 513–524. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.S.; Roundtree, I.A.; He, C. Post-Transcriptional Gene Regulation by mRNA Modifications. Nat. Rev. Mol. Cell Biol. 2017, 18, 31–42. [Google Scholar] [CrossRef]
- Jens, M.; Rajewsky, N. Competition between Target Sites of Regulators Shapes Post-Transcriptional Gene Regulation. Nat. Rev. Genet. 2015, 16, 113–126. [Google Scholar] [CrossRef]
- Regis, S.; Dondero, A.; Caliendo, F.; Bottino, C.; Castriconi, R. NK Cell Function Regulation by TGF-β-Induced Epigenetic Mechanisms. Front. Immunol. 2020, 11, 311. [Google Scholar] [CrossRef]
- Klinke, A.; Schubert, T.; Müller, M.; Legchenko, E.; Zelt, J.G.E.; Shimauchi, T.; Napp, L.C.; Rothman, A.M.K.; Bonnet, S.; Stewart, D.J.; et al. Emerging Therapies for Right Ventricular Dysfunction and Failure. Cardiovasc. Diagn. Ther. 2020, 10, 1735–1767. [Google Scholar] [CrossRef]
- Chakraborty, C.; Sharma, A.R.; Sharma, G.; Doss, C.G.P.; Lee, S.-S. Therapeutic miRNA and siRNA: Moving from Bench to Clinic as next Generation Medicine. Mol. Ther. Nucleic Acids 2017, 8, 132–143. [Google Scholar] [CrossRef]
- Chaudhary, R.; Scott, R.A.H.; Wallace, G.; Berry, M.; Logan, A.; Blanch, R.J. Inflammatory and Fibrogenic Factors in Proliferative Vitreoretinopathy Development. Transl. Vis. Sci. Technol. 2020, 9, 23. [Google Scholar] [CrossRef]
- Qiu, R.; Yang, M.; Wang, W.; Liu, J.; Yang, L.; Lei, B. The Protective Effects of VVN001 on LPS-Induced Inflammatory Responses in Human RPE Cells and in a Mouse Model of EIU. Inflammation 2021, 44, 780–794. [Google Scholar] [CrossRef]
- Klettner, A.; Roider, J. Retinal Pigment Epithelium Expressed Toll-like Receptors and Their Potential Role in Age-Related Macular Degeneration. Int. J. Mol. Sci. 2021, 22, 8387. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting Effective microRNA Target Sites in Mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, X. miRDB: An Online Database for Prediction of Functional microRNA Targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Wang, X. Prediction of Functional microRNA Targets by Integrative Modeling of microRNA Binding and Target Expression Data. Genome Biol. 2019, 20, 18. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING Database in 2023: Protein-Protein Association Networks and Functional Enrichment Analyses for Any Sequenced Genome of Interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
| mRNA | LPS | Tacrolimus | LPS + Tacrolimus | ||
|---|---|---|---|---|---|
| log2Fold Change | |||||
| H_6 vs. C | H_6 vs. C | H_6 vs. C | H_12 vs. C | H_24 vs. C | |
| TGF-β2 | +1.27 ± 0.17 * | +1.67 ± 0.65 * | +3.50 ± 0.39 * | +5.14 ± 0.11 * | +5.88 ± 0.27 * |
| TGF-β3 | +1.98 ± 0.54 * | +3.45 ± 0.43 * | +5.80 ± 0.41 * | +2.80 ± 0.35 * | +5.10 ± 0.43 * |
| TGF-βR1 | −2.13 ± 0.44 * | −3.12 ± 0.76 * | −4.93 ± 0.13 * | −4.06 ± 0.23 * | −5.76 ± 0.44 * |
| TGF-βR3 | +2.19 ± 0.31 * | +2.23 ± 0.34 * | +4.39 ± 0.24 * | +4.37 ± 0.30 * | +5.58 ± 0.10 * |
| SMAD2 | −1.98 ± 0.57 * | +2.54 ± 0.51 * | +2.62 ± 0.15 * | +2.19 ± 0.46 * | +4.39 ± 0.30 * |
| SMAD4 | −2.65 ± 0.81 * | −3.45 ± 0.67 * | −3.21 ± 0.37 * | +5.01 ± 0.28 * | +4.77 ± 0.39 * |
| MAPK3 | +2.09 ± 0.41 * | +5.17 ± 0.61 * | +5.12 ± 0.34 * | +3.68 ± 0.31 * | −2.90 ± 0.41 * |
| JAK1 | +3.45 ± 0.48 * | +3.21 ± 0.24 * | +3.44 ± 0.20 * | −2.99 ± 0.36 * | +5.45 ± 0.38 * |
| JAK2 | −2.34 ± 0.54 * | −3.99 ± 0.81 * | −4.21 ± 0.18 * | −3.51 ± 0.42 * | −2.75 ± 0.37 * |
| JAK3 | +2.54 ± 0.18 * | +3.18 ± 0.45 * | +3.98 ± 0.27 * | +4.85 ± 0.22 * | +5.63 ± 0.26 * |
| STAT3 | +2.11 ± 0.52 * | +4.77 ± 0.54 * | +5.60 ± 0.40 * | +4.14 ± 0.44 * | −2.85 ± 0.33 * |
| EGFR | −2.45 ± 0.78 * | +4.01 ± 0.62 * | +4.02 ± 0.33 * | −3.55 ± 0.20 * | +5.01 ± 0.37 * |
| SMAD7 | −4.56 ± 0.32 * | −2.81 ± 0.81 * | −2.80 ± 0.26 * | +3.98 ± 0.40 * | +5.22 ± 0.29 * |
| IFNG | −2.34 ± 0.38 * | +3.83 ± 0.18 * | +3.91 ± 0.32 * | +2.76 ± 0.45 * | +4.88 ± 0.41 * |
| IL-6 | −3.12 ± 0.71 * | +5.13 ± 0.42 * | +5.23 ± 0.28 * | +4.62 ± 0.19 * | −3.17 ± 0.36 * |
| CASP3 | +1.65 ± 0.13 * | −2.13 ± 0.18 * | −2.56 ± 0.41 * | +3.11 ± 0.43 * | +4.79 ± 0.32 * |
| IL-2 | +1.11 ± 0.44 * | +4.17 ± 0.71 * | +3.69 ± 0.29 * | +5.41 ± 0.23 * | +4.12 ± 0.39 * |
| CASP8 | +2.01 ± 0.14 * | +5.16 ± 0.43 * | +5.45 ± 0.31 * | −2.87 ± 0.38 * | +3.91 ± 0.42 * |
| KDR | −3.11 ± 0.43 * | +4.56 ± 0.76 * | +4.92 ± 0.37 * | +5.21 ± 0.21 * | −2.64 ± 0.28 * |
| PSAP | −2.34 ± 0.12 | −3.76 ± 0.19 * | −3.33 ± 0.30 * | +4.79 ± 0.24 * | +5.31 ± 0.35 * |
| mRNA | miRNA | Target Score | LPS | Tacrolimus | LPS + Tacrolimus | ||
|---|---|---|---|---|---|---|---|
| H_6 vs. C | H_6 vs. C | H_6 vs. C | H_12 vs. C | H_24 vs. C | |||
| TGF-β-2 | hsa-miR-200a-3p | 95 | +1.98 ± 0.76 * | +1.65 ± 0.61 * | −2.54 ± 0.19 * | −2.11 ± 0.13 * | −2.19 ± 0.19 * |
| SMAD4 | hsa-miR-589-3p | 99 | −2.12 ± 0.34 * | −2.98 ± 0.61 * | +3.21 ± 0.34 * | +3.22 ± 0.44 * | +2.16 ± 0.78 * |
| SMAD7 | hsa-miR-21 | 92 | +1.44 ± 0.17 * | +1.98 ± 0.31 * | +1.19 ± 0.19 | +1.87 ± 0.43 * | +2.02 ± 0.45 * |
| IL-2 | hsa-miR-27a-5p | 94 | +2.09 ± 0.65 * | +2.12 ± 0.19 * | −2.13 ± 0.43 * | −2.16 ± 0.22 * | −2.11 ± 0.18 * |
| Protein | Control | LPS | Tacrolimus | LPS + Tacrolimus | ||
|---|---|---|---|---|---|---|
| H_6 | H_6 | H_6 | H_12 | H_24 | ||
| TGF-β2 [pg/mL] | 124.11 ± 3.45 | 328.76 ± 8.91 * | 182.34 ± 5.22 * | 561.98 ± 9.12 * | 701.98 ± 10.11 * | 716.11 ± 6.71 * |
| SMAD4 [ng/mL] | 4.67 ± 0.98 | 6.12 ± 0.44 * | 6.89 ± 0.51 * | 3.45 ± 0.12 * | 3.11 ± 0.18 * | 2.99 ± 0.19 * |
| SMAD7 [ng/mL] | 6.19 ± 0.65 | 5.10 ± 0.35 * | 4.58 ± 0.42 * | 4.12 ± 0.19 * | 4.11 ± 0.76 * | 3.45 ± 0.19 * |
| IL-2 [pg/mL] | 176.87 ± 12.65 | 122.11 ± 9.12 * | 98.76 ± 8.45 * | 245.17 ± 19.91 * | 541.12 ± 12.67 * | 671.91 ± 13.45 * |
| mRNA | Oligonucleotide Sequence (5′-3′) |
|---|---|
| TGF-β2 | Forward: CAGCACACTCGATATGGACCA Reverse: CCTCGGGCTCAGGATAGTCT |
| TGF-β3 | Forward: CTGGATTGTGGTTCCATGCA Reverse: TCCCCGAATGCCTCACAT |
| TGF-βR1 | Forward: ACTGGCAGCTGTCATTGCTGGACCAG Reverse: CTGAGCCAGAACCTGACGTTGTCATATCA |
| TGF-βR3 | Forward: ACCGTGATGGGCATTGCGTTTGCA’ Reverse: GTGCTCTGCGTGCTGCCGATGCTGT |
| SMAD2 | Forward: TTGATCGACTGACAGGTGCT Reverse: GACCCCATCCTCACTCCAAA |
| SMAD4 | Forward: AAAGGTCTTTGATTTGCGTC Reverse: CTATTCCACCTACTGATCCTG |
| SMAD7 | Forward: CAGATTCCCAACTTCTTCTG Reverse: CTCTTGTTGTCCGAATTGAG |
| MAPK3 | Forward: AAGATCAGCCCCTTCGAACA Reverse: CCATCAGGTCCTGCACAATG |
| JAK1 | Forward: CTGCACCGACTTTGACAACA Reverse: ATCTGCTTCTTGAGGTGGCT |
| JAK2 | Forward: AGTAAAAGTCCACCAGCGGA Reverse: AGGAGGGGCGTTGATTTACA |
| JAK3 | Forward: CTCTTGGAACTGCTGGAGGA Reverse: AGCAGTGAAGGCATGAGTCT |
| STAT3 | Forward: AAAGCAGCAAAGAAGGAGGC Reverse: CTGGCCGACAATACTTTCCG |
| EGFR | Forward: AAAACCGGACTGAAGGAGCT Reverse: TGGATCACACTTTTGGCAGC |
| IL-6 | Forward: AGTCCTGATCCAGTTCCTGC CTACATTTGCCGAAGAGCCC |
| IL-2 | Forward: CCTCAACTCCTGCCACAATG Reverse: TGTGAGCATCCTGGTGAGTT |
| CASP3 | Forward: ACTGGACTGTGGCATTGAGA Reverse: GCACAAAGCGACTGGATGAA |
| CASP8 | Forward: GCAAAGGAAGCAAGAACCCA Reverse: CCTGGTGTCTGAAGTTCCCT |
| KDR | Forward: TTACTTGCAGGGGACAGAGG Reverse: TTCCCGGTAGAAGCACTTGT |
| PSAP | Forward: TTGAAAATTGGAGCCTGCCC Reverse: GATCTGTGCGTTCATTCCCC |
| GAPDH | Forward: GGTGAAGGTCGGAGTCAACGGA Reverse: GAGGGATCTCGCTCCTGGAAGA |
| ACTB | Forward: TCACCCACACTGTGCCCATCTACGA Reverse: CAGCGGAACCGCTCATTGCCAATGG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kiełbasińska, A.; Krysik, K.; Janiszewska-Bil, D.; Machaj, M.; Lelek, Z.; Sułkowska, J.; Nawotny-Czupryna, O.; Grabarek, B.O. Tacrolimus Modulates TGF-β Signaling–Related Genes and MicroRNAs in Human Retinal Pigment Epithelial Cells Activated by Lipopolysaccharide. Int. J. Mol. Sci. 2025, 26, 5402. https://doi.org/10.3390/ijms26115402
Kiełbasińska A, Krysik K, Janiszewska-Bil D, Machaj M, Lelek Z, Sułkowska J, Nawotny-Czupryna O, Grabarek BO. Tacrolimus Modulates TGF-β Signaling–Related Genes and MicroRNAs in Human Retinal Pigment Epithelial Cells Activated by Lipopolysaccharide. International Journal of Molecular Sciences. 2025; 26(11):5402. https://doi.org/10.3390/ijms26115402
Chicago/Turabian StyleKiełbasińska, Aleksandra, Katarzyna Krysik, Dominika Janiszewska-Bil, Martyna Machaj, Zuzanna Lelek, Joanna Sułkowska, Olga Nawotny-Czupryna, and Beniamin Oskar Grabarek. 2025. "Tacrolimus Modulates TGF-β Signaling–Related Genes and MicroRNAs in Human Retinal Pigment Epithelial Cells Activated by Lipopolysaccharide" International Journal of Molecular Sciences 26, no. 11: 5402. https://doi.org/10.3390/ijms26115402
APA StyleKiełbasińska, A., Krysik, K., Janiszewska-Bil, D., Machaj, M., Lelek, Z., Sułkowska, J., Nawotny-Czupryna, O., & Grabarek, B. O. (2025). Tacrolimus Modulates TGF-β Signaling–Related Genes and MicroRNAs in Human Retinal Pigment Epithelial Cells Activated by Lipopolysaccharide. International Journal of Molecular Sciences, 26(11), 5402. https://doi.org/10.3390/ijms26115402






