N-Glycosylation as a Key Requirement for the Positive Interaction of Integrin and uPAR in Glioblastoma
Abstract
1. Introduction
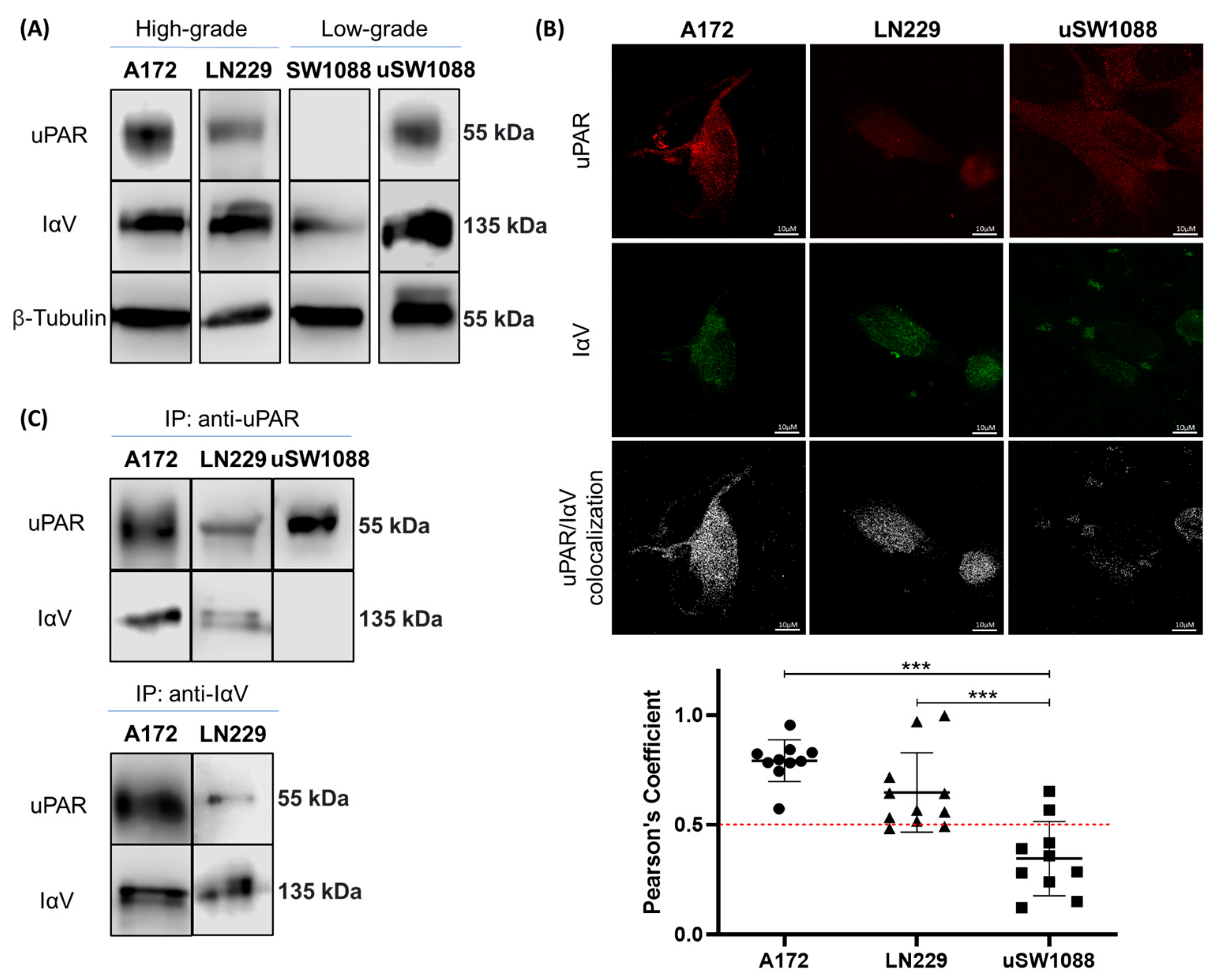
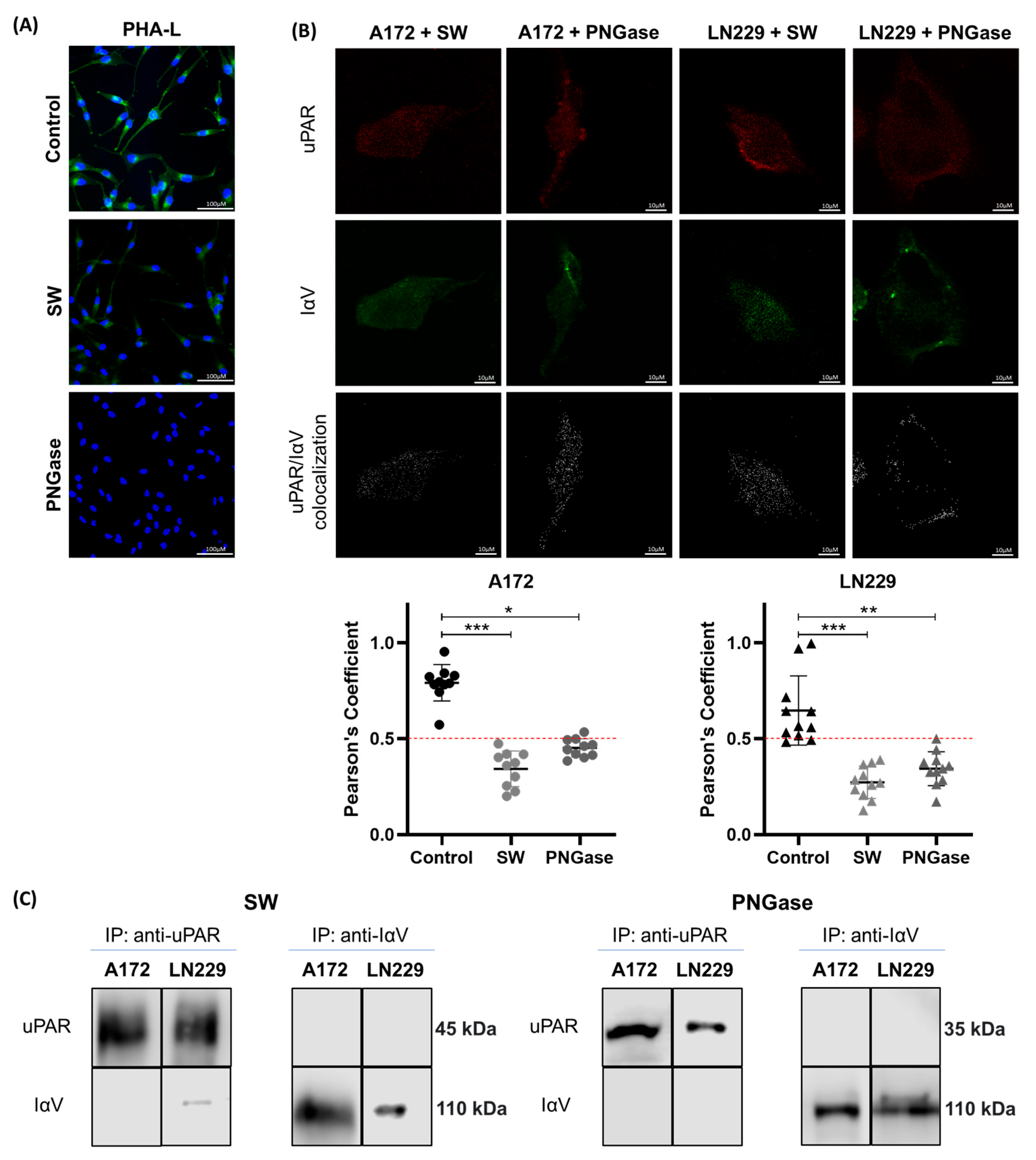
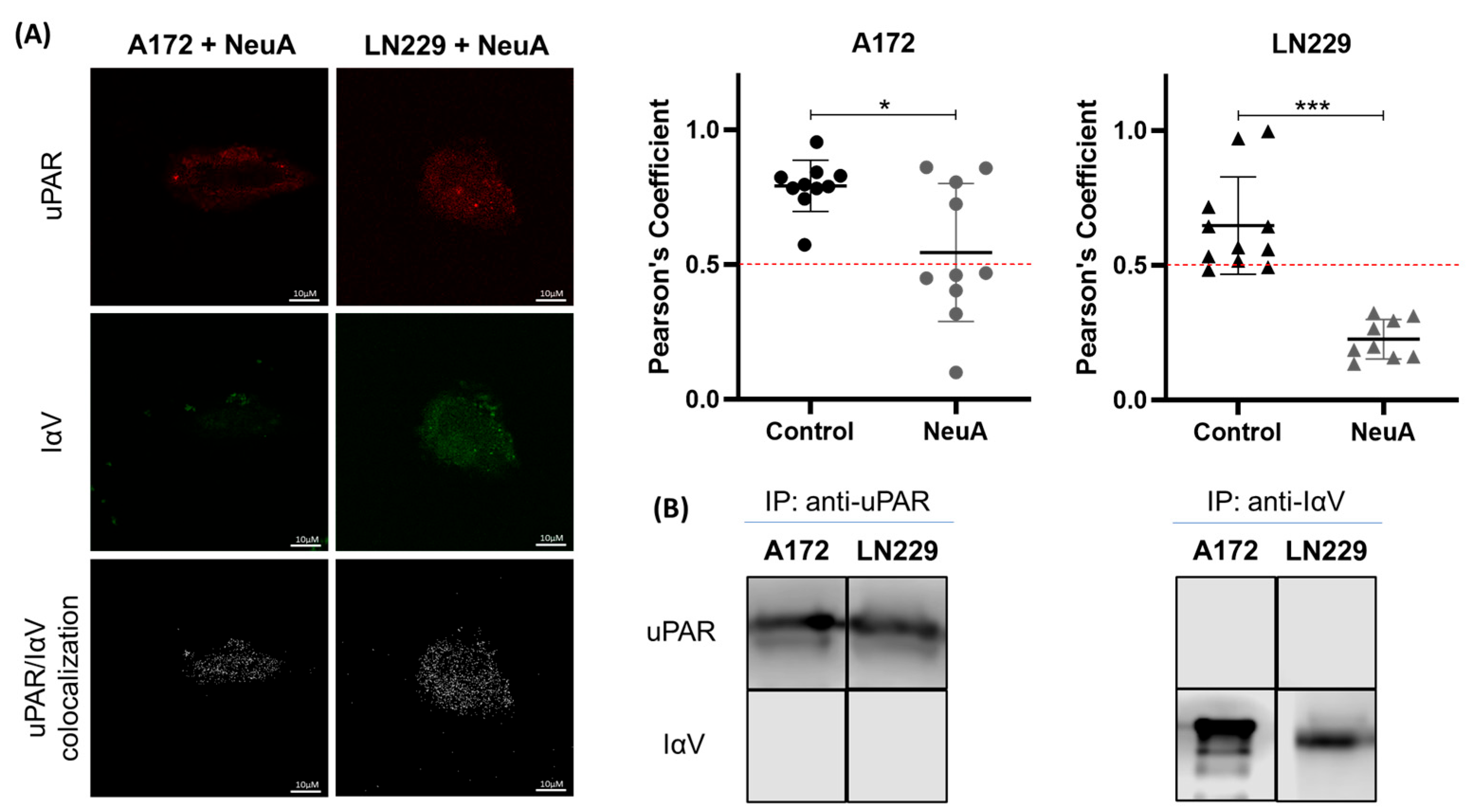
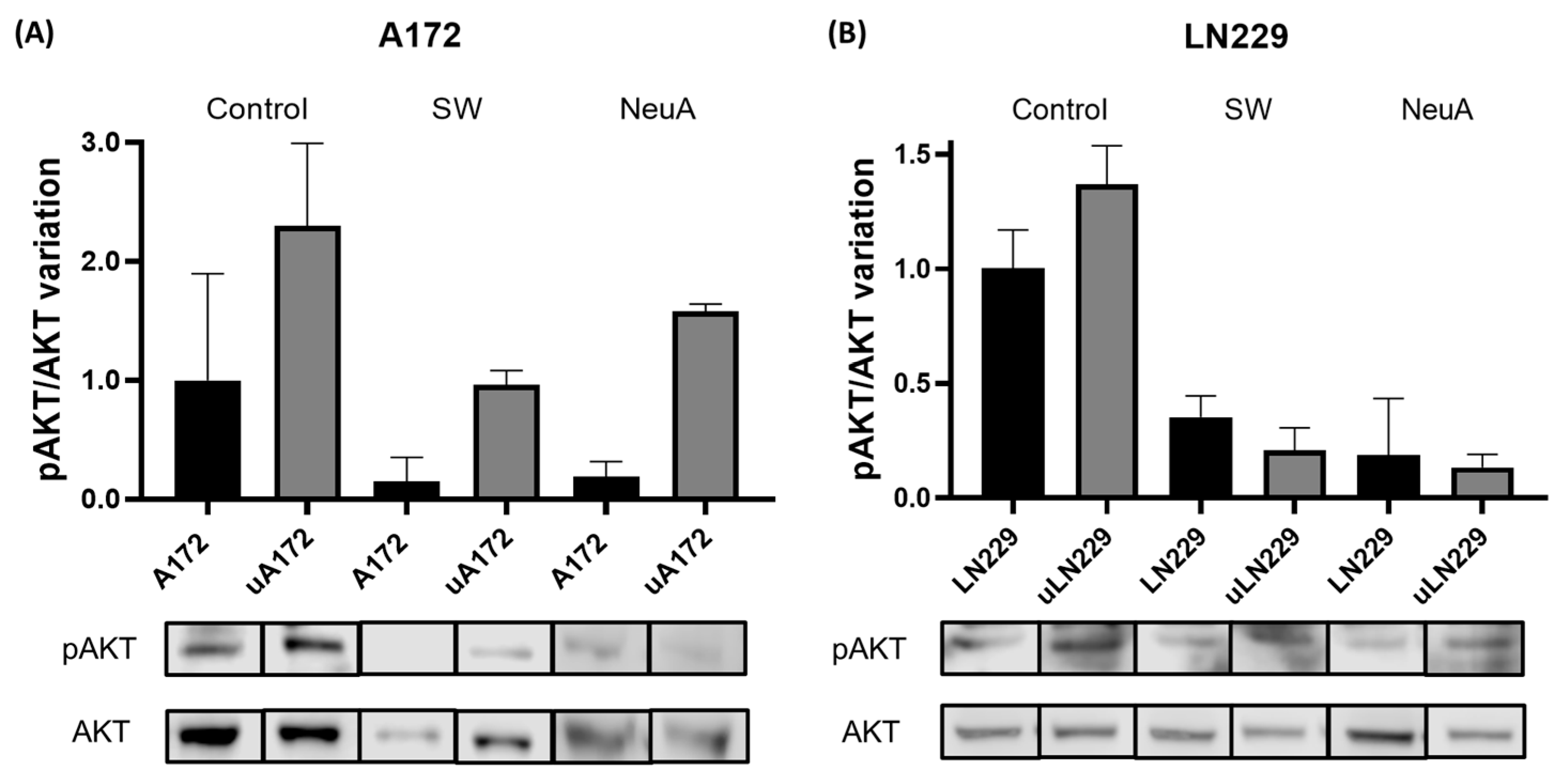
2. Results
3. Discussion
4. Materials and Methods
4.1. Cell Lines
4.2. Glycosylation Inhibition and Removal
4.3. Lectin Interference Assay
4.4. Plasmid DNA Transfection
4.5. Immunofluorescence
4.6. Confocal Microscopy
4.7. Protein Co-Immunoprecipitation
4.8. SDS-PAGE and Western Blot
4.9. Protein Concentration
4.10. Mass Spectrometry
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AB | Ammonium bicarbonate |
| ACN | Acetonitrile |
| CM | Confocal microscopy |
| Co-IP | Co-Immunoprecipitation |
| ConA | Concanavalin A |
| DAPI | 4′,6-diamidino-2-phenylindole |
| DMEM | Dulbecco’s Modified Eagle’s Medium |
| ECM | Extracellular Matrix |
| FAK | Focal adhesion kinase |
| FBS | Fetal bovine serum |
| GBM | Glioblastoma |
| IαV | Integrin Av |
| MAPK | Mitogen-activated protein kinase |
| MS | Mass spectrometry |
| NeuA | Neuraminidase A |
| NeuAc | N-Acetylneuraminic acid |
| NeuGc | N-Glycolylneuraminic acid |
| pAKT | Phospho-AKT |
| PBS | Phosphate-buffered saline |
| PHA-L | Phytohemagglutinin-L |
| PNGase | Peptide-N-Glycosidase F |
| RPMI | Roswell Park Memorial Institute |
| SW | Swainsonine |
| uPA | Urokinase-type plasminogen activator |
| uPAR | Urokinase-type plasminogen activator receptor |
| WB | Western blot |
References
- Mallick, S.; Benson, R.; Hakim, A.; Rath, G.K. Management of glioblastoma after recurrence: A changing paradigm. J. Egypt. Natl. Cancer Inst. 2016, 28, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Barczyk, M.; Carracedo, S.; Gullberg, D. Integrins. Cell Tissue Res. 2010, 339, 269–280. [Google Scholar] [CrossRef] [PubMed]
- Campbell, I.D.; Humphries, M.J. Integrin structure, activation, and interactions. Cold Spring Harb. Perspect. Biol. 2011, 3, a004994. [Google Scholar] [CrossRef]
- Peng, Z.; Hao, M.; Tong, H.; Yang, H.; Huang, B.; Zhang, Z.; Luo, K.Q. The interactions between integrin α5β1 of liver cancer cells and fibronectin of fibroblasts promote tumor growth and angiogenesis. Int. J. Biol. Sci. 2022, 18, 5019–5037. [Google Scholar] [CrossRef]
- Zhang, Y.; Reif, G.; Wallace, D.P. Extracellular matrix, integrins, and focal adhesion signaling in polycystic kidney disease. Cell. Signal. 2020, 72, 109646. [Google Scholar] [CrossRef]
- Wu, D.; Xu, Y.; Ding, T.; Zu, Y.; Yang, C.; Yu, L. Pairing of integrins with ECM proteins determines migrasome formation. Cell Res. 2017, 27, 1397–1400. [Google Scholar] [CrossRef]
- Lainé, A.; Labiad, O.; Hernandez-Vargas, H.; This, S.; Sanlaville, A.; Léon, S.; Dalle, S.; Sheppard, D.; Travis, M.A.; Paidassi, H.; et al. Regulatory T cells promote cancer immune-escape through integrin αvβ8-mediated TGF-β activation. Nat. Commun. 2021, 12, 6228. [Google Scholar] [CrossRef]
- Kumari, S.; Kumar, P. Identification and characterization of putative biomarkers and therapeutic axis in Glioblastoma multiforme microenvironment. Front. Cell Dev. Biol. 2023, 11, 1236271. [Google Scholar] [CrossRef]
- Suresh, A.; Biswas, A.; Perumal, S.; Khurana, S. Periostin and Integrin Signaling in Stem Cell Regulation. Adv. Exp. Med. Biol. 2019, 1132, 163–176. [Google Scholar] [CrossRef]
- Ellis, S.J.; Tanentzapf, G. Integrin-mediated adhesion and stem-cell-niche interactions. Cell Tissue Res. 2010, 339, 121–130. [Google Scholar] [CrossRef]
- Hoshino, A.; Costa-Silva, B.; Shen, T.-L.; Rodrigues, G.; Hashimoto, A.; Mark, M.T.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Shan, X.; Meng, X.; Gu, T.; Guo, L.; An, X.; Jiang, Q.; Ge, H.; Ning, X. Novel Integrin αvβ3-specific ligand for the sensitive diagnosis of glioblastoma. Mol. Pharm. 2019, 16, 3977–3984. [Google Scholar] [CrossRef]
- Nakada, M.; Nambu, E.; Furuyama, N.; Yoshida, Y.; Takino, T.; Hayashi, Y.; Sato, H.; Sai, Y.; Tsuji, T.; Miyamoto, K.-I.; et al. Integrin α3 is overexpressed in glioma stem-like cells and promotes invasion. Br. J. Cancer 2013, 108, 2516–2524. [Google Scholar] [CrossRef]
- Delamarre, E.; Taboubi, S.; Mathieu, S.; Bérenguer, C.; Rigot, V.; Lissitzky, J.-C.; Figarella-Branger, D.; Ouafik, L.; Luis, J. Expression of Integrin α6β1 Enhances Tumorigenesis in Glioma Cells. Am. J. Pathol. 2009, 175, 844–855. [Google Scholar] [CrossRef]
- Bello, L.; Francolini, M.; Marthyn, P.; Zhang, J.; Carroll, R.S.; Nikas, D.C.; Strasser, J.F.; Villani, R.; Cheresh, D.A.; Black, P.M. αvβ3 and αvβ5 Integrin Expression in Glioma Periphery. Neurosurgery 2001, 49, 380–390. [Google Scholar] [CrossRef]
- Gladson, C.L. Expression of integrin alpha v beta 3 in small blood vessels of glioblastoma tumors. J. Neuropathol. Exp. Neurol. 1996, 55, 1143–1149. [Google Scholar] [CrossRef]
- Schittenhelm, J.; Schwab, E.I.; Sperveslage, J.; Tatagiba, M.; Meyermann, R.; Fend, F.; Goodman, S.L.; Sipos, B. Longitudinal expression analysis of αv integrins in human gliomas reveals upregulation of integrin αvβ3 as a negative prognostic factor. J. Neuropathol. Exp. Neurol. 2013, 72, 194–210. [Google Scholar] [CrossRef]
- Gingras, M.-C.; Roussel, E.; Bruner, J.M.; Branch, C.D.; Moser, R.P. Comparison of cell adhesion molecule expression between glioblastoma multiforme and autologous normal brain tissue. J. Neuroimmunol. 1995, 57, 143–153. [Google Scholar] [CrossRef]
- Lorger, M.; Krueger, J.S.; O’Neal, M.; Staflin, K.; Felding-Habermann, B. Activation of tumor cell integrin αvβ3 controls angiogenesis and metastatic growth in the brain. Proc. Natl. Acad. Sci. USA 2009, 106, 10666–10671. [Google Scholar] [CrossRef]
- Martin, S.; Janouskova, H.; Dontenwill, M. Integrins and p53 pathways in glioblastoma resistance to temozolomide. Front. Oncol. 2012, 2, 157. [Google Scholar] [CrossRef]
- Stanzani, E.; Pedrosa, L.; Bourmeau, G.; Anezo, O.; Noguera-Castells, A.; Esteve-Codina, A.; Passoni, L.; Matteoli, M.; de la Iglesia, N.; Seano, G.; et al. Dual role of integrin alpha-6 in glioblastoma: Supporting stemness in proneural stem-like cells while inducing radioresistance in mesenchymal stem-like cells. Cancers 2021, 13, 3055. [Google Scholar] [CrossRef] [PubMed]
- Casey, J.R.; Petranka, J.G.; Kottra, J.; Fleenor, D.E.; Rosse, W.F. The structure of the urokinase-type plasminogen activator receptor gene. Blood 1994, 84, 1151–1156. [Google Scholar] [CrossRef] [PubMed]
- Ghiso, J.A.A. Inhibition of FAK signaling activated by urokinase receptor induces dormancy in human carcinoma cells in vivo. Oncogene 2002, 21, 2513–2524. [Google Scholar] [CrossRef]
- Vial, E.; Sahai, E.; Marshall, C.J. ERK-MAPK signaling coordinately regulates activity of Rac1 and RhoA for tumor cell motility. Cancer Cell 2003, 4, 67–79. [Google Scholar] [CrossRef]
- Liu, D.; Ghiso, J.A.; Estrada, Y.; Ossowski, L. EGFR is a transducer of the urokinase receptor initiated signal that is required for in vivo growth of a human carcinoma. Cancer Cell 2002, 1, 445–457. [Google Scholar] [CrossRef]
- Kjøller, L.; Hall, A. Rac mediates cytoskeletal rearrangements and increased cell motility induced by urokinase-type plasminogen activator receptor binding to vitronectin. J. Cell Biol. 2001, 152, 1145–1158. [Google Scholar] [CrossRef]
- de Bock, C.E.; Wang, Y. Clinical significance of urokinase-type plasminogen activator receptor (uPAR) expression in cancer. Med. Res. Rev. 2004, 24, 13–39. [Google Scholar] [CrossRef]
- Persson, M.; Nedergaard, M.K.; Brandt-Larsen, M.; Skovgaard, D.; Jørgensen, J.T.; Michaelsen, S.R.; Madsen, J.; Lassen, U.; Poulsen, H.S.; Kjaer, A. Urokinase-Type Plasminogen Activator Receptor as a Potential PET Biomarker in Glioblastoma. J. Nucl. Med. 2016, 57, 272–278. [Google Scholar] [CrossRef]
- Gilder, A.S.; Natali, L.; Van Dyk, D.M.; Zalfa, C.; Banki, M.A.; Pizzo, D.P.; Wang, H.; Klemke, R.L.; Mantuano, E.; Gonias, S.L. The Urokinase Receptor Induces a Mesenchymal Gene Expression Signature in Glioblastoma Cells and Promotes Tumor Cell Survival in Neurospheres. Sci. Rep. 2018, 8, 2982. [Google Scholar] [CrossRef]
- Smith, H.W.; Marshall, C.J. Regulation of cell signalling by uPAR. Nat. Rev. Mol. Cell Biol. 2010, 11, 23–36. [Google Scholar] [CrossRef]
- Wei, Y.; Lukashev, M.; Simon, D.I.; Bodary, S.C.; Rosenberg, S.; Doyle, M.V.; Chapman, H.A. Regulation of Integrin Function by the Urokinase Receptor. Science 1996, 273, 1551–1555. [Google Scholar] [CrossRef] [PubMed]
- Sowmya, G.; Khan, J.M.; Anand, S.; Ahn, S.B.; Baker, M.S.; Ranganathan, S. A site for direct integrin αvβ6·uPAR interaction from structural modelling and docking. J. Struct. Biol. 2014, 185, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.B.; Mohamedali, A.; Anand, S.; Cheruku, H.R.; Birch, D.; Sowmya, G.; Cantor, D.; Ranganathan, S.; Inglis, D.W.; Frank, R.; et al. Characterization of the interaction between heterodimeric αvβ6 integrin and urokinase plasminogen activator receptor (uPAR) using functional proteomics. J. Proteome Res. 2014, 13, 5956–5964. [Google Scholar] [CrossRef]
- Janik, M.E.; Przybyło, M.; Pocheć, E.; Pokrywka, M.; Lityńska, A. Effect of alpha3beta1 and alphavbeta3 integrin glycosylation on interaction of melanoma cells with vitronectin. Acta Biochim. Pol. 2010, 57, 55–61. [Google Scholar] [CrossRef]
- Munkley, J.; Elliott, D.J. Hallmarks of glycosylation in cancer. Oncotarget 2016, 7, 35478–35489. [Google Scholar] [CrossRef]
- Hakomori, S. Tumor malignancy defined by aberrant glycosylation and sphingo(glyco)lipid metabolism. Cancer Res. 1996, 56, 5309–5318. [Google Scholar]
- Kannagi, R.; Yin, J.; Miyazaki, K.; Izawa, M. Current relevance of incomplete synthesis and neo-synthesis for cancer-associated alteration of carbohydrate determinants—Hakomori’s concepts revisited. Biochim. Biophys. Acta 2008, 1780, 525–531. [Google Scholar] [CrossRef]
- Lehmann, M.; El Battari, A.; Abadie, B.; Martin, J.M.; Marvaldi, J. Role of alpha v beta 5 and alpha v beta 6 integrin glycosylation in the adhesion of a colonic adenocarcinoma cell line (HT29-D4). J. Cell. Biochem. 1996, 61, 266–277. [Google Scholar] [CrossRef]
- Pocheć, E.; Bubka, M.; Rydlewska, M.; Janik, M.; Pokrywka, M.; Lityńska, A. Aberrant glycosylation of αvβ3 integrin is associated with melanoma progression. Anticancer Res. 2015, 35, 2093–2103. [Google Scholar]
- Behrendt, N.; Rønne, E.; Ploug, M.; Petri, T.; Løber, D.; Nielsen, L.S.; Schleuning, W.D.; Blasi, F.; Appella, E.; Danø, K. The human receptor for urokinase plasminogen activator. NH2-terminal amino acid sequence and glycosylation variants. J. Biol. Chem. 1990, 265, 6453–6460. [Google Scholar] [CrossRef]
- Kjaergaard, M.; Hansen, L.V.; Jacobsen, B.; Gardsvoll, H.; Ploug, M. Structure and ligand interactions of the urokinase receptor (uPAR). Front. Biosci. 2008, 13, 5441–5461. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, M.M.; Bond, M.R.; Abramowitz, L.K.; Biesbrock, D.; Woodroofe, C.C.; Kim, E.J.; Swenson, R.E.; Hanover, J.A. Tools and tactics to define specificity of metabolic chemical reporters. Front. Mol. Biosci. 2023, 10, 1286690. [Google Scholar] [CrossRef]
- Mukherjee, M.M.; Biesbrock, D.; Abramowitz, L.K.; Pavan, M.; Kumar, B.; Walter, P.J.; Azadi, P.; Jacobson, K.A.; Hanover, J.A. Selective bioorthogonal probe for N-glycan hybrid structures. Nat. Chem. Biol. 2025, 21, 681–692. [Google Scholar] [CrossRef]
- Elbein, A.D. Inhibitors of the biosynthesis and processing of N-linked oligosaccharide chains. Annu. Rev. Biochem. 1987, 56, 497–534. [Google Scholar] [CrossRef]
- Maley, F.; Trimble, R.B.; Tarentino, A.L.; Plummer, T.H., Jr. Characterization of glycoproteins and their associated oligosaccharides through the use of endoglycosidases. Anal. Biochem. 1989, 180, 195–204. [Google Scholar] [CrossRef]
- Gabri, M.R.; Otero, L.L.; Gomez, D.E.; Alonso, D.F. Exogenous incorporation of neugc-rich mucin augments n-glycolyl sialic acid content and promotes malignant phenotype in mouse tumor cell lines. J. Exp. Clin. Cancer Res. 2009, 28, 146. [Google Scholar] [CrossRef]
- Dhar, C.; Sasmal, A.; Varki, A. From “Serum Sickness” to “Xenosialitis”: Past, Present, and Future Significance of the Non-human Sialic Acid Neu5Gc. Front. Immunol. 2019, 10, 807. [Google Scholar] [CrossRef]
- Blasi, F. uPA, uPAR, PAI-I: Key intersection of proteolytic, adhesive and chemotacfic highways? Immunol. Today 1997, 18, 415–417. [Google Scholar] [CrossRef]
- Sidenius, N.; Blasi, F. The urokinase plasminogen activator system in cancer: Recent advances and implication for prognosis and therapy. Cancer Metastasis Rev. 2003, 22, 205–222. [Google Scholar] [CrossRef]
- Desgrosellier, J.S.; Cheresh, D.A. Integrins in cancer: Biological implications and therapeutic opportunities. Nat. Rev. Cancer 2010, 10, 9–22. [Google Scholar] [CrossRef]
- Hamidi, H.; Ivaska, J. Every step of the way: Integrins in cancer progression and metastasis. Nat. Rev. Cancer 2018, 18, 533–548. [Google Scholar] [CrossRef] [PubMed]
- Cáceres-Calle, D.; Torre-Cea, I.; Marcos-Zazo, L.; Carrera-Aguado, I.; Guerra-Paes, E.; Berlana-Galán, P.; Muñoz-Félix, J.M.; Sánchez-Juanes, F. Integrins as Key Mediators of Metastasis. Int. J. Mol. Sci. 2025, 26, 904. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Peng, C.; Liang, B.; Shahbaz, M.; Liu, S.; Wang, B.; Sun, Q.; Niu, Z.; Niu, W.; Liu, E.; et al. β6 integrin induces the expression of metalloproteinase-3 and metalloproteinase-9 in colon cancer cells via ERK-ETS1 pathway. Cancer Lett. 2014, 354, 427–437. [Google Scholar] [CrossRef]
- Cantor, D.I.; Cheruku, H.R.; Nice, E.C.; Baker, M.S. Integrin αvβ6 sets the stage for colorectal cancer metastasis. Cancer Metastasis Rev. 2015, 34, 715–734. [Google Scholar] [CrossRef]
- Annis, M.G.; Ouellet, V.; Rennhack, J.P.; L’esperance, S.; Rancourt, C.; Mes-Masson, A.-M.; Andrechek, E.R.; Siegel, P.M. Integrin-uPAR signaling leads to FRA-1 phosphorylation and enhanced breast cancer invasion. Breast Cancer Res. 2018, 20, 9. [Google Scholar] [CrossRef]
- Veeravalli, K.K.; Chetty, C.; Ponnala, S.; Gondi, C.S.; Lakka, S.S.; Fassett, D.; Klopfenstein, J.D.; Dinh, D.H.; Gujrati, M.; Rao, J.S. MMP-9, uPAR and cathepsin B silencing downregulate integrins in human glioma xenograft cells in vitro and in vivo in nude mice. PLoS ONE 2010, 5, e11583. [Google Scholar] [CrossRef]
- Veeravalli, K.K.; Rao, J.S. MMP-9 and uPAR regulated glioma cell migration. Cell Adhes. Migr. 2012, 6, 509–512. [Google Scholar] [CrossRef]
- Cuello, H.A.; Segatori, V.I.; Albertó, M.; Gulino, C.A.; Aschero, R.; Camarero, S.; Mutti, L.G.; Madauss, K.; Alonso, D.F.; Lubieniecki, F.; et al. Aberrant O-glycosylation modulates aggressiveness in neuroblastoma. Oncotarget 2018, 9, 34176–34188. [Google Scholar] [CrossRef]
- Cuello, H.A.; Ferreira, G.M.; Gulino, C.A.; Toledo, A.G.; Segatori, V.I.; Gabri, M.R. Terminally sialylated and fucosylated complex N-glycans are involved in the malignant behavior of high-grade glioma. Oncotarget 2020, 11, 4822–4835. [Google Scholar] [CrossRef]
- Pinho, S.S.; Reis, C.A. Glycosylation in cancer: Mechanisms and clinical implications. Nat. Rev. Cancer 2015, 15, 540–555. [Google Scholar] [CrossRef]
- Gu, J.; Taniguchi, N. Regulation of integrin functions by N-glycans. Glycoconj. J. 2004, 21, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Isaji, T.; Sato, Y.; Kariya, Y.; Fukuda, T. Importance of N-Glycosylation on α5β1 Integrin for Its Biological Functions. Biol. Pharm. Bull. 2009, 32, 780–785. [Google Scholar] [CrossRef] [PubMed]
- Janik, M.E.; Lityńska, A.; Vereecken, P. Cell migration—The role of integrin glycosylation. Biochim. Biophys. Acta 2010, 1800, 545–555. [Google Scholar] [CrossRef]
- Hang, Q.; Isaji, T.; Hou, S.; Wang, Y.; Fukuda, T.; Gu, J. A Key Regulator of Cell Adhesion: Identification and Characterization of Important N-Glycosylation Sites on Integrin α5 for Cell Migration. Mol. Cell. Biol. 2017, 37, e00558-16. [Google Scholar] [CrossRef]
- Cai, X.; Thinn, A.M.M.; Wang, Z.; Shan, H.; Zhu, J. The importance of N-glycosylation on β3 integrin ligand binding and conformational regulation. Sci. Rep. 2017, 7, 4656. [Google Scholar] [CrossRef]
- Simon, D.I.; Wei, Y.; Zhang, L.; Rao, N.K.; Xu, H.; Chen, Z.; Liu, Q.; Rosenberg, S.; Chapman, H.A. Identification of a Urokinase Receptor-Integrin Interaction Site. J. Biol. Chem. 2000, 275, 10228–10234. [Google Scholar] [CrossRef]
- Zhang, F.; Tom, C.C.; Kugler, M.C.; Ching, T.-T.; Kreidberg, J.A.; Wei, Y.; Chapman, H.A. Distinct ligand binding sites in integrin α3β1 regulate matrix adhesion and cell–cell contact. J. Cell Biol. 2003, 163, 177–188. [Google Scholar] [CrossRef]
- Chaurasia, P.; Aguirre-Ghiso, J.A.; Liang, O.D.; Gardsvoll, H.; Ploug, M.; Ossowski, L. A region in urokinase plasminogen receptor domain III controlling a functional association with α5β1 integrin and tumor growth. J. Biol. Chem. 2006, 281, 14852–14863. [Google Scholar] [CrossRef]
- Degryse, B.; Resnati, M.; Czekay, R.-P.; Loskutoff, D.J.; Blasi, F. Domain 2 of the urokinase receptor contains an integrin-interacting epitope with intrinsic signaling activity: Generation of a new integrin inhibitor. J. Biol. Chem. 2005, 280, 24792–24803. [Google Scholar] [CrossRef]
- Sato, Y.; Isaji, T.; Tajiri, M.; Yoshida-Yamamoto, S.; Yoshinaka, T.; Somehara, T.; Fukuda, T.; Wada, Y.; Gu, J. An N-glycosylation site on the beta-propeller domain of the integrin alpha5 subunit plays key roles in both its function and site-specific modification by beta1,4-N-acetylglucosaminyltransferase III. J. Biol. Chem. 2009, 284, 11873–11881. [Google Scholar] [CrossRef]
- Cao, L.; Wu, Y.; Wang, X.; Li, X.; Tan, Z.; Guan, F. Role of Site-Specific Glycosylation in the I-Like Domain of Integrin β1 in Small Extracellular Vesicle-Mediated Malignant Behavior and FAK Activation. Int. J. Mol. Sci. 2021, 22, 1770. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Xiao, W.; Sun, S.; Sohrabi, A.; Liang, J.; Seidlits, S.K. Extracellular Matrix Proteins Confer Cell Adhesion-Mediated Drug Resistance Through Integrin αv in Glioblastoma Cells. Front. Cell Dev. Biol. 2021, 9, 616580. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.-H.; Hill, M.L.; Brumwell, A.N.; Chapman, H.A.; Wei, Y. Signaling through urokinase and urokinase receptor in lung cancer cells requires interactions with β1 integrins. J. Cell Sci. 2008, 121, 3747–3756. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferreira, G.M.; Cuello, H.A.; Nogueira, A.C.; Castillo, J.O.; Rojo, S.; Gulino, C.A.; Segatori, V.I.; Gabri, M.R. N-Glycosylation as a Key Requirement for the Positive Interaction of Integrin and uPAR in Glioblastoma. Int. J. Mol. Sci. 2025, 26, 5310. https://doi.org/10.3390/ijms26115310
Ferreira GM, Cuello HA, Nogueira AC, Castillo JO, Rojo S, Gulino CA, Segatori VI, Gabri MR. N-Glycosylation as a Key Requirement for the Positive Interaction of Integrin and uPAR in Glioblastoma. International Journal of Molecular Sciences. 2025; 26(11):5310. https://doi.org/10.3390/ijms26115310
Chicago/Turabian StyleFerreira, Gretel Magalí, Hector Adrian Cuello, Aylen Camila Nogueira, Jeremias Omar Castillo, Selene Rojo, Cynthia Antonella Gulino, Valeria Inés Segatori, and Mariano Rolando Gabri. 2025. "N-Glycosylation as a Key Requirement for the Positive Interaction of Integrin and uPAR in Glioblastoma" International Journal of Molecular Sciences 26, no. 11: 5310. https://doi.org/10.3390/ijms26115310
APA StyleFerreira, G. M., Cuello, H. A., Nogueira, A. C., Castillo, J. O., Rojo, S., Gulino, C. A., Segatori, V. I., & Gabri, M. R. (2025). N-Glycosylation as a Key Requirement for the Positive Interaction of Integrin and uPAR in Glioblastoma. International Journal of Molecular Sciences, 26(11), 5310. https://doi.org/10.3390/ijms26115310






