Mutant p53 Associates with Human Equilibrative Nucleoside 1 Upregulation and Better Response to Adjuvant Gemcitabine in Intrahepatic Cholangiocarcinoma Patients
Abstract
1. Introduction
2. Results
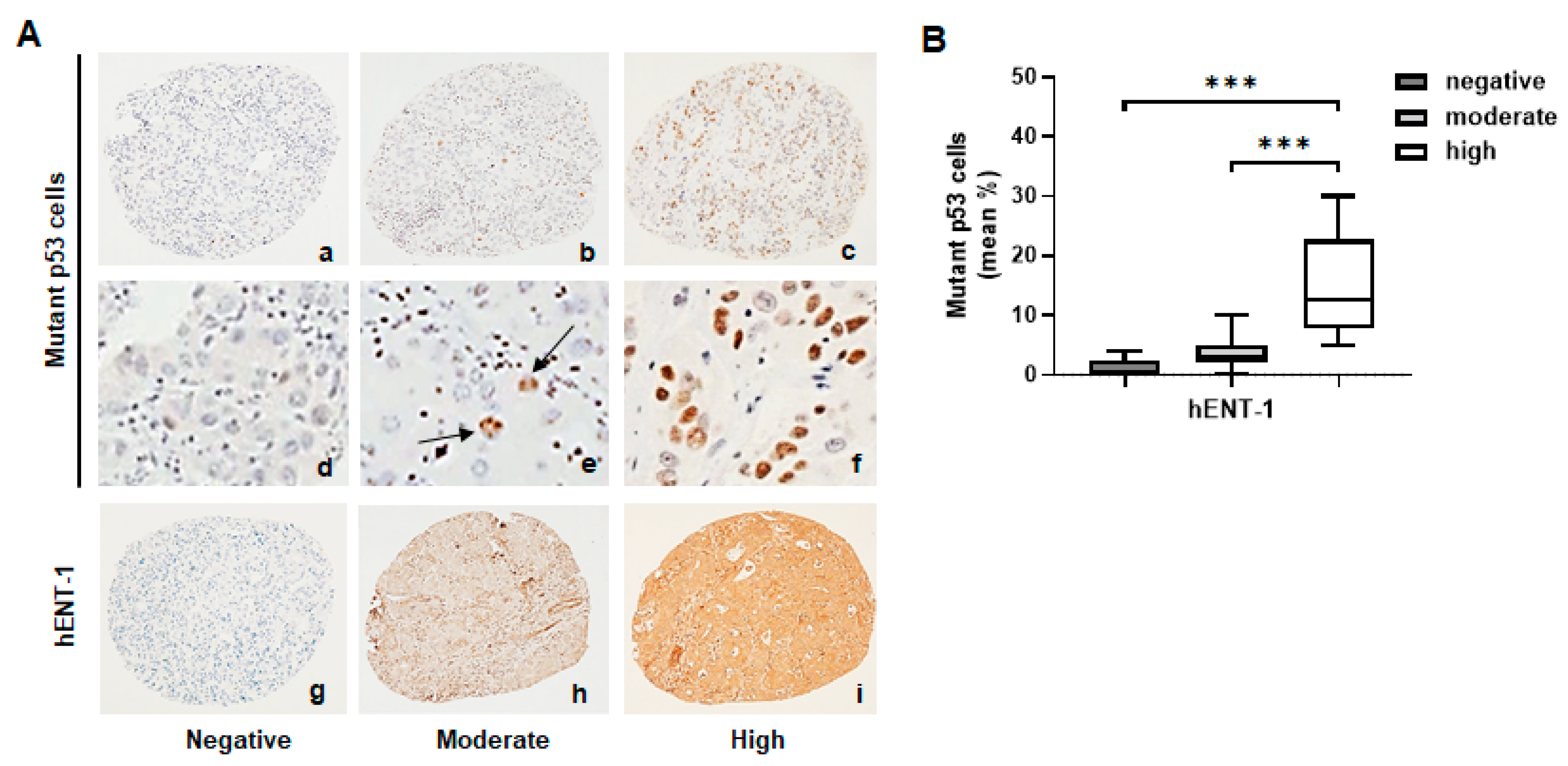
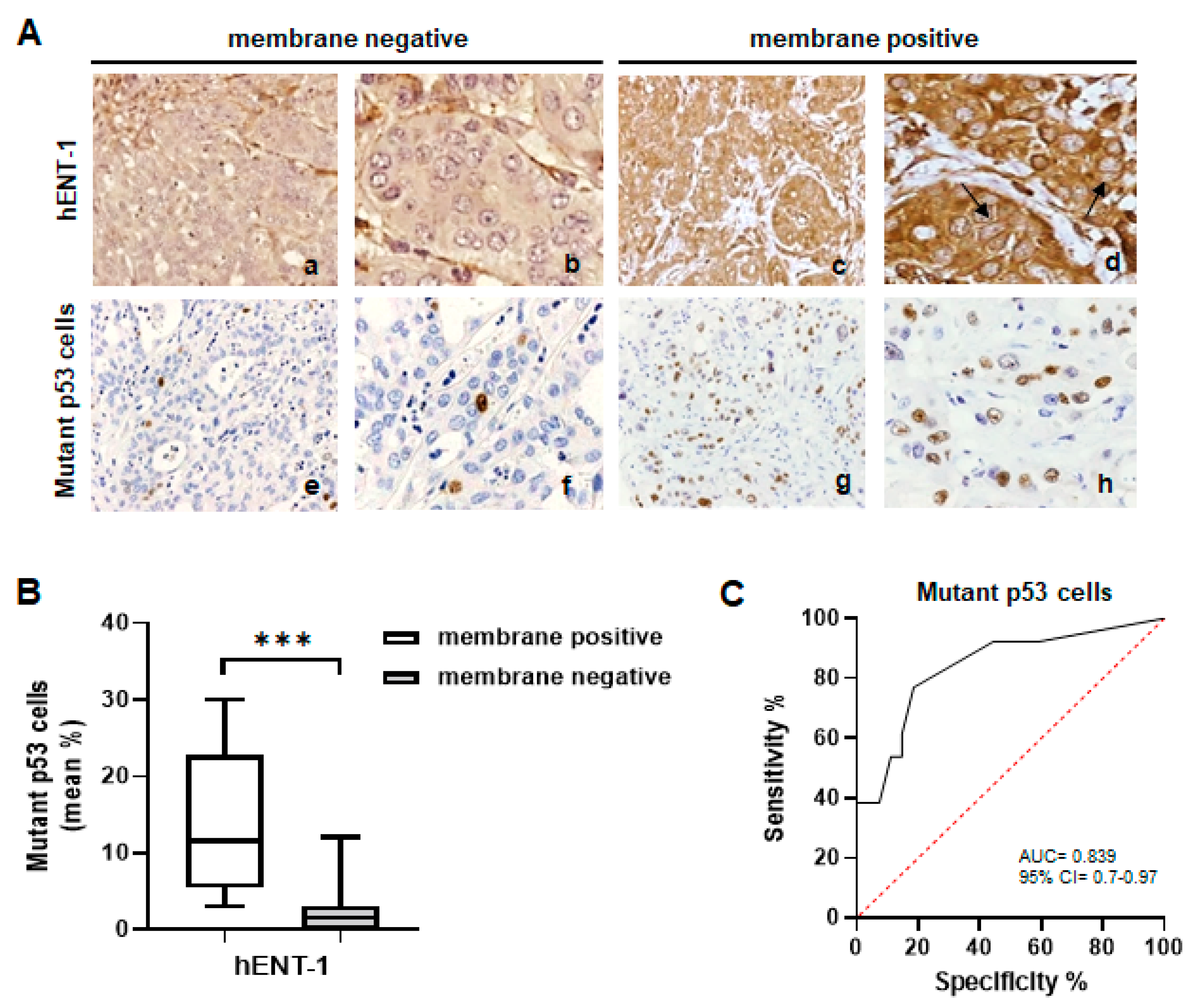
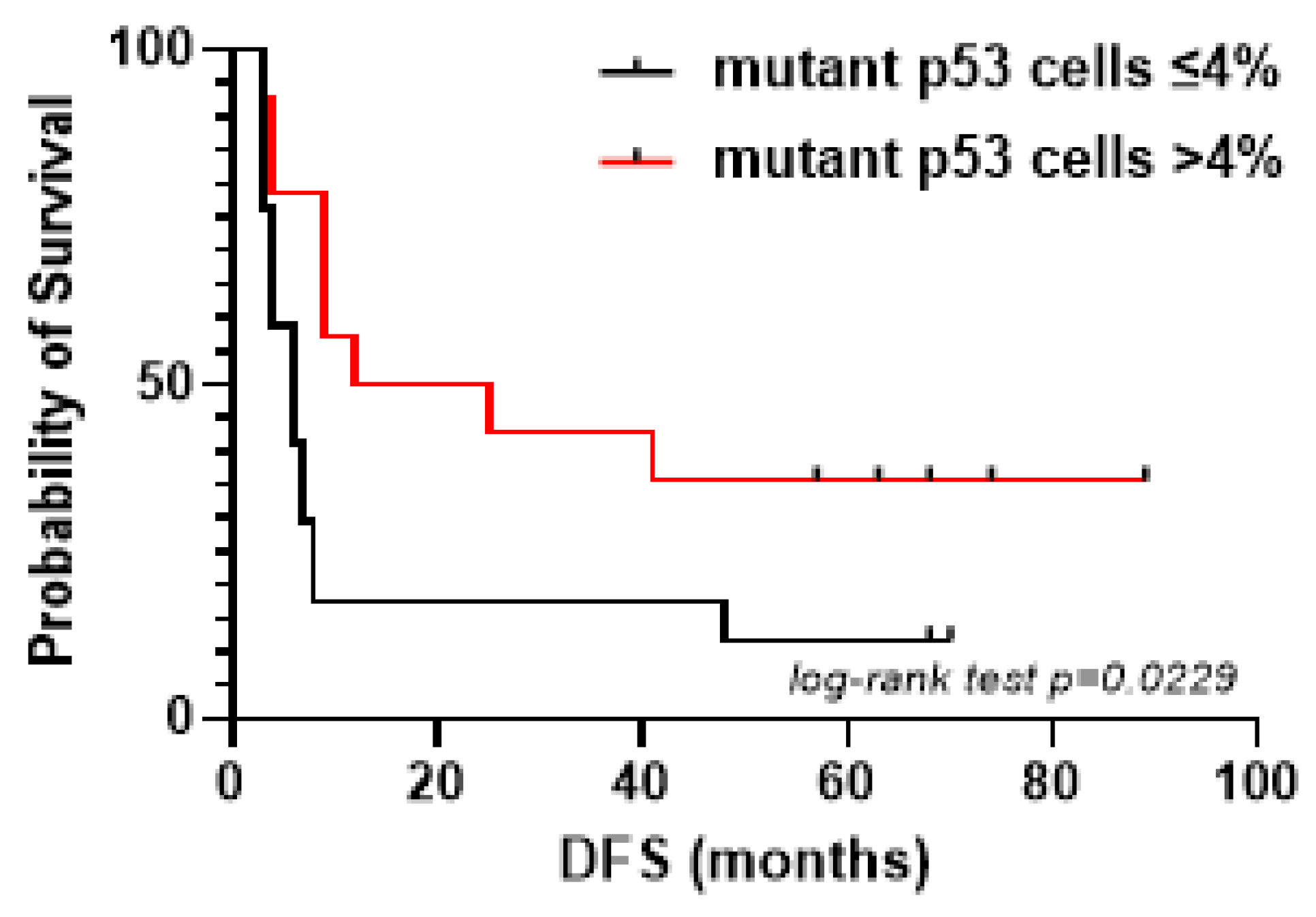
2.1. Mutant p53 Associates with High hENT-1 Expression and Membrane Localization in iCCA Tissue Samples
2.2. hENT-1 Is Upregulated in Mutant p53 iCCA Cell Lines
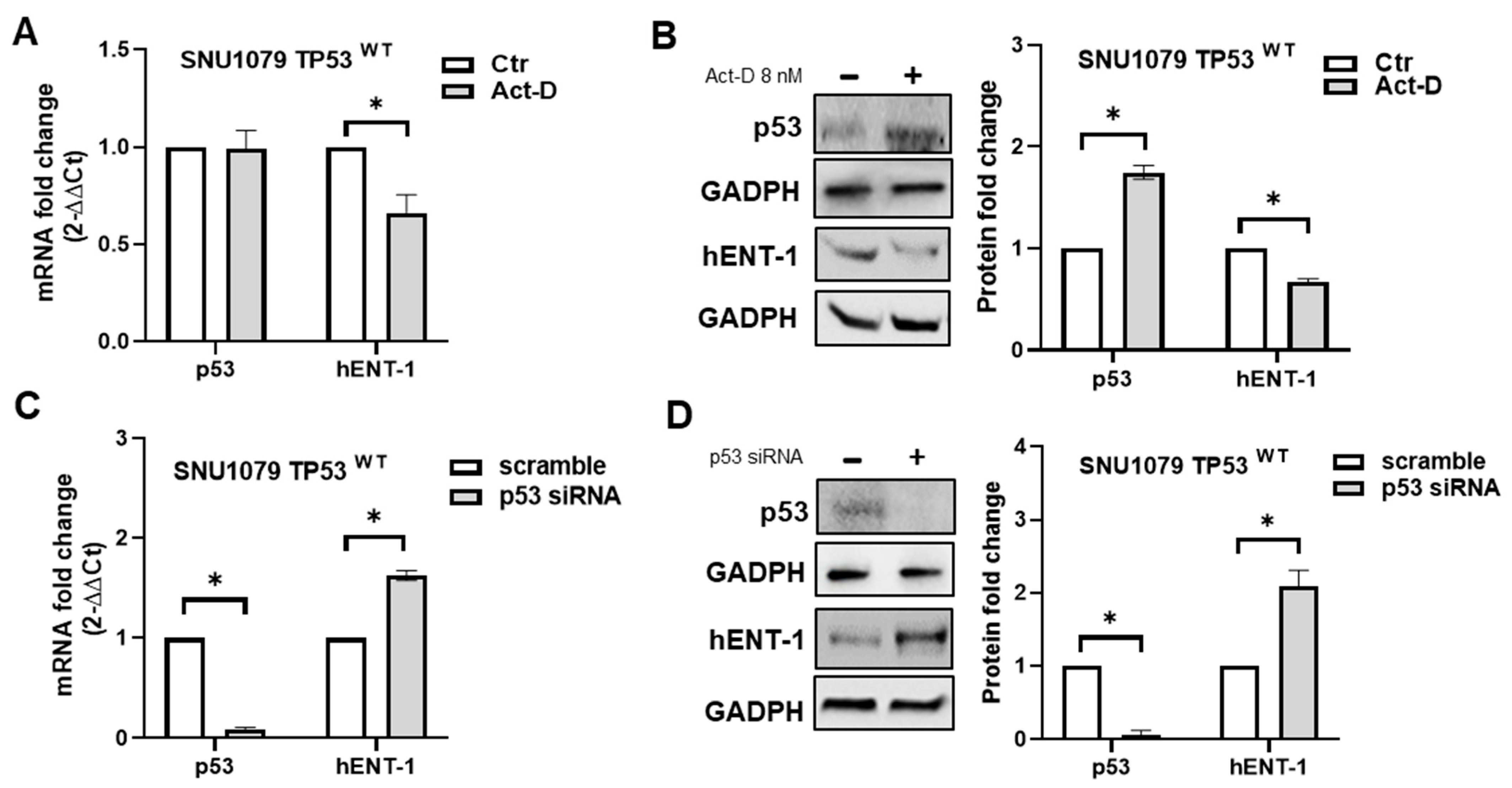
2.3. p53 Status Regulates hENT-1 Expression in iCCA Cell Lines
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. Tissue Microarray and Immunohistochemistry
4.3. Cell Lines
4.4. p53 siRNA Transfection
4.5. RNA Isolation and qRT-PCR
4.6. Western Blotting Analysis
4.7. Confocal Microscopy
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jung, D.H.; Hwang, S.; Kim, K.H.; Hong, S.M.; Lee, Y.J.; Ahn, C.S.; Moon, D.B.; Ha, T.Y.; Song, G.W.; Park, G.C.; et al. Clinicopathological Features and Post-resection Prognosis of Double Primary Hepatocellular Carcinoma and Intrahepatic Cholangiocarcinoma. World J. Surg. 2017, 41, 825–834. [Google Scholar] [CrossRef]
- Valle, J.; Wasan, H.; Palmer, D.H.; Cunningham, D.; Anthoney, A.; Maraveyas, A.; Madhusudan, S.; Iveson, T.; Hughes, S.; Pereira, S.P.; et al. Cisplatin plus gemcitabine versus gemcitabine for biliary tract cancer. N. Engl. J. Med. 2010, 362, 1273–1281. [Google Scholar] [CrossRef]
- Oh, D.Y.; He, A.R.; Bouattour, M.; Okusaka, T.; Qin, S.; Chen, L.T.; Kitano, M.; Lee, C.K.; Kim, J.W.; Chen, M.H.; et al. Durvalumab or placebo plus gemcitabine and cisplatin in participants with advanced biliary tract cancer (TOPAZ-1): Updated overall survival from a randomised phase 3 study. Lancet Gastroenterol. Hepatol. 2024, 9, 694–704. [Google Scholar] [CrossRef] [PubMed]
- Brandi, G.; Deserti, M.; Vasuri, F.; Farioli, A.; Degiovanni, A.; Palloni, A.; Frega, G.; Barbera, M.A.; de Lorenzo, S.; Garajova, I.; et al. Membrane Localization of Human Equilibrative Nucleoside Transporter 1 in Tumor Cells May Predict Response to Adjuvant Gemcitabine in Resected Cholangiocarcinoma Patients. Oncologist 2016, 21, 600–607. [Google Scholar] [CrossRef] [PubMed]
- Tavolari, S.; Deserti, M.; Vasuri, F.; Curti, S.; Palloni, A.; Pinna, A.D.; Cescon, M.; Frega, G.; De Lorenzo, S.; Barbera, M.A.; et al. Membrane human equilibrative nucleoside transporter 1 is associated with a high proliferation rate and worse survival in resected intrahepatic cholangiocarcinoma patients not receiving adjuvant treatments. Eur. J. Cancer. 2019, 106, 160–170. [Google Scholar] [CrossRef] [PubMed]
- Wright, N.J.; Lee, S.Y. Toward a Molecular Basis of Cellular Nucleoside Transport in Humans. Chem. Rev. 2021, 121, 5336–5358. [Google Scholar] [CrossRef]
- Andraus, W.; Tustumi, F.; de Meira Junior, J.D.; Pinheiro, R.S.N.; Waisberg, D.R.; Lopes, L.D.; Arantes, R.M.; Rocha Santos, V.; de Martino, R.B.; Carneiro D’Albuquerque, L.A. Molecular Profile of Intrahepatic Cholangiocarcinoma. Int. J. Mol. Sci. 2023, 25, 461. [Google Scholar] [CrossRef]
- Bouaoun, L.; Sonkin, D.; Ardin, M.; Hollstein, M.; Byrnes, G.; Zavadil, J.; Olivier, M. TP53 Variations in Human Cancers: New Lessons from the IARC TP53 Database and Genomics Data. Hum. Mutat. 2016, 37, 865–876. [Google Scholar] [CrossRef]
- Muller, P.A.; Vousden, K.H. Mutant p53 in cancer: New functions and therapeutic opportunities. Cancer Cell 2014, 25, 304–317. [Google Scholar] [CrossRef]
- Kollareddy, M.; Dimitrova, E.; Vallabhaneni, K.C.; Chan, A.; Le, T.; Chauhan, K.M.; Carrero, Z.I.; Ramakrishnan, G.; Watabe, K.; Haupt, Y.; et al. Regulation of nucleotide metabolism by mutant p53 contributes to its gain-of-function activities. Nat. Commun. 2015, 6, 7389. [Google Scholar] [CrossRef]
- Vijayakumaran, R.; Tan, K.H.; Miranda, P.J.; Haupt, S.; Haupt, Y. Regulation of Mutant p53 Protein Expression. Front. Oncol. 2015, 5, 284. [Google Scholar] [CrossRef] [PubMed]
- Choong, M.L.; Yang, H.; Lee, M.A.; Lane, D.P. Specific activation of the p53 pathway by low dose actinomycin D: A new route to p53 based cyclotherapy. Cell Cycle. 2009, 8, 2810–2818. [Google Scholar] [CrossRef] [PubMed]
- Perera, S.; Jang, G.H.; Wang, Y.; Kelly, D.; Allen, M.; Zhang, A.; Denroche, R.E.; Dodd, A.; Ramotar, S.; Hutchinson, S.; et al. hENT1 Expression Predicts Response to Gemcitabine and Nab-Paclitaxel in Advanced Pancreatic Ductal Adenocarcinoma. Clin. Cancer Res. 2022, 28, 5115–5120. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Zhao, F.; Luo, W.; Yang, G.; Wang, Y.; Qiu, J.; Liu, Y.; You, L.; Zheng, L.; Zhang, T. Human Equilibrative Nucleoside Transporter 1: Novel Biomarker and Prognostic Indicator for Patients with Gemcitabine-Treated Pancreatic Cancer. Cancer Manag. Res. 2024, 16, 651–661. [Google Scholar] [CrossRef]
- Santini, D.; Perrone, G.; Vincenzi, B.; Lai, R.; Cass, C.; Alloni, R.; Rabitti, C.; Antinori, A.; Vecchio, F.; Morini, S.; et al. Human equilibrative nucleoside transporter 1 (hENT1) protein is associated with short survival in resected ampullary cancer. Ann. Oncol. 2008, 19, 724–728. [Google Scholar] [CrossRef]
- Vincenzi, B.; Stacchiotti, S.; Collini, P.; Pantano, F.; Rabitti, C.; Perrone, G.; Iuliani, M.; Baldi, A.; Badalamenti, G.; Sanfilippo, R.; et al. Human equilibrative nucleoside transporter 1 gene expression is associated with gemcitabine efficacy in advanced leiomyosarcoma and angiosarcoma. Br. J. Cancer 2017, 117, 340–346. [Google Scholar] [CrossRef]
- Santini, D.; Vincenzi, B.; Fratto, M.E.; Perrone, G.; Lai, R.; Catalano, V.; Cass, C.; Ruffini, P.A.; Spoto, C.; Muretto, P.; et al. Prognostic role of human equilibrative transporter 1 (hENT1) in patients with resected gastric cancer. J. Cell. Physiol. 2010, 223, 384–388. [Google Scholar] [CrossRef]
- Leisewitz, A.V.; Zimmerman, E.I.; Huang, M.; Jones, S.Z.; Yang, J.; Graves, L.M. Regulation of ENT1 expression and ENT1-dependent nucleoside transport by c-Jun N-terminal kinase. Biochem. Biophys. Res. Commun. 2011, 404, 370–375. [Google Scholar] [CrossRef]
- Eltzschig, H.K.; Abdulla, P.; Hoffman, E.; Hamilton, K.E.; Daniels, D.; Schönfeld, C.; Löffler, M.; Reyes, G.; Duszenko, M.; Karhausen, J.; et al. HIF-1-dependent repression of equilibrative nucleoside transporter (ENT) in hypoxia. J. Exp. Med. 2005, 202, 1493–1505. [Google Scholar] [CrossRef]
- Petrovic, V.; Teng, S.; Piquette-Miller, M. Regulation of drug transporters during infection and inflammation. Mol. Interv. 2007, 7, 99–111. [Google Scholar] [CrossRef]
- Sinn, M.; Sinn, B.V.; Treue, D.; Keilholz, U.; Damm, F.; Schmuck, R.; Lohneis, P.; Klauschen, F.; Striefler, J.K.; Bahra, M.; et al. TP53 Mutations Predict Sensitivity to Adjuvant Gemcitabine in Patients with Pancreatic Ductal Adenocarcinoma: Next-Generation Sequencing Results from the CONKO-001 Trial. Clin. Cancer Res. 2020, 26, 3732–3739. [Google Scholar] [CrossRef] [PubMed]


| Characteristics | Patients (n = 38) |
| Age at surgery (years), median (range) | 61 (33–79) |
Gender, n (%)
| 20 (52.6) 18 (47.4) |
Histological grade, n (%)
| 9 (23.7) 8 (21) 13 (34.2) 3 (7.9) 5 (13.1) |
Size and extent (T), n (%)
| 11 (29) 19 (50) 4 (10.5) 4 (10.5) 0 (0) |
Regional lymph nodes (N), n (%)
| 20 (52.6) 14 (36.8) 4 (10.6) |
Distant metastases (M), n (%)
| 36 (94.7) 2 (5.3) 0 (0) |
Resection margins, n (%)
| 37 (97.3) 1 (2.7) 0 (0) |
| Mutant p53 Cells ≤ 4% | Mutant p53 Cells > 4% | p-Value * | |
| Membrane hENT-1 positive | 3 | 10 | 0.0013 |
| Membrane hENT-1 negative | 10 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deserti, M.; Relli, V.; Palloni, A.; Vasuri, F.; Malvi, D.; Degiovanni, A.; Rimedio, S.; Delbaldo, C.; Deiana, C.; Brandi, G.; et al. Mutant p53 Associates with Human Equilibrative Nucleoside 1 Upregulation and Better Response to Adjuvant Gemcitabine in Intrahepatic Cholangiocarcinoma Patients. Int. J. Mol. Sci. 2025, 26, 5259. https://doi.org/10.3390/ijms26115259
Deserti M, Relli V, Palloni A, Vasuri F, Malvi D, Degiovanni A, Rimedio S, Delbaldo C, Deiana C, Brandi G, et al. Mutant p53 Associates with Human Equilibrative Nucleoside 1 Upregulation and Better Response to Adjuvant Gemcitabine in Intrahepatic Cholangiocarcinoma Patients. International Journal of Molecular Sciences. 2025; 26(11):5259. https://doi.org/10.3390/ijms26115259
Chicago/Turabian StyleDeserti, Marzia, Valeria Relli, Andrea Palloni, Francesco Vasuri, Deborah Malvi, Alessio Degiovanni, Simone Rimedio, Chiara Delbaldo, Chiara Deiana, Giovanni Brandi, and et al. 2025. "Mutant p53 Associates with Human Equilibrative Nucleoside 1 Upregulation and Better Response to Adjuvant Gemcitabine in Intrahepatic Cholangiocarcinoma Patients" International Journal of Molecular Sciences 26, no. 11: 5259. https://doi.org/10.3390/ijms26115259
APA StyleDeserti, M., Relli, V., Palloni, A., Vasuri, F., Malvi, D., Degiovanni, A., Rimedio, S., Delbaldo, C., Deiana, C., Brandi, G., & Tavolari, S. (2025). Mutant p53 Associates with Human Equilibrative Nucleoside 1 Upregulation and Better Response to Adjuvant Gemcitabine in Intrahepatic Cholangiocarcinoma Patients. International Journal of Molecular Sciences, 26(11), 5259. https://doi.org/10.3390/ijms26115259






