Design, Co-Expression, and Evaluation for Assembly of the Structural Proteins from Thermophilic Bacteriophage ΦIN93
Abstract
1. Introduction
2. Results
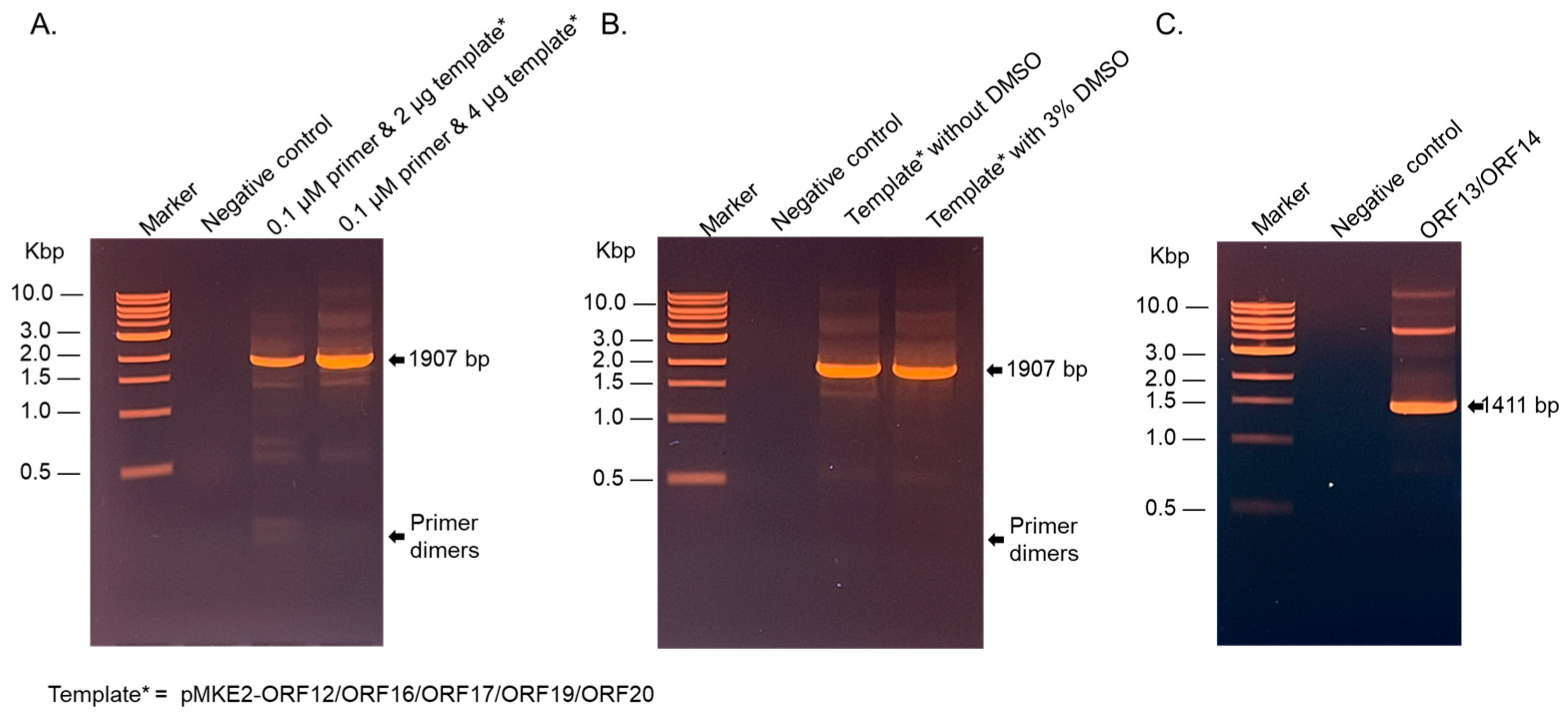
2.1. The DNA of the Coat Proteins and Membrane-Associated Proteins (Non-Codon Optimized) Were Successfully Amplified by PCR in the Presence of 3% Dimethyl Sulfoxide (DMSO)
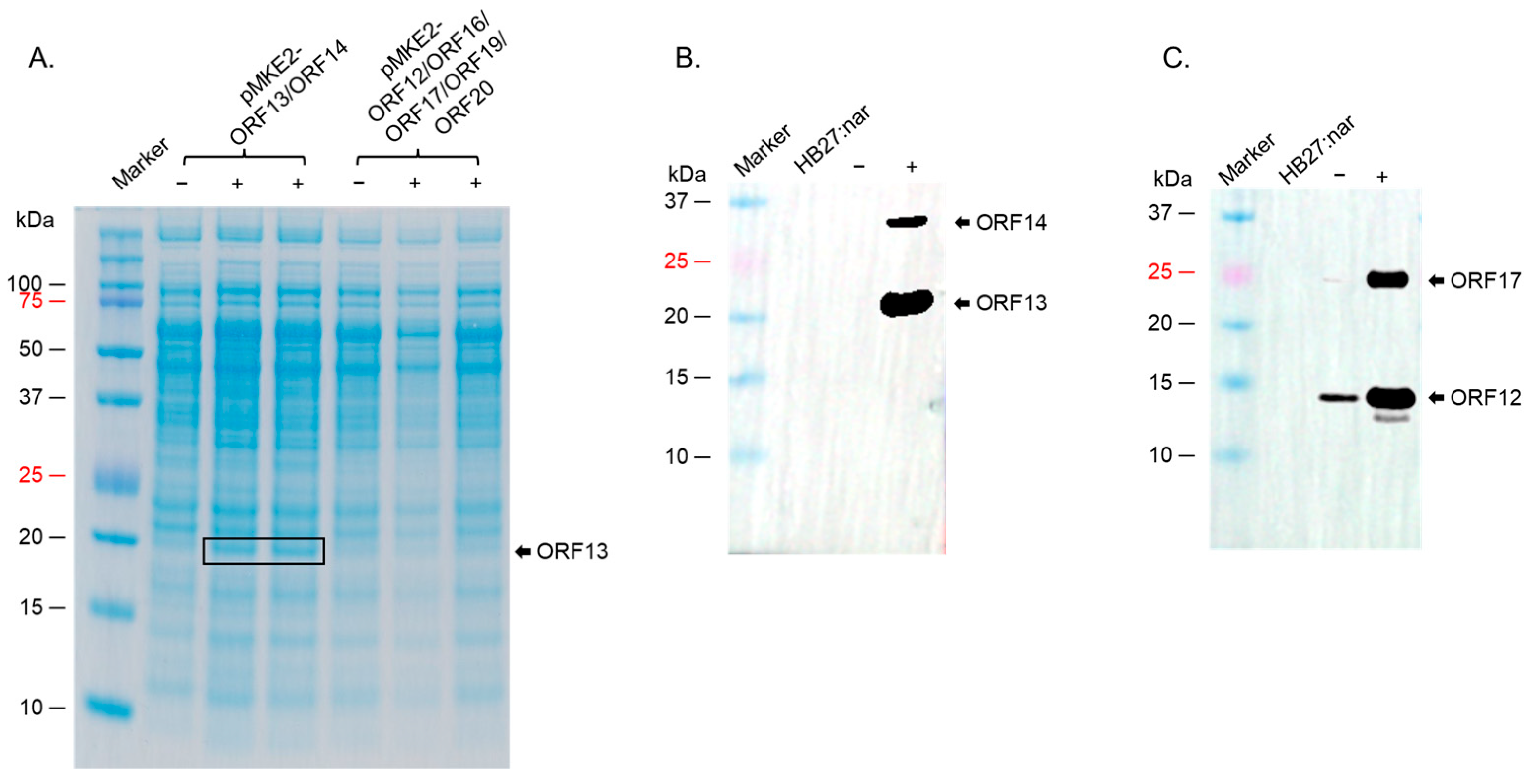
2.2. ORF13/ORF14 Coat Proteins and ORF12/ORF17 Membrane-Associated Proteins Were Successfully Expressed in T. thermophilus HB27:nar but at Different Levels
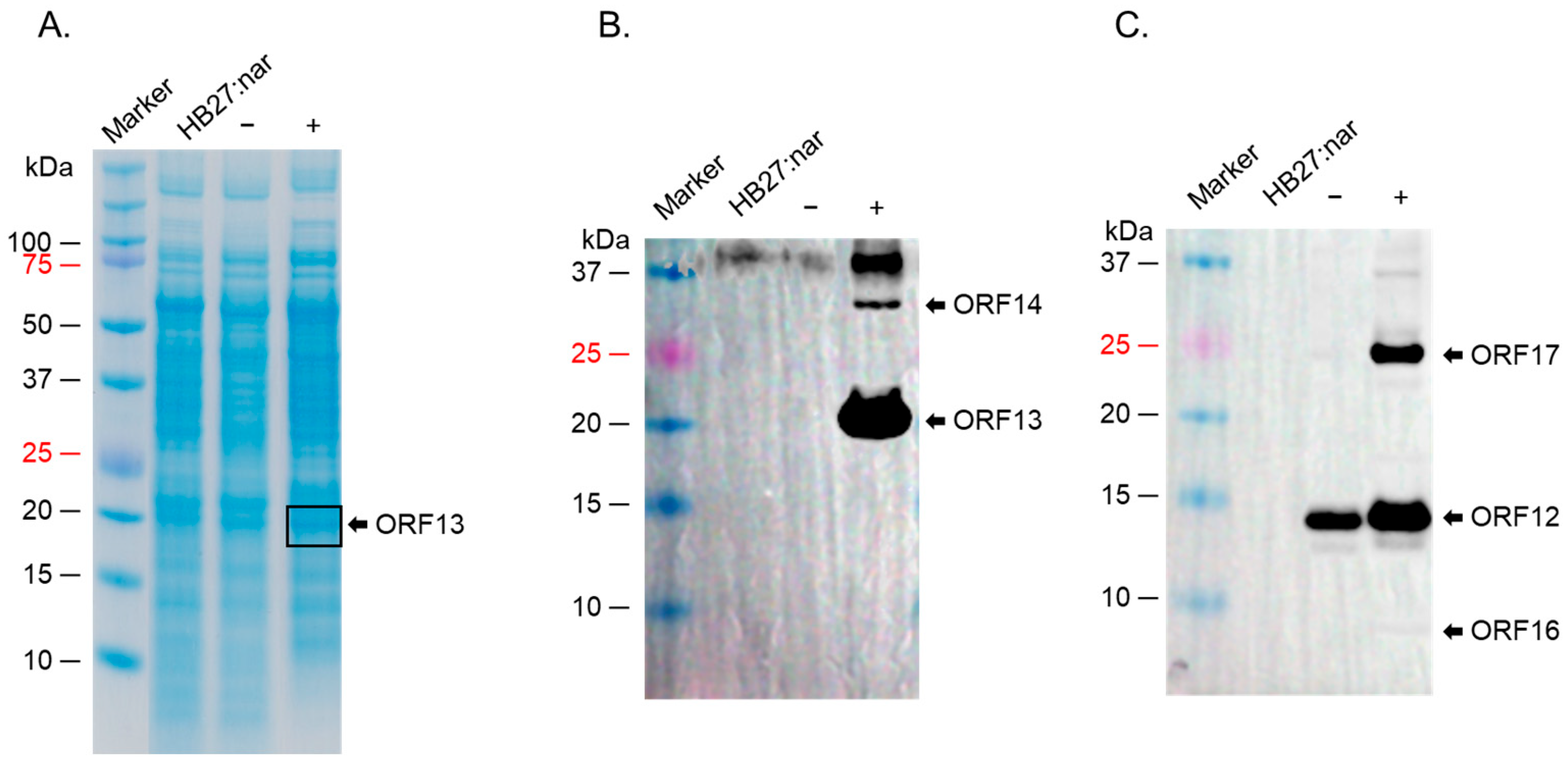
2.3. ORF12, ORF13, ORF14, ORF16, and ORF17 Were Successfully Co-Expressed in a Strain of E. coli (BL21 Star)
3. Discussion
4. Materials and Methods
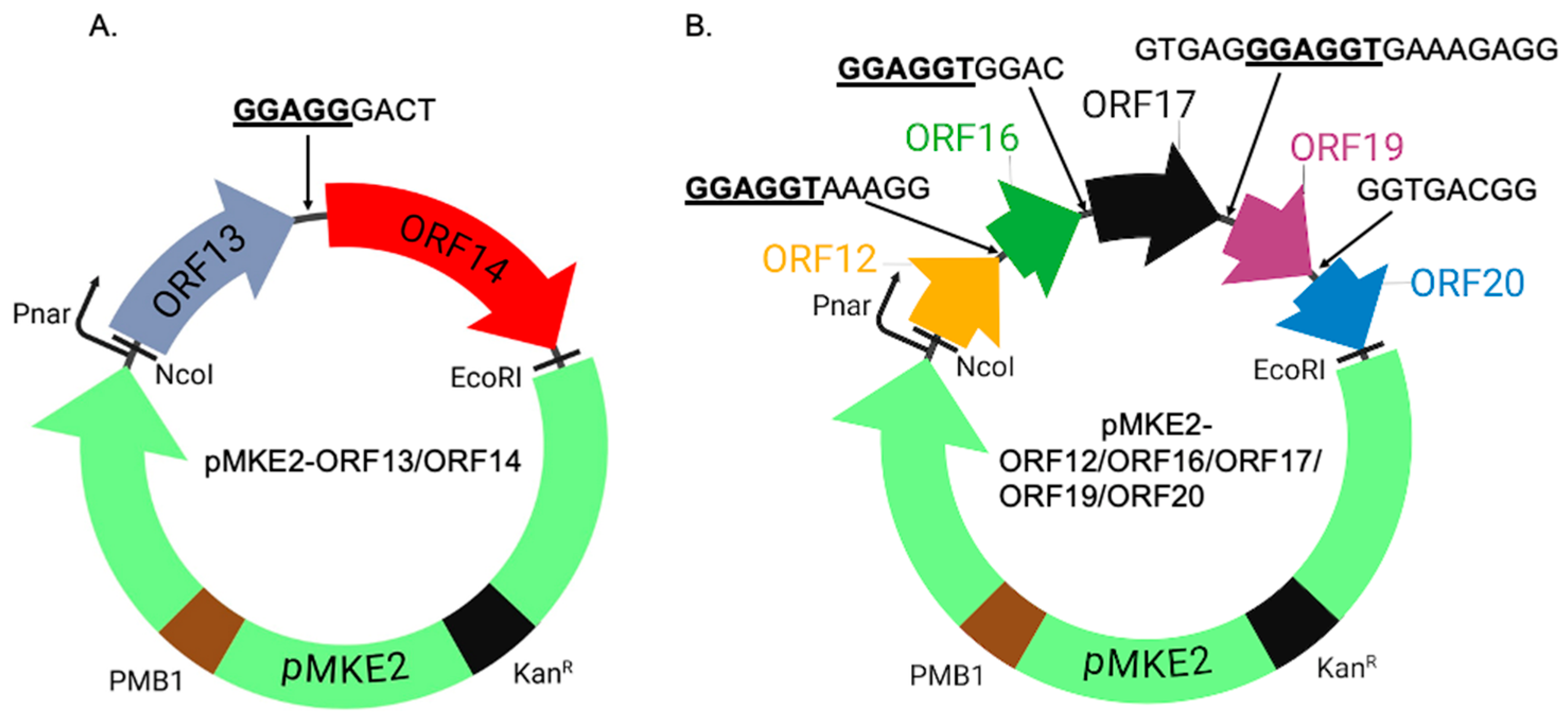
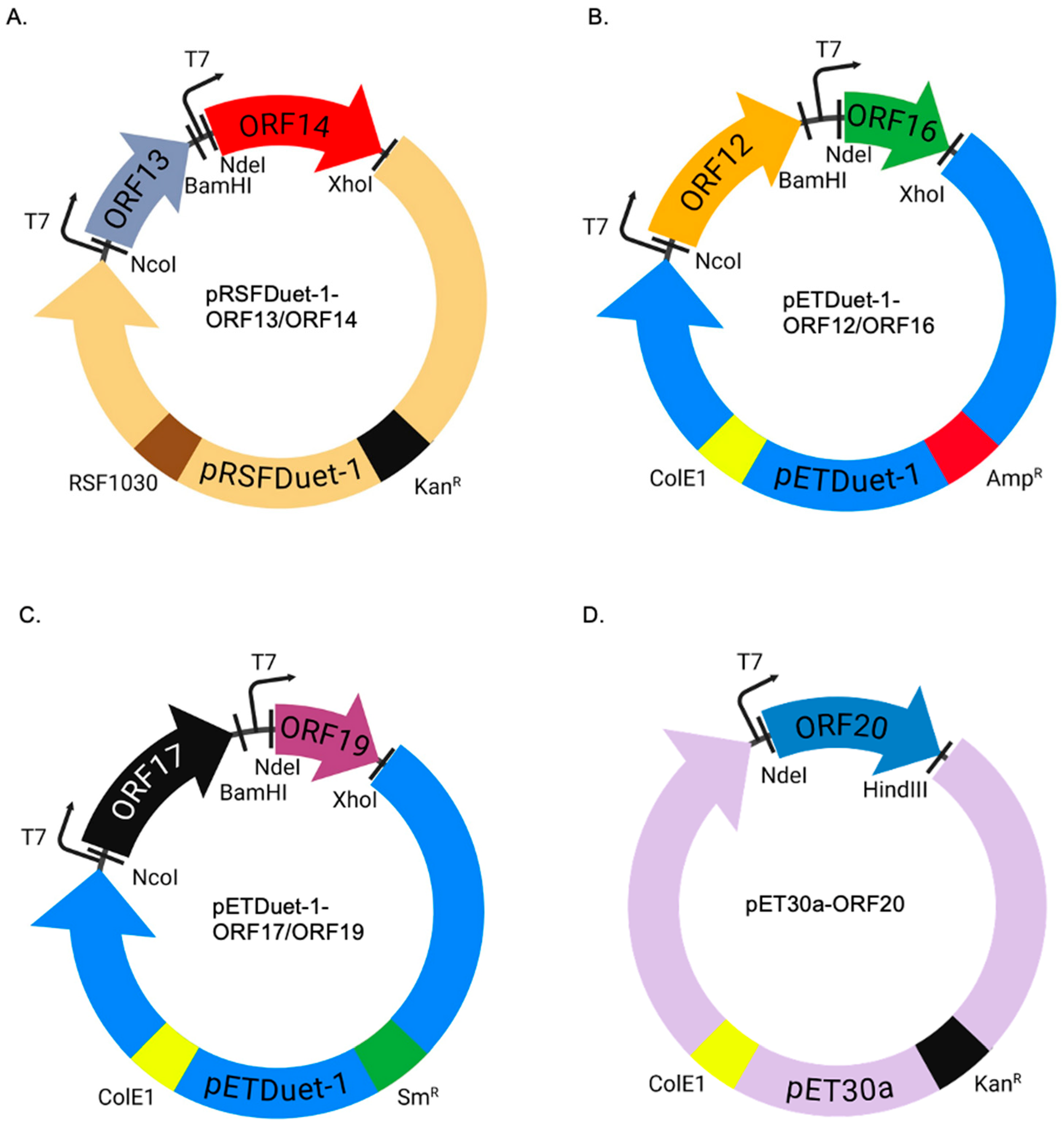
4.1. Cloning of Coat Proteins and Membrane-Associated Proteins in Expression Vectors
4.2. Co-Expression of Coat Proteins and Membrane-Associated Proteins in a Thermophilic Bacterium (HB27:nar) and in E. coli
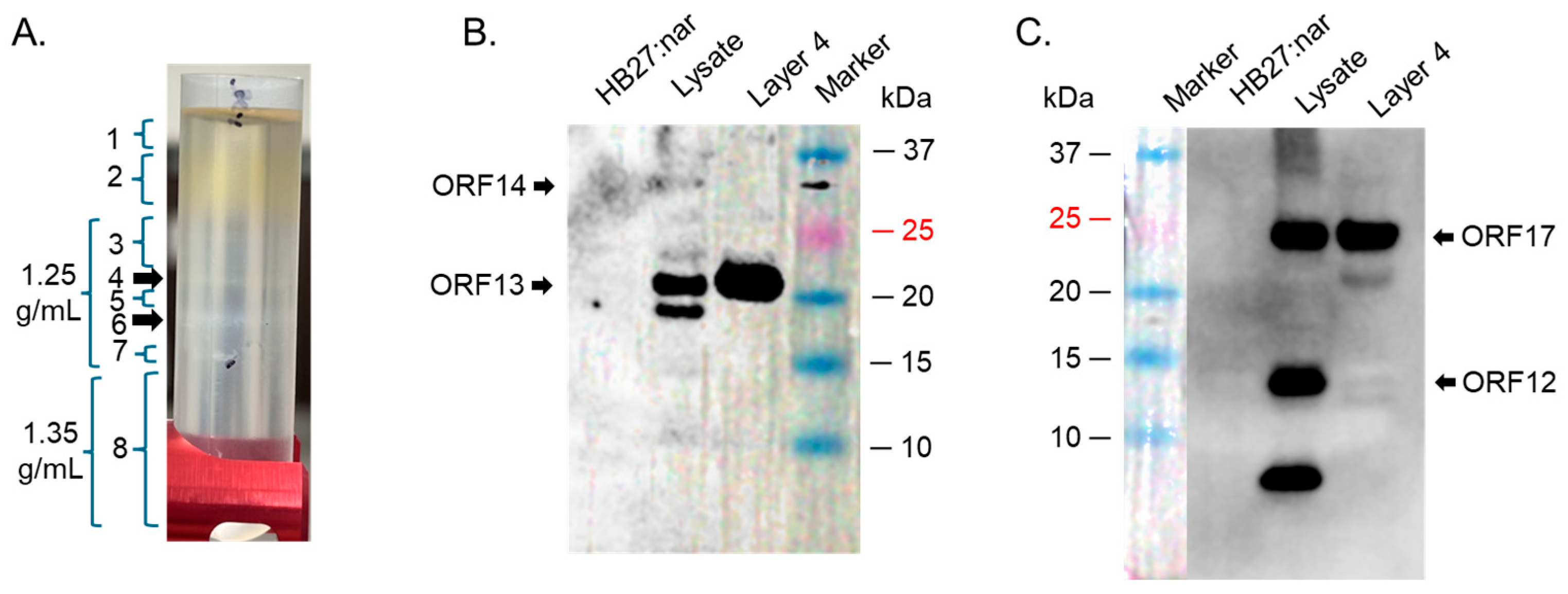
4.3. Ultracentrifugation of Co-Expressed Proteins to Assess the Formation of VLPs
4.4. Western Blot on Bacterial Lysates and on Bands from Ultracentrifugation
4.5. Transmission Electron Microscopy (TEM)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Matsushita, I.; Yamashita, N. Isolation and Characterization of Bacteriophage Induced from a New Isolate of Thermus aquaticus. Microbiol. Cult. Coll. 1995, 11, 133–138. [Google Scholar]
- Pawlowski, A.; Rissanen, I.; Bamford, J.K.; Krupovic, M.; Jalasvuori, M. Gammasphaerolipovirus, a newly proposed bacteriophage genus, unifies viruses of halophilic archaea and thermophilic bacteria within the novel family Sphaerolipoviridae. Arch. Virol. 2014, 159, 1541–1554. [Google Scholar] [CrossRef] [PubMed]
- Jalasvuori, M.; Jaatinen, S.T.; Laurinavicius, S.; Ahola-Iivarinen, E.; Kalkkinen, N.; Bamford, D.H.; Bamford, J.K. The closest relatives of icosahedral viruses of thermophilic bacteria are among viruses and plasmids of the halophilic archaea. J. Virol. 2009, 83, 9388–9397. [Google Scholar] [CrossRef]
- Zhai, L.; Anderson, D.; Bruckner, E.; Tumban, E. Novel expression of coat proteins from thermophilic bacteriophage PhiIN93 and evaluation for assembly into virus-like particles. Protein Expr. Purif. 2021, 187, 105932. [Google Scholar] [CrossRef]
- Kheirvari, M.; Liu, H.; Tumban, E. Virus-like Particle Vaccines and Platforms for Vaccine Development. Viruses 2023, 15, 1109. [Google Scholar] [CrossRef]
- Pumpens, P.; Pushko, P. Virus-like Particles: A Comprehensive Guide; CRC Pr I Llc: Boca Raton, FL, USA, 2022. [Google Scholar]
- Rissanen, I.; Grimes, J.M.; Pawlowski, A.; Mantynen, S.; Harlos, K.; Bamford, J.K.; Stuart, D.I. Bacteriophage p23-77 capsid protein structures reveal the archetype of an ancient branch from a major virus lineage. Structure 2013, 21, 718–726. [Google Scholar] [CrossRef]
- Jaatinen, S.T.; Happonen, L.J.; Laurinmaki, P.; Butcher, S.J.; Bamford, D.H. Biochemical and structural characterisation of membrane-containing icosahedral dsDNA bacteriophages infecting thermophilic Thermus thermophilus. Virology 2008, 379, 10–19. [Google Scholar] [CrossRef]
- Zak, D.E.; Andersen-Nissen, E.; Peterson, E.R.; Sato, A.; Hamilton, M.K.; Borgerding, J.; Krishnamurty, A.T.; Chang, J.T.; Adams, D.J.; Hensley, T.R.; et al. Merck Ad5/HIV induces broad innate immune activation that predicts CD8(+) T-cell responses but is attenuated by preexisting Ad5 immunity. Proc. Natl. Acad. Sci. USA 2012, 109, E3503–E3512. [Google Scholar] [CrossRef]
- Knuchel, M.C.; Marty, R.R.; Morin, T.N.; Ilter, O.; Zuniga, A.; Naim, H.Y. Relevance of a pre-existing measles immunity prior immunization with a recombinant measles virus vector. Hum. Vaccin. Immunother. 2013, 9, 599–606. [Google Scholar] [CrossRef]
- Zarnitsyna, V.I.; Lavine, J.; Ellebedy, A.; Ahmed, R.; Antia, R. Multi-epitope Models Explain How Pre-existing Antibodies Affect the Generation of Broadly Protective Responses to Influenza. PLoS Pathog. 2016, 12, e1005692. [Google Scholar] [CrossRef]
- Voysey, M.; Kelly, D.F.; Fanshawe, T.R.; Sadarangani, M.; O’Brien, K.L.; Perera, R.; Pollard, A.J. The Influence of Maternally Derived Antibody and Infant Age at Vaccination on Infant Vaccine Responses: An Individual Participant Meta-analysis. JAMA Pediatr. 2017, 171, 637–646. [Google Scholar] [CrossRef] [PubMed]
- Masat, E.; Pavani, G.; Mingozzi, F. Humoral immunity to AAV vectors in gene therapy: Challenges and potential solutions. Discov. Med. 2013, 15, 379–389. [Google Scholar] [PubMed]
- McCluskie, M.J.; Evans, D.M.; Zhang, N.; Benoit, M.; McElhiney, S.P.; Unnithan, M.; DeMarco, S.C.; Clay, B.; Huber, C.; Deora, A.; et al. The effect of preexisting anti-carrier immunity on subsequent responses to CRM197 or Qb-VLP conjugate vaccines. Immunopharmacol. Immunotoxicol. 2016, 38, 184–196. [Google Scholar] [CrossRef]
- Demina, T.A.; Pietila, M.K.; Svirskaite, J.; Ravantti, J.J.; Atanasova, N.S.; Bamford, D.H.; Oksanen, H.M. HCIV-1 and Other Tailless Icosahedral Internal Membrane-Containing Viruses of the Family Sphaerolipoviridae. Viruses 2017, 9, 32. [Google Scholar] [CrossRef]
- Matsushita, I.; Yanase, H. The genomic structure of thermus bacteriophage phiIN93. J. Biochem. 2009, 146, 775–785. [Google Scholar] [CrossRef]
- Liu, H.; Kheirvari, M.; Tumban, E. Potential Applications of Thermophilic Bacteriophages in One Health. Int. J. Mol. Sci. 2023, 24, 8222. [Google Scholar] [CrossRef]
- Travassos, R.; Martins, S.A.; Fernandes, A.; Correia, J.D.G.; Melo, R. Tailored Viral-like Particles as Drivers of Medical Breakthroughs. Int. J. Mol. Sci. 2024, 25, 6699. [Google Scholar] [CrossRef]
- Samandoulgou, I.; Hammami, R.; Morales Rayas, R.; Fliss, I.; Jean, J. Stability of Secondary and Tertiary Structures of Virus-Like Particles Representing Noroviruses: Effects of pH, Ionic Strength, and Temperature and Implications for Adhesion to Surfaces. Appl. Environ. Microbiol. 2015, 81, 7680–7686. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, L.; Zhang, C.; Lu, Y. Exploration on the expression and assembly of virus-like particles. Biotechnol. Notes 2021, 2, 51–58. [Google Scholar] [CrossRef]
- Le, D.T.; Müller, K.M. In Vitro Assembly of Virus-Like Particles and Their Applications. Life 2021, 11, 334. [Google Scholar] [CrossRef]
- Ramirez-Arcos, S.; Fernandez-Herrero, L.A.; Marin, I.; Berenguer, J. Anaerobic growth, a property horizontally transferred by an Hfr-like mechanism among extreme thermophiles. J. Bacteriol. 1998, 180, 3137–3143. [Google Scholar] [CrossRef] [PubMed]
- Biolabs, N.E. Guidelines for PCR Optimization with Phusion High-Fidelity DNA Polymerase. 2023. Available online: https://www.neb.com/en-us/protocols/2012/06/01/guidelines-for-pcr-optimization-with-phusion-high-fidelity-dna-polymerase (accessed on 1 June 2012).
- Liu, Y.; Zhou, J. The P124A mutation of SRP14 alters its migration on SDS-PAGE without impacting its function. Acta Biochim. Biophys. Sin. 2024, 56, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Kirkland, T.N.; Finley, F.; Orsborn, K.I.; Galgiani, J.N. Evaluation of the proline-rich antigen of Coccidioides immitis as a vaccine candidate in mice. Infect. Immun. 1998, 66, 3519–3522. [Google Scholar] [CrossRef]
- Stainier, I.; Bleves, S.; Josenhans, C.; Karmani, L.; Kerbourch, C.; Lambermont, I.; Totemeyer, S.; Boyd, A.; Cornelis, G.R. YscP, a Yersinia protein required for Yop secretion that is surface exposed, and released in low Ca2+. Mol. Microbiol. 2000, 37, 1005–1018. [Google Scholar] [CrossRef]
- Jones, K.L.; Kim, S.-W.; Keasling, J.D. Low-Copy Plasmids can Perform as Well as or Better Than High-Copy Plasmids for Metabolic Engineering of Bacteria. Metab. Eng. 2000, 2, 328–338. [Google Scholar] [CrossRef]
- Silva, F.; Queiroz, J.A.; Domingues, F.C. Evaluating metabolic stress and plasmid stability in plasmid DNA production by Escherichia coli. Biotechnol. Adv. 2012, 30, 691–708. [Google Scholar] [CrossRef]
- Lozano Terol, G.; Gallego-Jara, J.; Sola Martínez, R.A.; Martínez Vivancos, A.; Cánovas Díaz, M.; de Diego Puente, T. Impact of the Expression System on Recombinant Protein Production in Escherichia coli BL21. Front. Microbiol. 2021, 12, 682001. [Google Scholar] [CrossRef]
- Glick, B.R. Metabolic load and heterologous gene expression. Biotechnol. Adv. 1995, 13, 247–261. [Google Scholar] [CrossRef]
- Hayat, S.M.G.; Farahani, N.; Golichenari, B.; Sahebkar, A. Recombinant Protein Expression in Escherichia coli (E. coli): What We Need to Know. Curr. Pharm. Des. 2018, 24, 718–725. [Google Scholar] [CrossRef]
- Moreno, R.; Haro, A.; Castellanos, A.; Berenguer, J. High-level overproduction of His-tagged Tth DNA polymerase in Thermus thermophilus. Appl. Environ. Microbiol. 2005, 71, 591–593. [Google Scholar] [CrossRef]
- Wu, W.L.; Liao, J.H.; Lin, G.H.; Lin, M.H.; Chang, Y.C.; Liang, S.Y.; Yang, F.L.; Khoo, K.H.; Wu, S.H. Phosphoproteomic analysis reveals the effects of PilF phosphorylation on type IV pilus and biofilm formation in Thermus thermophilus hb27. Mol. Cell Proteomics 2013, 12, 2701–2713. [Google Scholar] [CrossRef] [PubMed]
- Zhai, L.; Peabody, J.; Pang, Y.S.; Schiller, J.; Chackerian, B.; Tumban, E. A novel candidate HPV vaccine: MS2 phage VLP displaying a tandem HPV L2 peptide offers similar protection in mice to Gardasil-9. Antiviral. Res. 2017, 147, 116–123. [Google Scholar] [CrossRef] [PubMed]
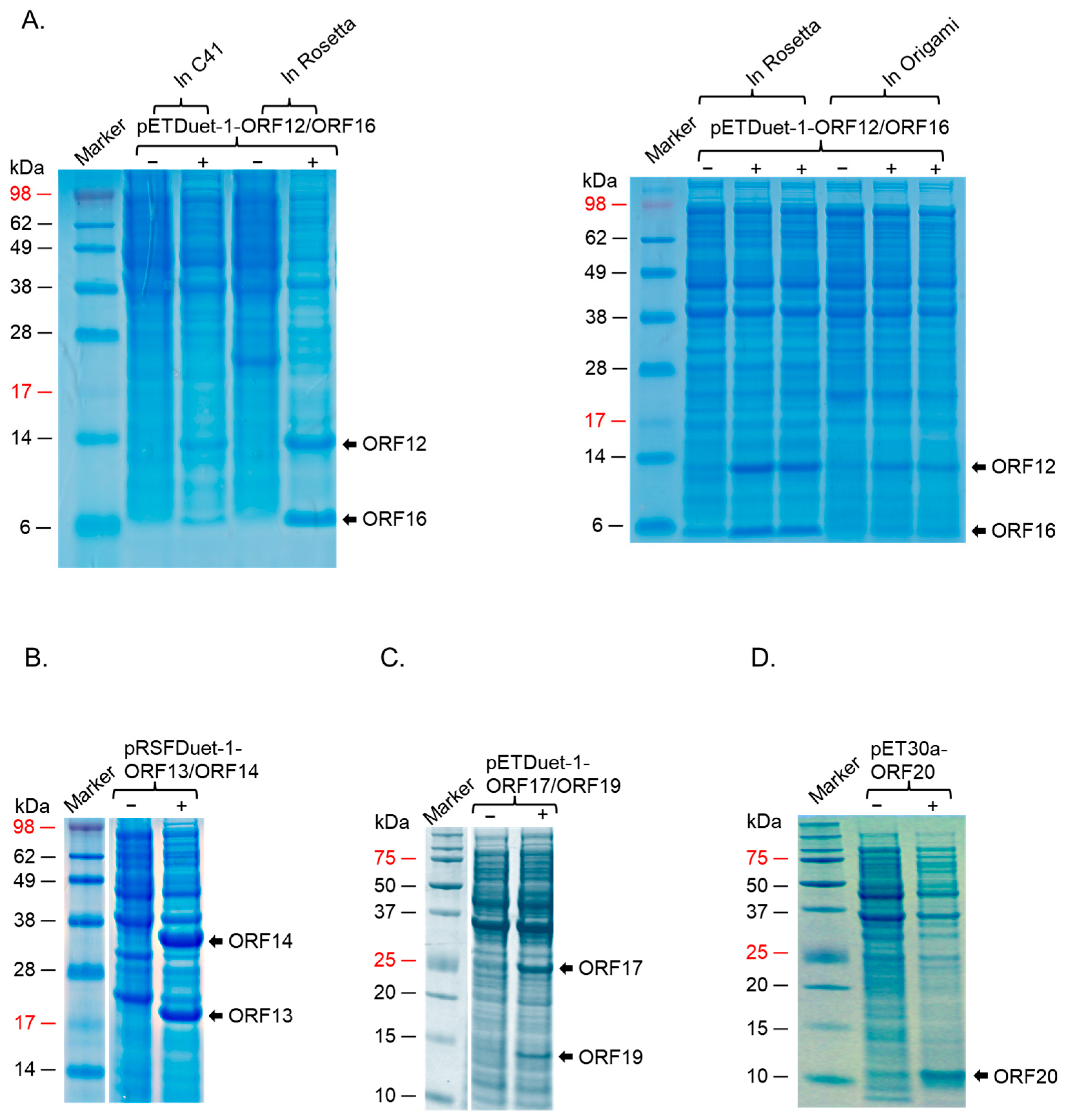
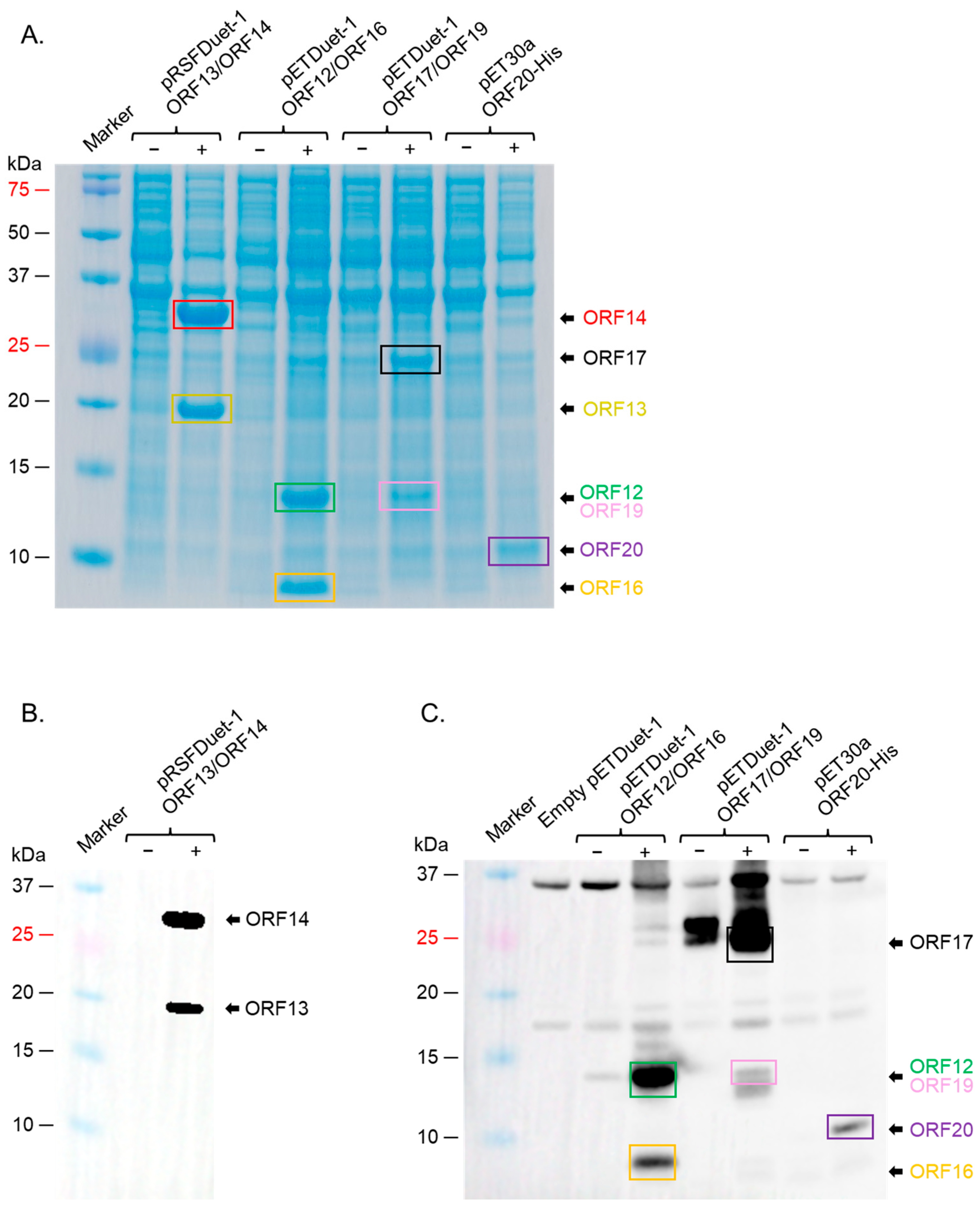
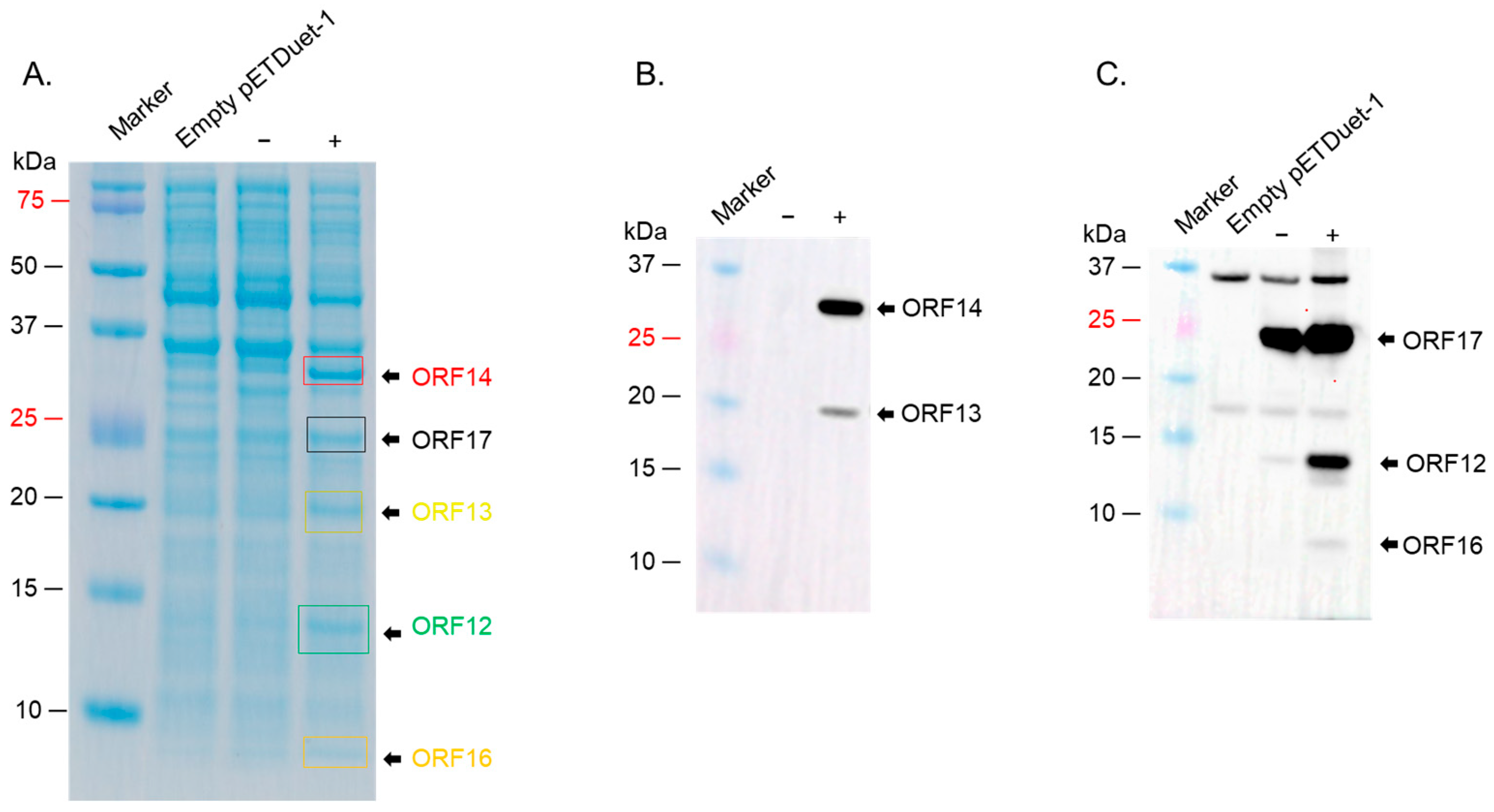
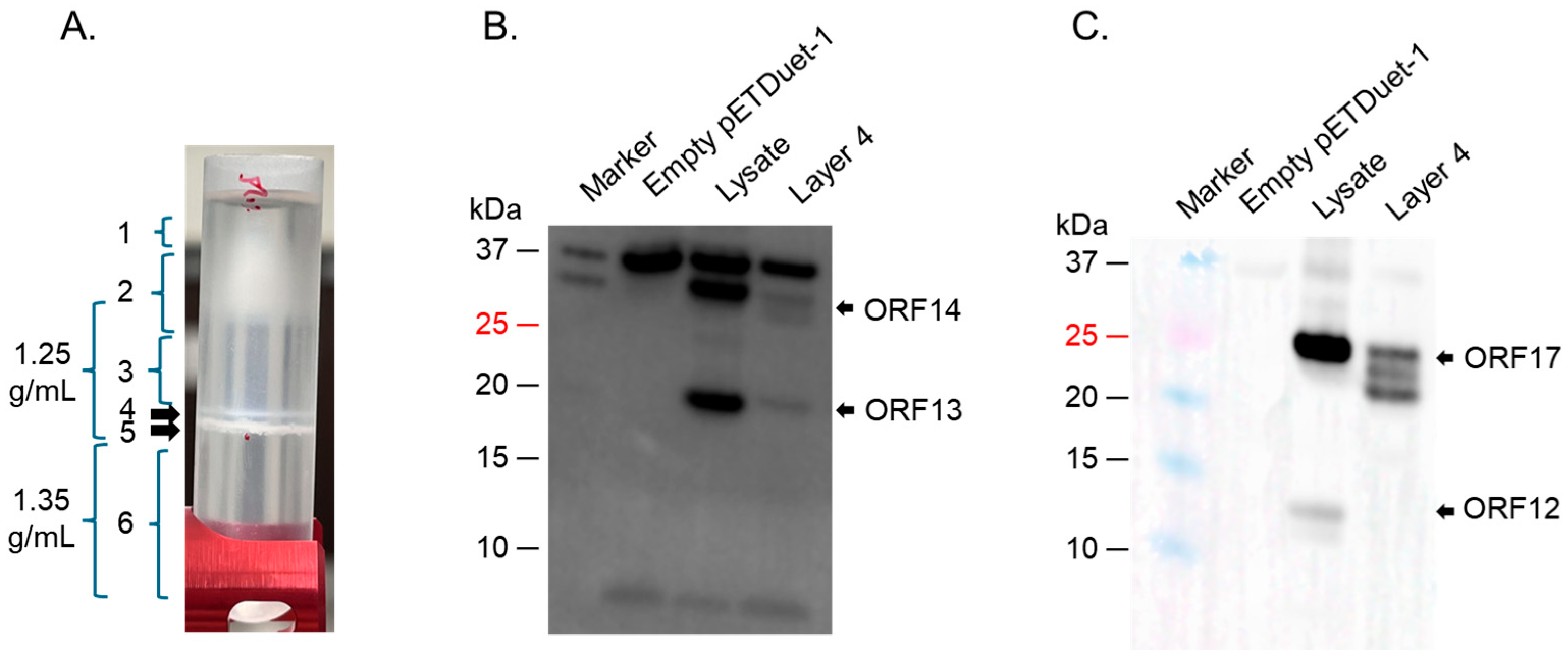
| ΦIN93 Protein | Predicted Molecular Weight (KDa) * | Predicted Function | Homolog in P23-77 (% Homology) |
|---|---|---|---|
| ORF12 | 14.6 | Membrane-associate protein | VP15 (~83%) |
| ORF13 | 19.3 | capsid protein | VP16 (~80%) |
| ORF14 | 32.0 | capsid protein | VP17 (73%) |
| ORF16 | 8.9 | Membrane-associate protein | VP19 (~68%) |
| ORF17 | 23.3 | Membrane-associate protein | VP20 (~69%) |
| ORF19 | 10.3 | Membrane-associate protein | VP22 (~51%) |
| ORF20 | 7.7 | Membrane-associate protein | VP23 (~56%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.; Kheirvari, M.; Tumban, E. Design, Co-Expression, and Evaluation for Assembly of the Structural Proteins from Thermophilic Bacteriophage ΦIN93. Int. J. Mol. Sci. 2025, 26, 5201. https://doi.org/10.3390/ijms26115201
Liu H, Kheirvari M, Tumban E. Design, Co-Expression, and Evaluation for Assembly of the Structural Proteins from Thermophilic Bacteriophage ΦIN93. International Journal of Molecular Sciences. 2025; 26(11):5201. https://doi.org/10.3390/ijms26115201
Chicago/Turabian StyleLiu, Hong, Milad Kheirvari, and Ebenezer Tumban. 2025. "Design, Co-Expression, and Evaluation for Assembly of the Structural Proteins from Thermophilic Bacteriophage ΦIN93" International Journal of Molecular Sciences 26, no. 11: 5201. https://doi.org/10.3390/ijms26115201
APA StyleLiu, H., Kheirvari, M., & Tumban, E. (2025). Design, Co-Expression, and Evaluation for Assembly of the Structural Proteins from Thermophilic Bacteriophage ΦIN93. International Journal of Molecular Sciences, 26(11), 5201. https://doi.org/10.3390/ijms26115201





