Novel lncRNA UGGT1-AS1 Regulates UGGT1 Expression in Breast Cancer Cell Line
Abstract
1. Introduction
2. Results
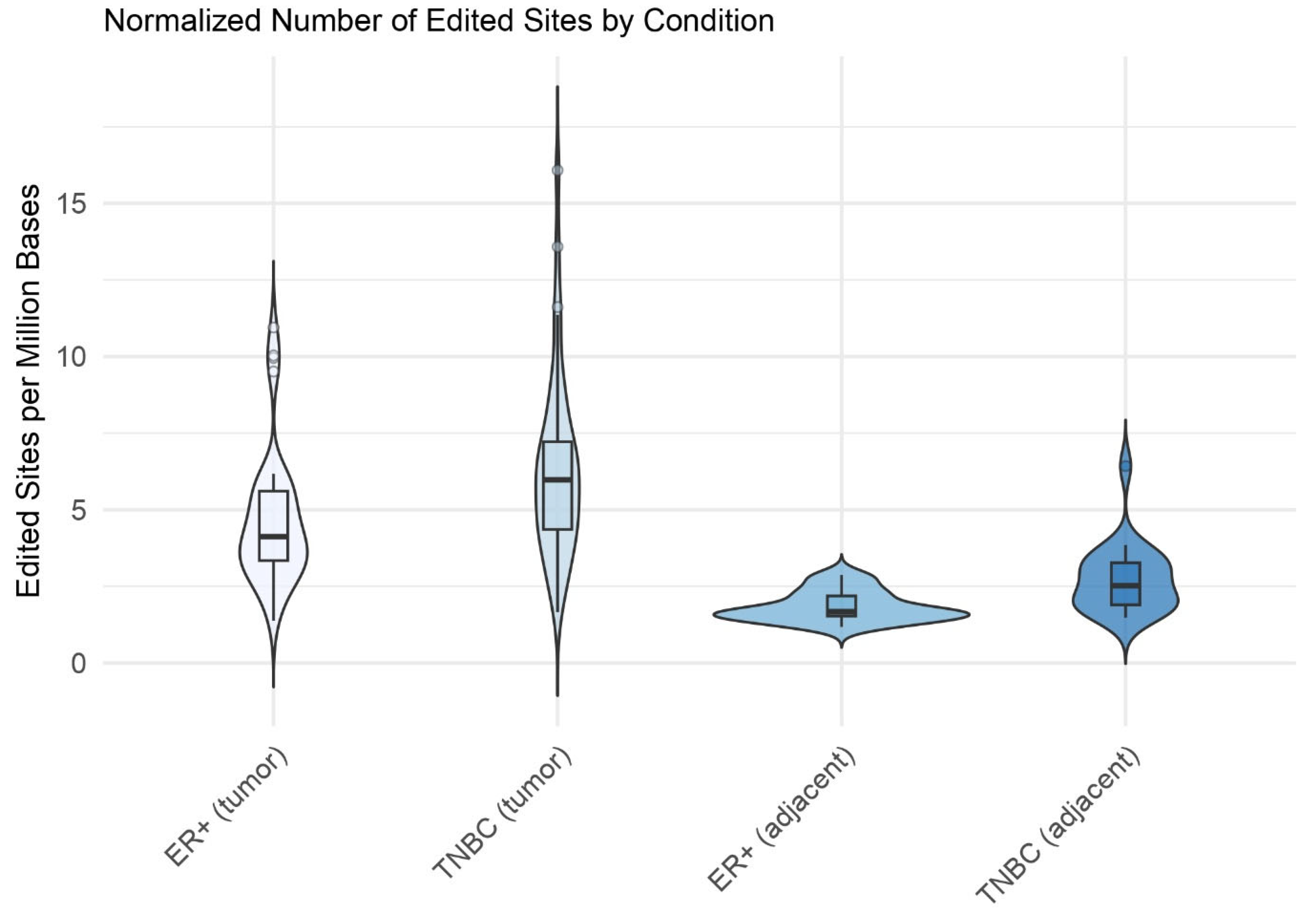
2.1. A-to-I Editing Events in Breast Cancer
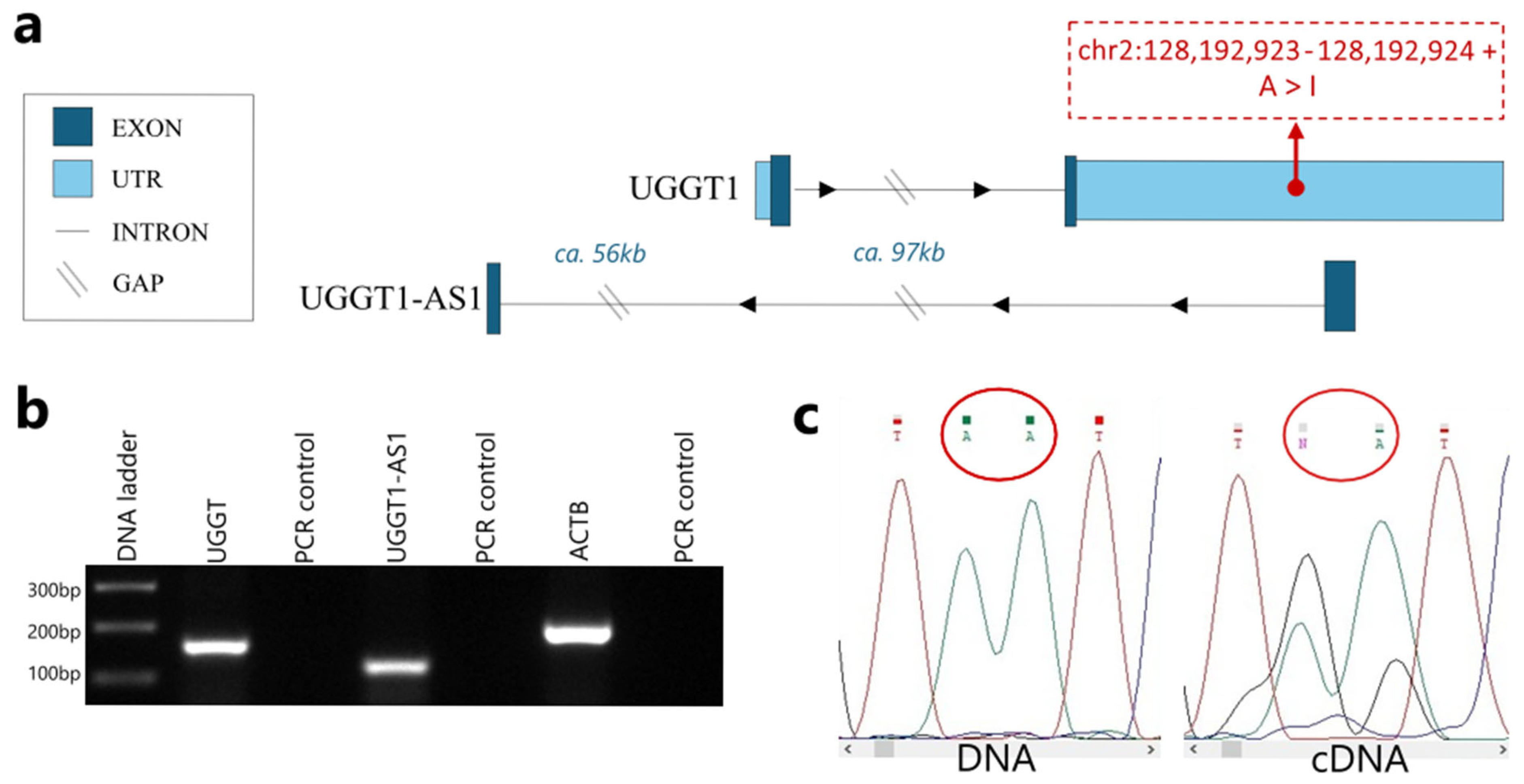
2.2. UGGT1-AS1 Is a Natural Antisense Transcript of the UGGT1 Gene
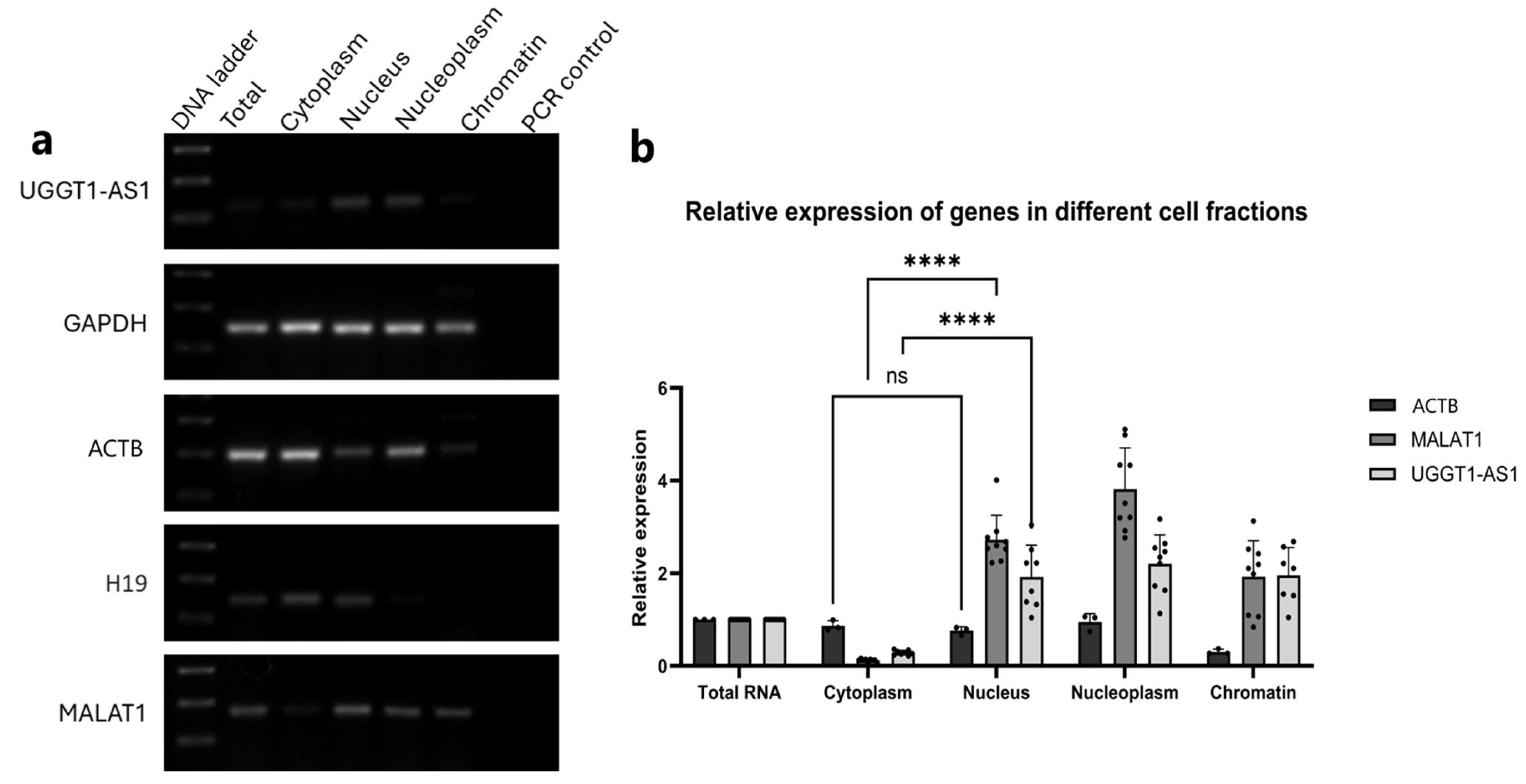
2.3. UGGT1-AS1 Transcripts Are Located in the Cell Nucleus
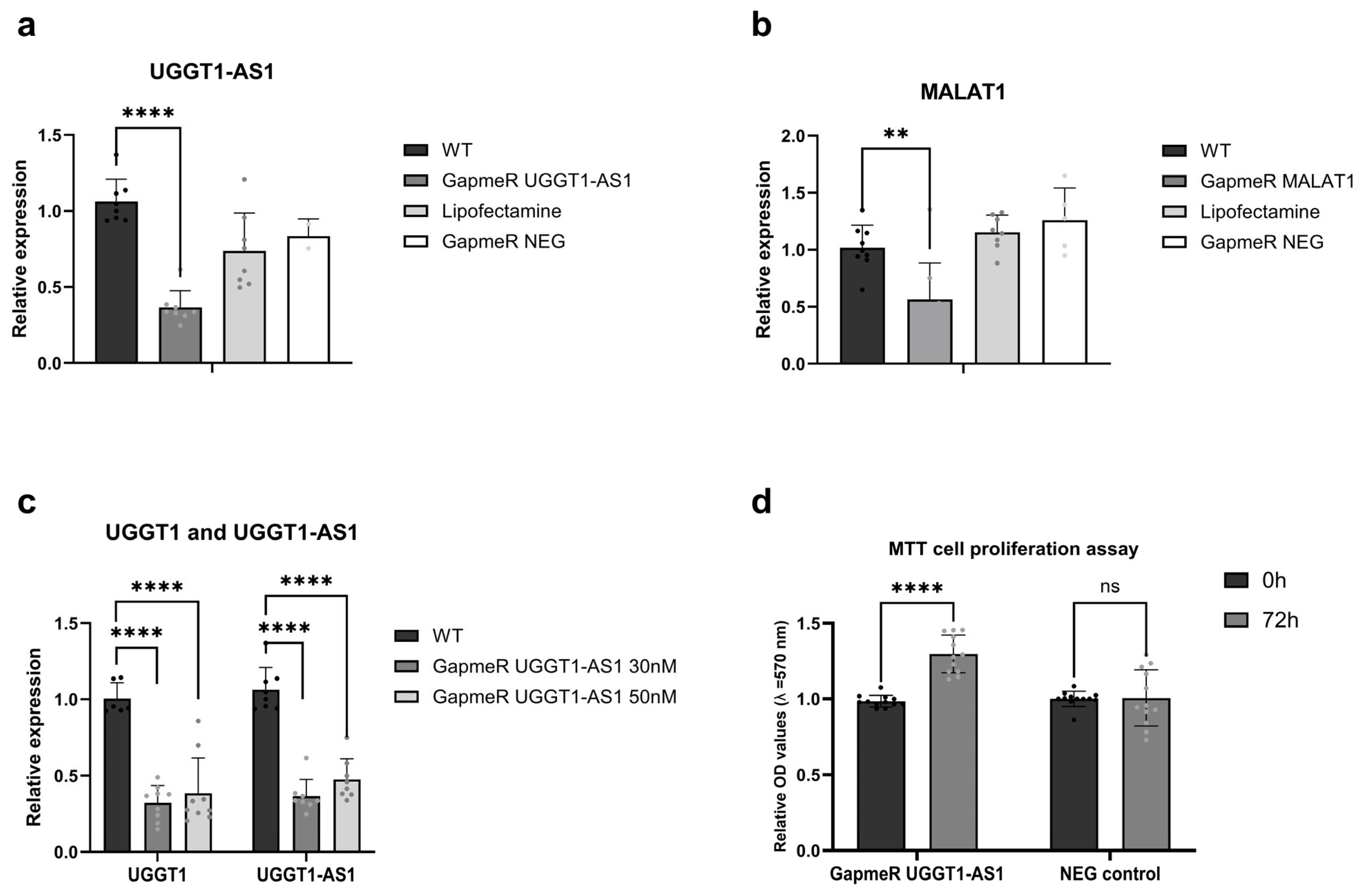
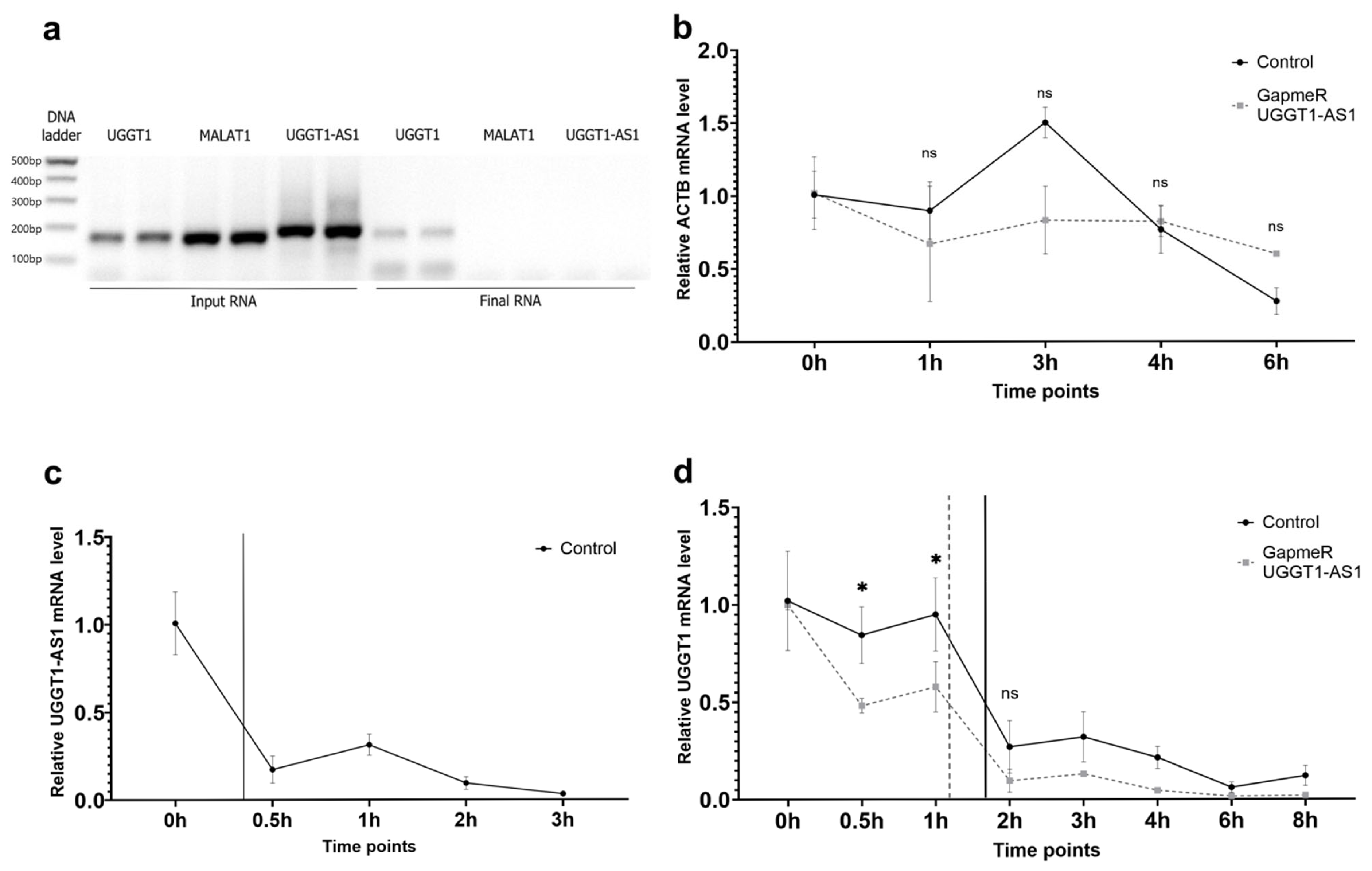
2.4. Cellular Roles for UGGT1-AS1
3. Discussion
4. Materials and Methods
4.1. Pre-Processing of RNA-Seq Data
4.2. Search for Adenosine-to-Inosine Editing Sites
4.3. Cell Culturing
4.4. Subcellular Fractionation
4.5. DNA and RNA Isolation
4.6. RT-PCR and Sanger Sequencing
4.7. Quantitative Real-Time PCR
4.8. Gene Silencing with LNA GapmeRs
4.9. MTT Assay
4.10. RNA Stability Assay
4.11. RNA Antisense Purification (RAP-RNA)
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barwal, T.S.; Sharma, U.; Bazala, S.; Singh, I.; Jain, M.; Prakash, H.; Shekhar, S.; Sandberg, E.N.; Bishayee, A.; Jain, A. MicroRNAs and Long Noncoding RNAs as Novel Therapeutic Targets in Estrogen Receptor-Positive Breast and Ovarian Cancers. Int. J. Mol. Sci. 2021, 22, 4072. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zitello, E.; Guo, R.; Deng, Y. The function of LncRNAs and their role in the prediction, diagnosis, and prognosis of lung cancer. Clin. Transl. Med. 2021, 11, e367. [Google Scholar] [CrossRef] [PubMed]
- Dong, A.; Preusch, C.B.; So, W.-K.; Lin, K.; Luan, S.; Yi, R.; Wong, J.W.; Wu, Z.; Cheung, T.H. A long noncoding RNA, LncMyoD, modulates chromatin accessibility to regulate muscle stem cell myogenic lineage progression. Proc. Natl. Acad. Sci. USA 2020, 117, 32464–32475. [Google Scholar] [CrossRef]
- Ouyang, J.; Zhong, Y.; Zhang, Y.; Yang, L.; Wu, P.; Hou, X.; Xiong, F.; Li, X.; Zhang, S.; Gong, Z.; et al. Long non-coding RNAs are involved in alternative splicing and promote cancer progression. Br. J. Cancer 2022, 126, 1113–1124. [Google Scholar] [CrossRef] [PubMed]
- Faghihi, M.A.; Wahlestedt, C. Regulatory roles of natural antisense transcripts. Nat. Rev. Mol. Cell Biol. 2009, 10, 637–643. [Google Scholar] [CrossRef]
- Rosikiewicz, W.; Makałowska, I. Biological Functions of Natural Antisense Transcripts. Acta Biochim. Pol. 2016, 63, 665–673. [Google Scholar] [CrossRef]
- Nishikura, K. A-to-I editing of coding and non-coding RNAs by ADARs. Nat. Rev. Mol. Cell Biol. 2016, 17, 83–96. [Google Scholar] [CrossRef]
- Wang, M.-Q.; Zhu, W.-J.; Gao, P. New insights into long non-coding RNAs in breast cancer: Biological functions and therapeutic prospects. Exp. Mol. Pathol. 2021, 120, 104640. [Google Scholar] [CrossRef]
- Gumireddy, K.; Li, A.; Kossenkov, A.V.; Sakurai, M.; Yan, J.; Li, Y.; Xu, H.; Wang, J.; Zhang, P.J.; Zhang, L.; et al. The mRNA-edited form of GABRA3 suppresses GABRA3-mediated Akt activation and breast cancer metastasis. Nat. Commun. 2016, 7, 10715. [Google Scholar] [CrossRef]
- Hsiao, Y.-H.E.; Bahn, J.H.; Yang, Y.; Lin, X.; Tran, S.; Yang, E.-W.; Quinones-Valdez, G.; Xiao, X. RNA editing in nascent RNA affects pre-mRNA splicing. Genome Res. 2018, 28, 812–823. [Google Scholar] [CrossRef]
- Paz, N.; Levanon, E.Y.; Amariglio, N.; Heimberger, A.B.; Ram, Z.; Constantini, S.; Barbash, Z.S.; Adamsky, K.; Safran, M.; Hirschberg, A.; et al. Altered adenosine-to-inosine RNA editing in human cancer. Genome Res. 2007, 17, 1586–1595. [Google Scholar] [CrossRef] [PubMed]
- Wanowska, E.; Samorowska, K.; Szcześniak, M.W. Emerging Roles of Long Noncoding RNAs in Breast Cancer Epigenetics and Epitranscriptomics. Front. Cell Dev. Biol. 2022, 10, 922351. [Google Scholar] [CrossRef] [PubMed]
- Fumagalli, D.; Gacquer, D.; Rothé, F.; Lefort, A.; Libert, F.; Brown, D.; Kheddoumi, N.; Shlien, A.; Konopka, T.; Salgado, R.; et al. Principles Governing A-to-I RNA Editing in the Breast Cancer Transcriptome. Cell Rep. 2015, 13, 277–289. [Google Scholar] [CrossRef]
- Sagredo, E.A.; Blanco, A.; Sagredo, A.I.; Pérez, P.; Sepúlveda-Hermosilla, G.; Morales, F.; Müller, B.; Verdugo, R.; Marcelain, K.; Harismendy, O.; et al. ADAR1-mediated RNA-editing of 3′UTRs in breast cancer. Biol. Res. 2018, 51, 36. [Google Scholar] [CrossRef] [PubMed]
- Kurkowiak, M.; Arcimowicz, Ł.; Chruściel, E.; Urban-Wójciuk, Z.; Papak, I.; Keegan, L.; O’connell, M.; Kowalski, J.; Hupp, T.; Marek-Trzonkowska, N. The effects of RNA editing in cancer tissue at different stages in carcinogenesis. RNA Biol. 2021, 18, 1524–1539. [Google Scholar] [CrossRef]
- Ito, Y.; Takeda, Y.; Seko, A.; Izumi, M.; Kajihara, Y. Functional analysis of endoplasmic reticulum glucosyltransferase (UGGT): Synthetic chemistry’s initiative in glycobiology. Semin. Cell Dev. Biol. 2015, 41, 90–98. [Google Scholar] [CrossRef]
- Wang, B.; Tian, P.; Sun, Q.; Zhang, H.; Han, L.; Zhu, B. A novel, effective machine learning-based RNA editing profile for predicting the prognosis of lower-grade gliomas. Heliyon 2023, 9, e18075. [Google Scholar] [CrossRef]
- Böttger, F.; Schaaij-Visser, T.B.; de Reus, I.; Piersma, S.R.; Pham, T.V.; Nagel, R.; Brakenhoff, R.H.; Thunnissen, E.; Smit, E.F.; Jimenez, C.R. Proteome analysis of non-small cell lung cancer cell line secretomes and patient sputum reveals biofluid biomarker candidates for cisplatin response prediction. J. Proteom. 2019, 196, 106–119. [Google Scholar] [CrossRef]
- Santos, F.; Capela, A.M.; Mateus, F.; Nóbrega-Pereira, S.; de Jesus, B.B. Non-coding antisense transcripts: Fine regulation of gene expression in cancer. Comput. Struct. Biotechnol. J. 2022, 20, 5652–5660. [Google Scholar] [CrossRef]
- Katayama, S.; Tomaru, Y.; Kasukawa, T.; Waki, K.; Nakanishi, M.; Nakamura, M.; Nishida, H.; Yap, C.C.; Suzuki, M.; Kawai, J.; et al. Antisense Transcription in the Mammalian Transcriptome. Science 2005, 309, 1564–1566. [Google Scholar] [CrossRef]
- Szcześniak, M.W.; Wanowska, E.; Mukherjee, N.; Ohler, U.; Makałowska, I. Towards a deeper annotation of human lncRNAs. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194385. [Google Scholar] [CrossRef] [PubMed]
- Wulff, B.-E.; Nishikura, K. Substitutional A-to-I RNA editing. Wiley Interdiscip. Rev. RNA 2010, 1, 90–101. [Google Scholar] [CrossRef] [PubMed]
- Wight, M.; Werner, A. The functions of natural antisense transcripts. Essays Biochem. 2013, 54, 91–101. [Google Scholar] [PubMed]
- Ratnadiwakara, M.; Änkö, M.-L. mRNA Stability Assay Using transcription inhibition by Actinomycin D in Mouse Pluripotent Stem Cells. Bio Protoc. 2018, 8, e3072. [Google Scholar] [CrossRef]
- Jadaliha, M.; Gholamalamdari, O.; Tang, W.; Zhang, Y.; Petracovici, A.; Hao, Q.; Tariq, A.; Kim, T.G.; Holton, S.E.; Singh, D.K.; et al. A natural antisense lncRNA controls breast cancer progression by promoting tumor suppressor gene mRNA stability. PLoS Genet. 2018, 14, e1007802. [Google Scholar] [CrossRef]
- Balbin, O.A.; Malik, R.; Dhanasekaran, S.M.; Prensner, J.R.; Cao, X.; Wu, Y.-M.; Robinson, D.; Wang, R.; Chen, G.; Beer, D.G.; et al. The landscape of antisense gene expression in human cancers. Genome Res. 2015, 25, 1068–1079. [Google Scholar] [CrossRef]
- Latgé, G.; Poulet, C.; Bours, V.; Josse, C.; Jerusalem, G. Natural Antisense Transcripts: Molecular Mechanisms and Implications in Breast Cancers. Int. J. Mol. Sci. 2018, 19, 123. [Google Scholar] [CrossRef]
- Wenric, S.; ElGuendi, S.; Caberg, J.-H.; Bezzaou, W.; Fasquelle, C.; Charloteaux, B.; Karim, L.; Hennuy, B.; Frères, P.; Collignon, J.; et al. Transcriptome-wide analysis of natural antisense transcripts shows their potential role in breast cancer. Sci. Rep. 2017, 7, 17452. [Google Scholar] [CrossRef]
- Szcześniak, M.W.; Makałowska, I. lncRNA-RNA Interactions across the Human Transcriptome. PLoS ONE 2016, 11, e0150353. [Google Scholar] [CrossRef]
- Eggington, J.M.; Greene, T.; Bass, B.L. Predicting sites of ADAR editing in double-stranded RNA. Nat. Commun. 2011, 2, 319. [Google Scholar] [CrossRef]
- Nishikura, K. Functions and Regulation of RNA Editing by ADAR Deaminases. Annu. Rev. Biochem. 2010, 79, 321–349. [Google Scholar] [CrossRef] [PubMed]
- Salameh, A.; Lee, A.K.; Cardó-Vila, M.; Nunes, D.N.; Efstathiou, E.; Staquicini, F.I.; Dobroff, A.S.; Marchiò, S.; Navone, N.M.; Hosoya, H.; et al. PRUNE2 is a human prostate cancer suppressor regulated by the intronic long noncoding RNA PCA3. Proc. Natl. Acad. Sci. USA 2015, 112, 8403–8408. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Lu, Y.; Yan, S.; Xing, Q.; Tian, W. SPRINT: An SNP-free toolkit for identifying RNA editing sites. Bioinformatics 2017, 33, 3538–3548. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef]
- Gagnon, K.T.; Li, L.; A Janowski, B.; Corey, D.R. Analysis of nuclear RNA interference in human cells by subcellular fractionation and Argonaute loading. Nat. Protoc. 2014, 9, 2045–2060. [Google Scholar] [CrossRef]
- Rao, X.; Huang, X.; Zhou, Z.; Lin, X. An improvement of the 2ˆ(–delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat. Bioinform. Biomath. 2013, 3, 71–85. [Google Scholar]
- Engreitz, J.; Lander, E.S.; Guttman, M. RNA antisense purification (RAP) for mapping RNA interactions with chromatin. Methods Mol. Biol. 2015, 1262, 183–197. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Samorowska, K.; Wanowska, E.; Szcześniak, M.W. Novel lncRNA UGGT1-AS1 Regulates UGGT1 Expression in Breast Cancer Cell Line. Int. J. Mol. Sci. 2025, 26, 5108. https://doi.org/10.3390/ijms26115108
Samorowska K, Wanowska E, Szcześniak MW. Novel lncRNA UGGT1-AS1 Regulates UGGT1 Expression in Breast Cancer Cell Line. International Journal of Molecular Sciences. 2025; 26(11):5108. https://doi.org/10.3390/ijms26115108
Chicago/Turabian StyleSamorowska, Klaudia, Elżbieta Wanowska, and Michał Wojciech Szcześniak. 2025. "Novel lncRNA UGGT1-AS1 Regulates UGGT1 Expression in Breast Cancer Cell Line" International Journal of Molecular Sciences 26, no. 11: 5108. https://doi.org/10.3390/ijms26115108
APA StyleSamorowska, K., Wanowska, E., & Szcześniak, M. W. (2025). Novel lncRNA UGGT1-AS1 Regulates UGGT1 Expression in Breast Cancer Cell Line. International Journal of Molecular Sciences, 26(11), 5108. https://doi.org/10.3390/ijms26115108




