Cyclic GMP-AMP Synthase (cGAS) Deletion Promotes Less Prominent Inflammatory Macrophages and Sepsis Severity in Catheter-Induced Infection and LPS Injection Models
Abstract
1. Introduction
2. Results
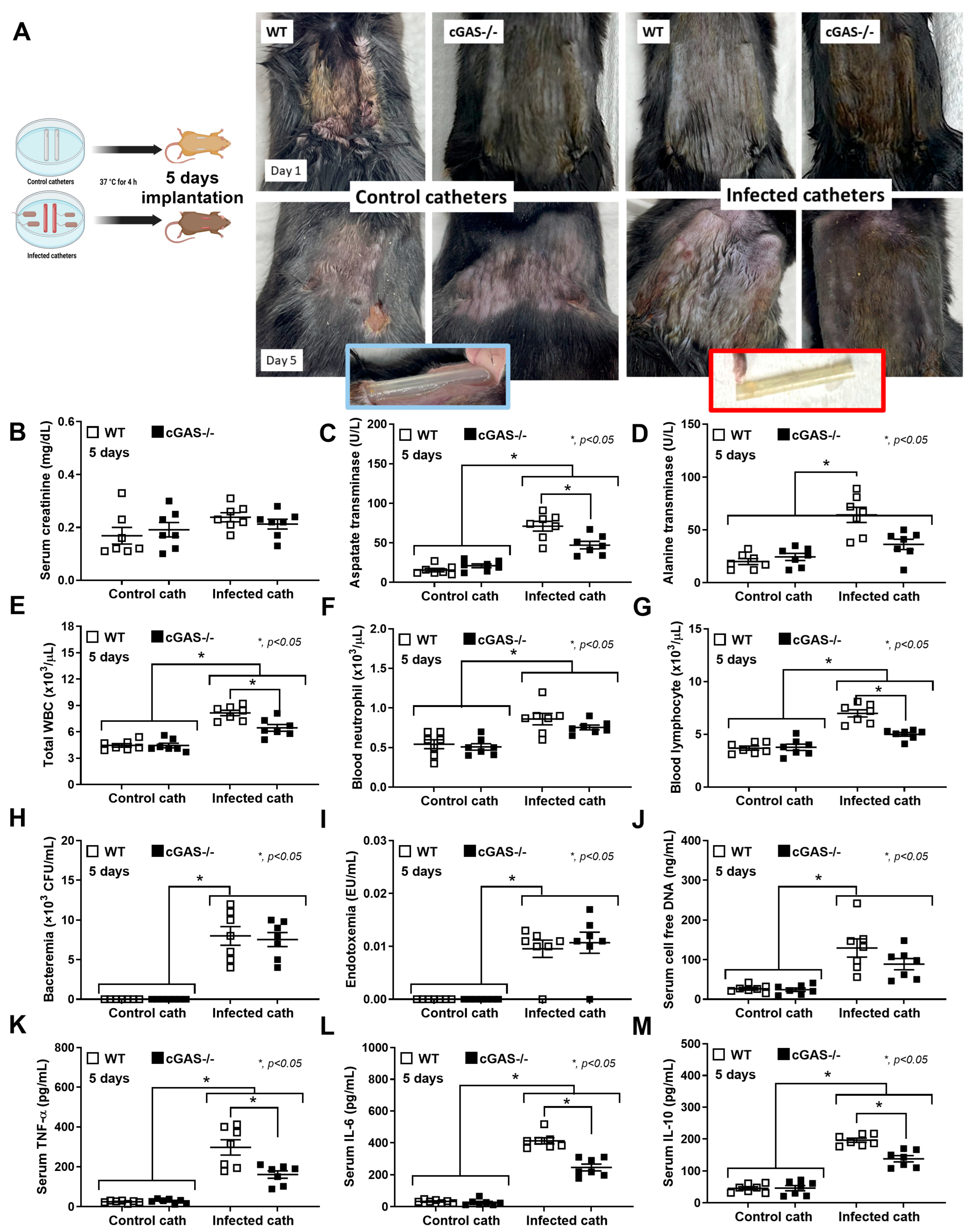
2.1. Deletion of cGAS Reduces Disease Severity in a Catheter-Induced Sepsis Mouse Model
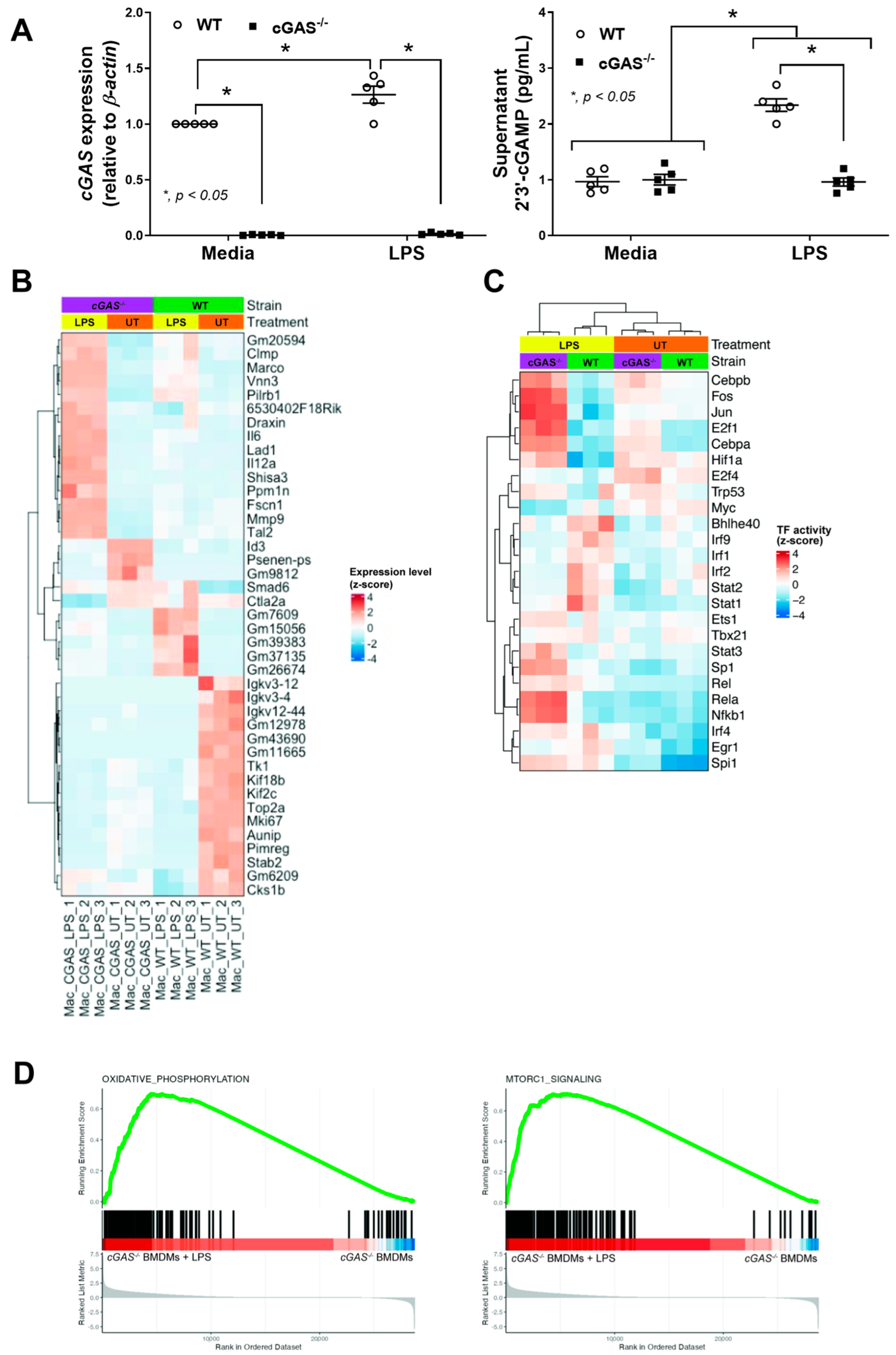
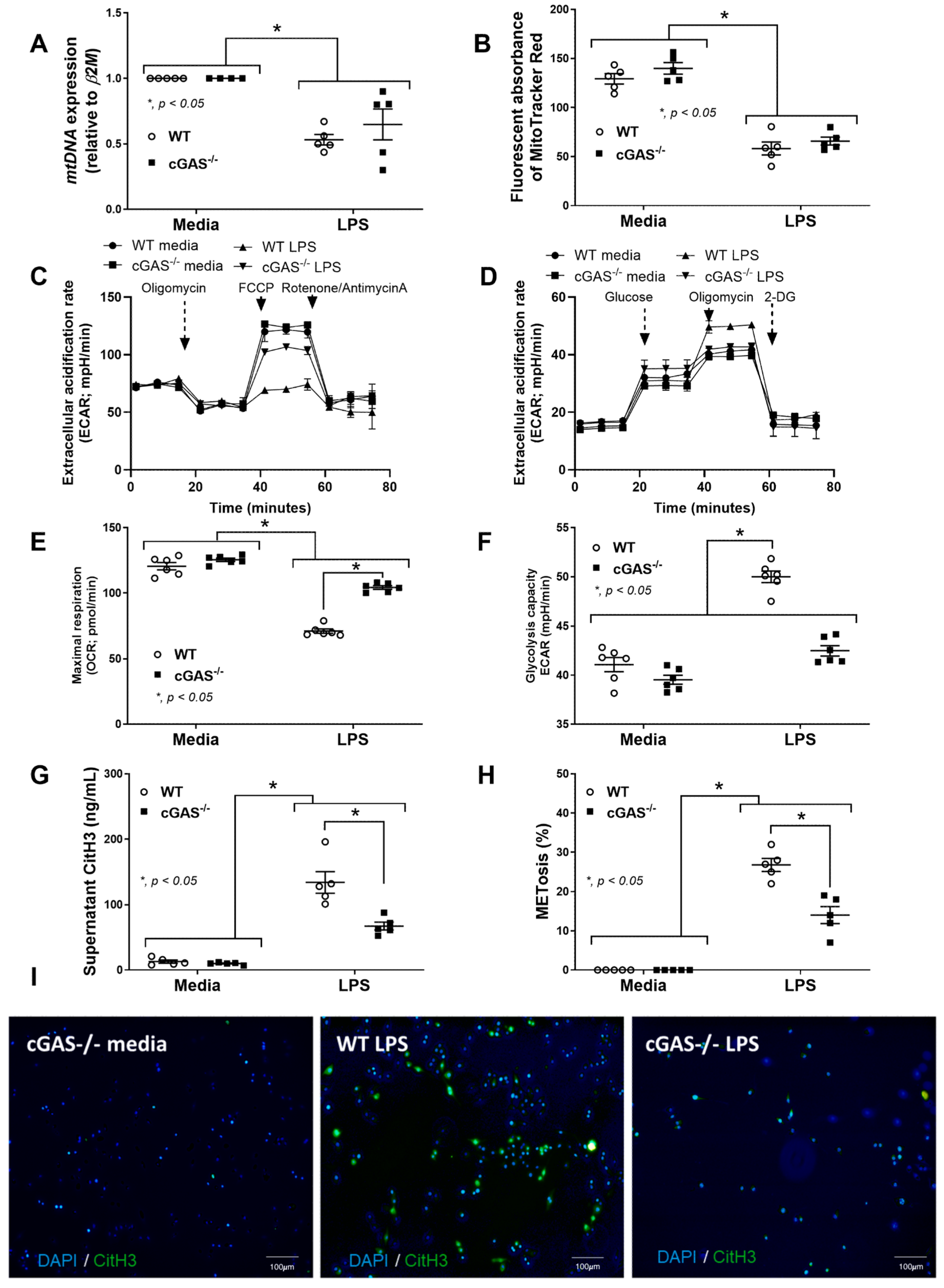
2.2. Transcriptomic Analysis Reveals Diminished IRF and JAK/STAT Activity, but Enriched OXPHOS and mTORC1 Pathways in cGAS-Deficient Macrophages
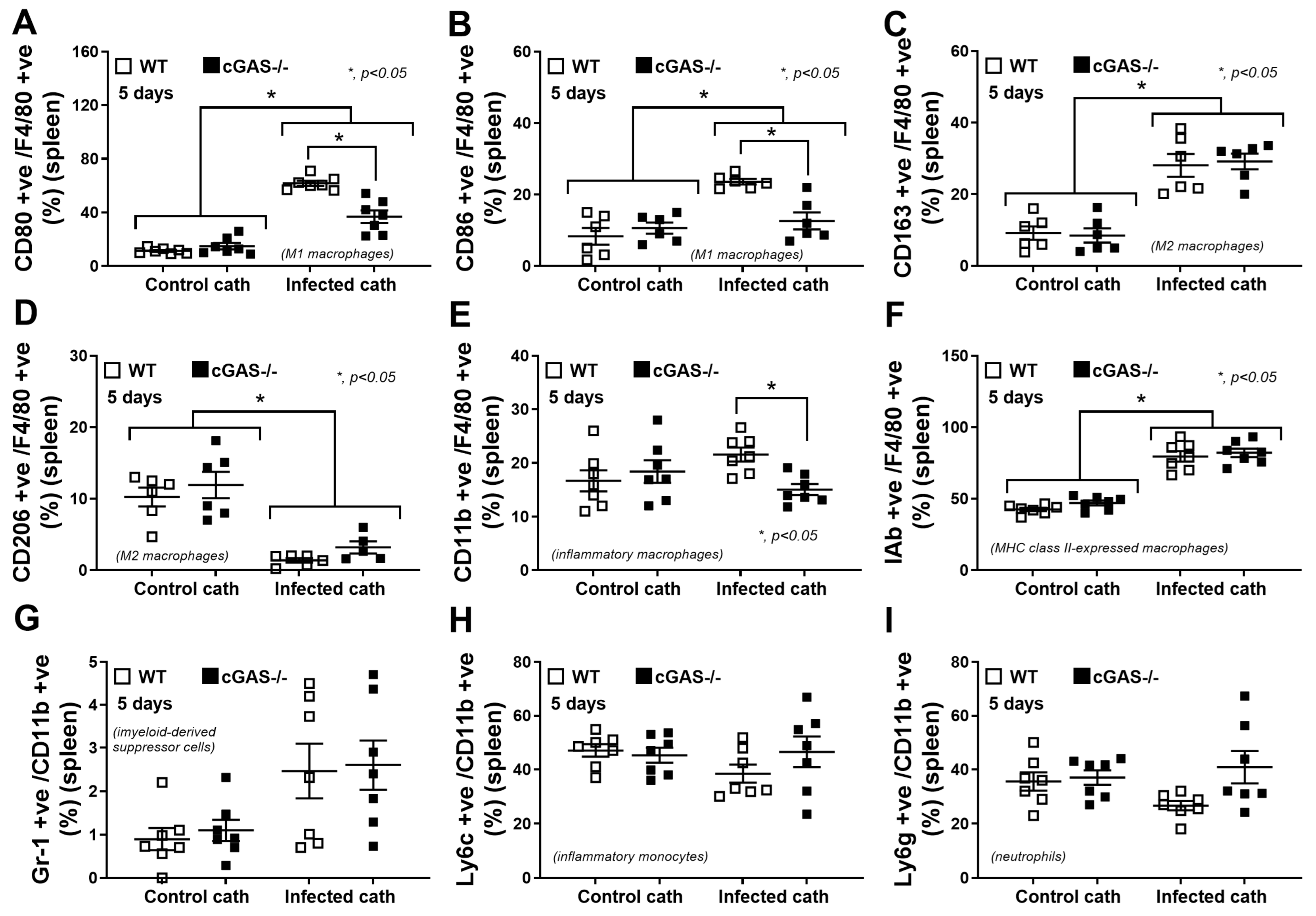
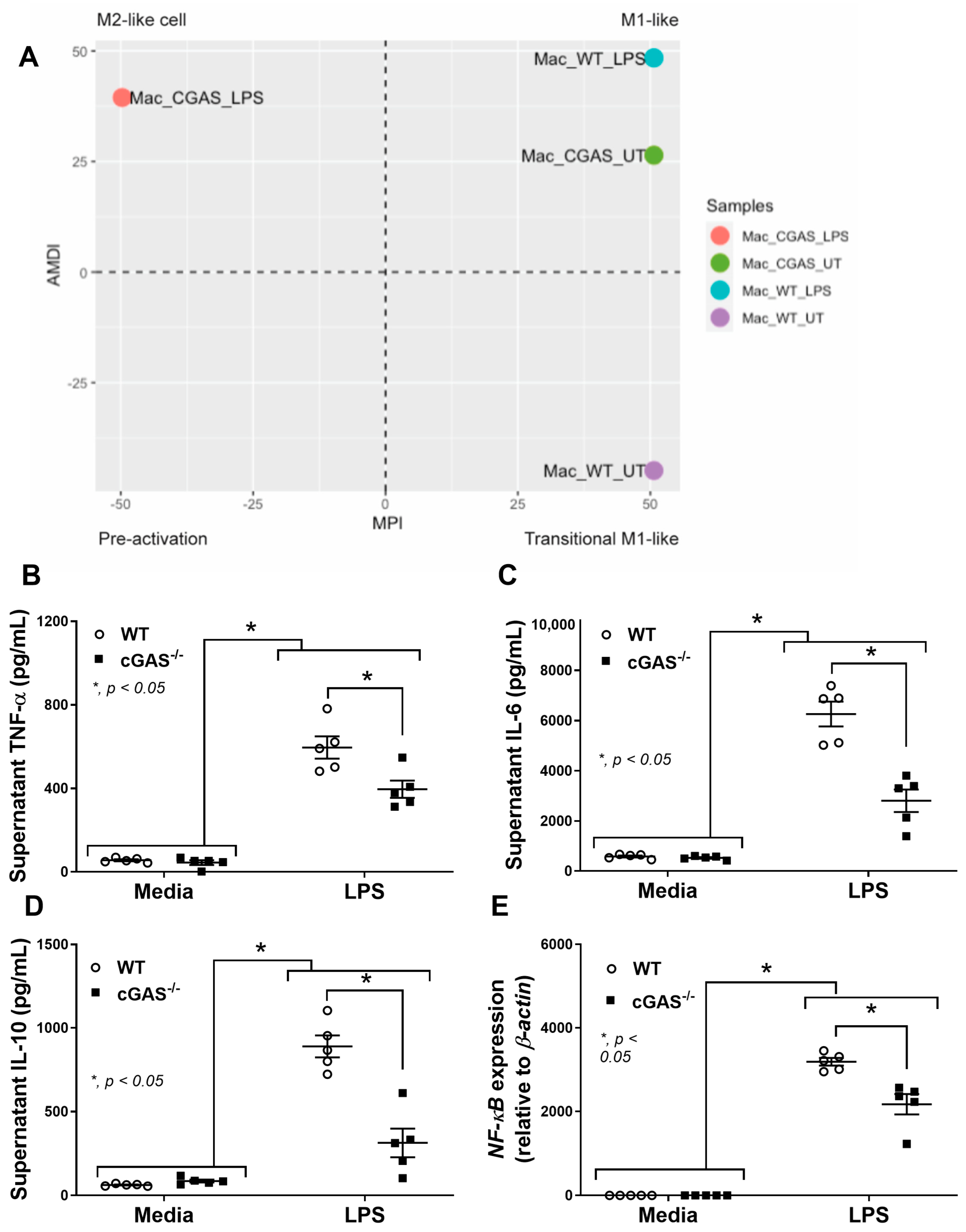
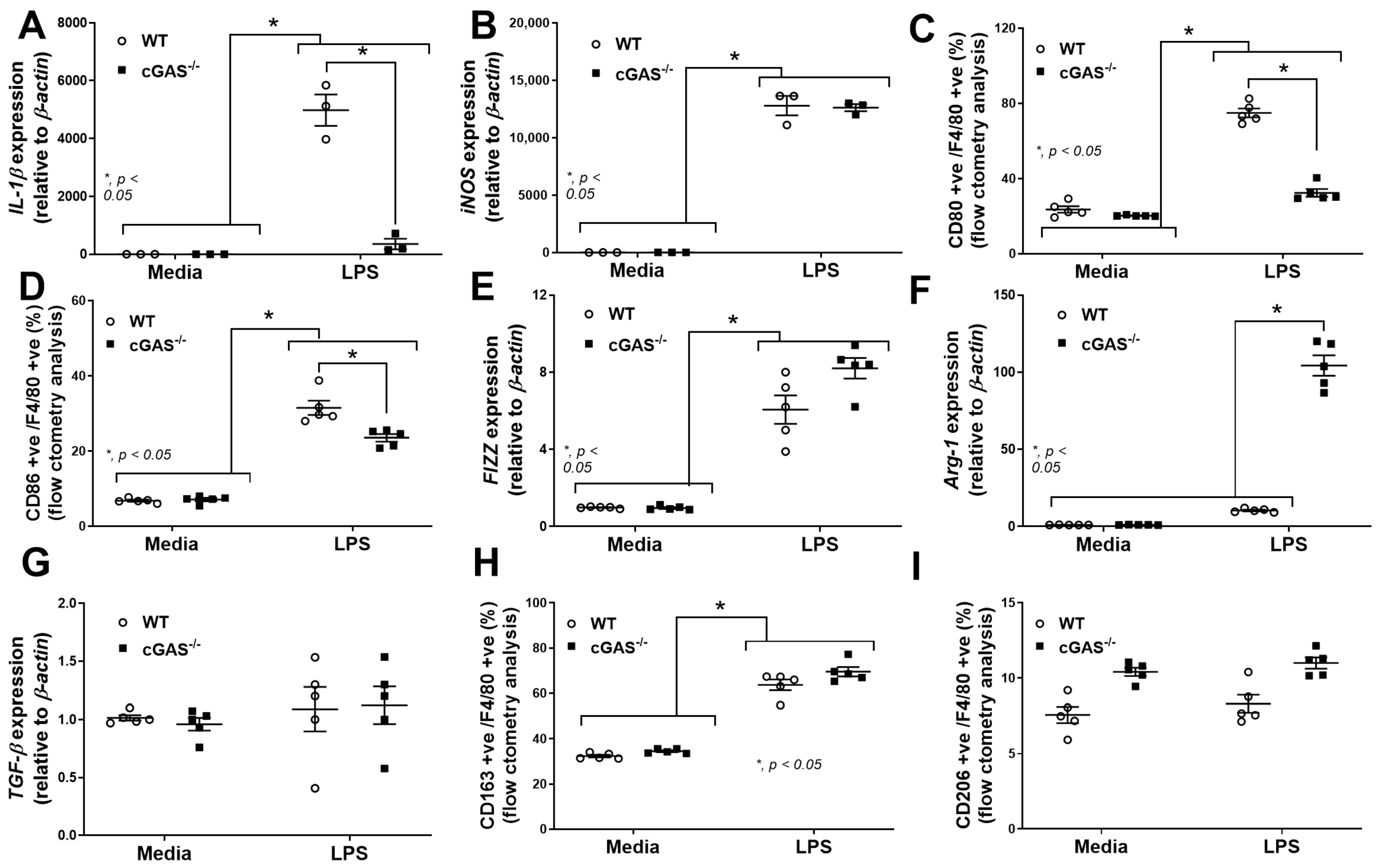
2.3. cGAS Restricts Macrophages to M1-like Subtype
3. Discussion
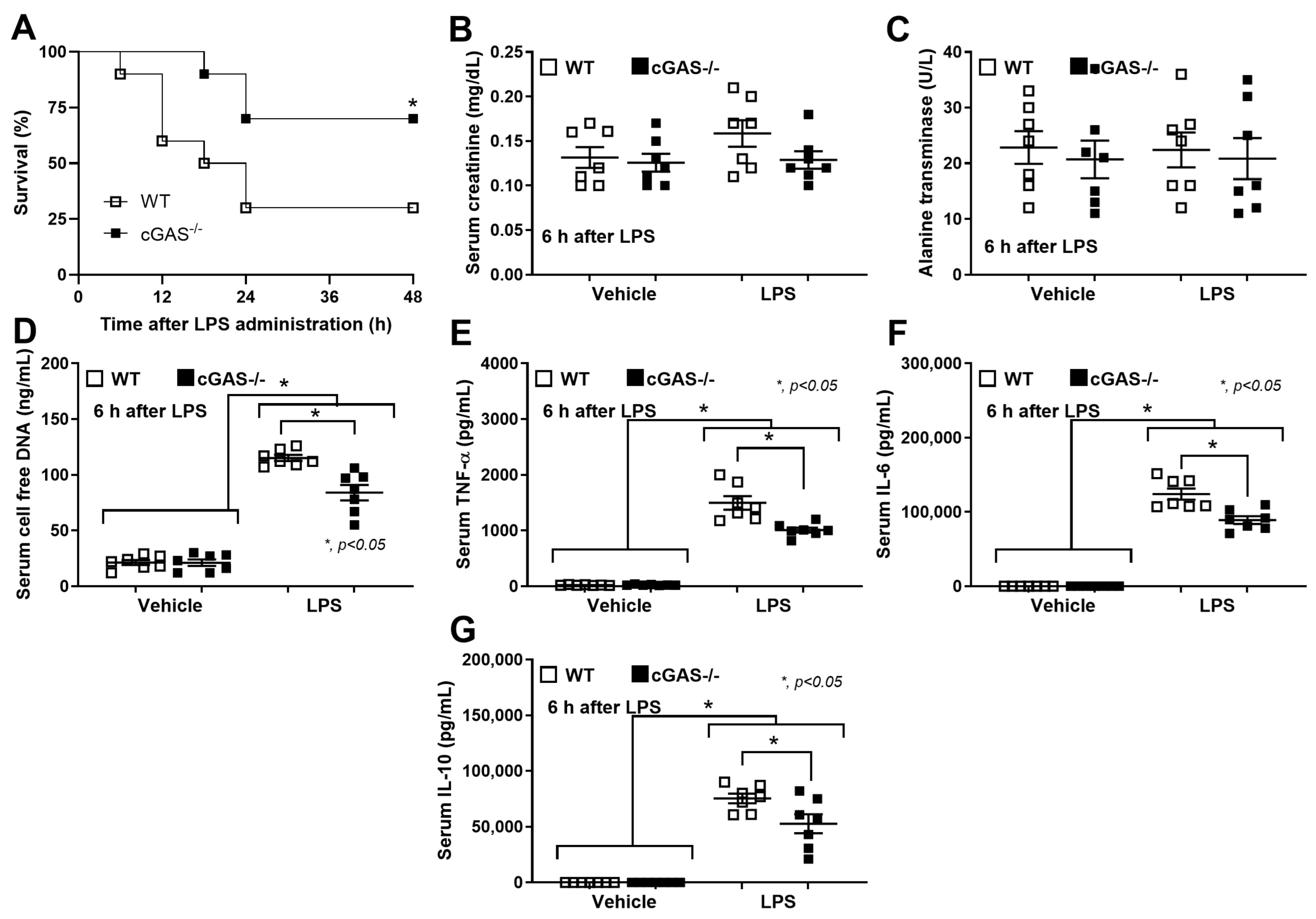
3.1. Sepsis-Induced Mitochondrial Injury and the Less Prominent Catheter-Induced Sepsis and LPS Injection Models in cGAS−/− Mice Compared with WT
3.2. Macrophage Anti-Inflammatory Direction in cGAS−/− Compared with WT Cells
4. Materials and Methods
4.1. Animals and Animal Model
4.2. Flow Cytometry Analysis
4.3. The Transcriptomic Analysis
4.4. The In Vitro Analysis
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pinitchun, C.; Panpetch, W.; Bhunyakarnjanarat, T.; Udompornpitak, K.; Do, H.T.; Visitchanakun, P.; Wannigama, D.L.; Udomkarnjananun, S.; Sukprasansap, M.; Tencomnao, T.; et al. Aging-induced dysbiosis worsens sepsis severity but is attenuated by probiotics in D-galactose-administered mice with cecal ligation and puncture model. PLoS ONE 2024, 19, e0311774. [Google Scholar] [CrossRef] [PubMed]
- Kunanopparat, A.; Leelahavanichkul, A.; Visitchanakun, P.; Kueanjinda, P.; Phuengmaung, P.; Sae-khow, K.; Boonmee, A.; Benjaskulluecha, S.; Palaga, T.; Hirankarn, N. The Regulatory Roles of Ezh2 in Response to Lipopolysaccharide (LPS) in Macrophages and Mice with Conditional Ezh2 Deletion with LysM-Cre System. Int. J. Mol. Sci. 2023, 24, 5363. [Google Scholar] [CrossRef] [PubMed]
- Sae-khow, K.; Phuengmaung, P.; Issara-Amphorn, J.; Makjaroen, J.; Visitchanakun, P.; Boonmee, A.; Benjaskulluecha, S.; Palaga, T.; Leelahavanichkul, A. Less Severe Polymicrobial Sepsis in Conditional mgmt-Deleted Mice Using LysM-Cre System, Impacts of DNA Methylation and MGMT Inhibitor in Sepsis. Int. J. Mol. Sci. 2023, 24, 10175. [Google Scholar] [CrossRef] [PubMed]
- Phuengmaung, P.; Khiewkamrop, P.; Makjaroen, J.; Issara-Amphorn, J.; Boonmee, A.; Benjaskulluecha, S.; Ritprajak, P.; Nita-Lazar, A.; Palaga, T.; Hirankarn, N.; et al. Less Severe Sepsis in Cecal Ligation and Puncture Models with and without Lipopolysaccharide in Mice with Conditional Ezh2-Deleted Macrophages (LysM-Cre System). Int. J. Mol. Sci. 2023, 24, 8517. [Google Scholar] [CrossRef]
- Taratummarat, S.; Sangphech, N.; Vu, C.T.B.; Palaga, T.; Ondee, T.; Surawut, S.; Sereemaspun, A.; Ritprajak, P.; Leelahavanichkul, A. Gold nanoparticles attenuates bacterial sepsis in cecal ligation and puncture mouse model through the induction of M2 macrophage polarization. BMC Microbiol. 2018, 18, 85. [Google Scholar] [CrossRef]
- Dang, C.P.; Issara-Amphorn, J.; Charoensappakit, A.; Udompornpitak, K.; Bhunyakarnjanarat, T.; Saisorn, W.; Sae-Khow, K.; Leelahavanichkul, A. BAM15, a Mitochondrial Uncoupling Agent, Attenuates Inflammation in the LPS Injection Mouse Model: An Adjunctive Anti-Inflammation on Macrophages and Hepatocytes. J. Innate Immun. 2021, 13, 359–375. [Google Scholar] [CrossRef]
- Li, X.D.; Wu, J.; Gao, D.; Wang, H.; Sun, L.; Chen, Z.J. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science 2013, 341, 1390–1394. [Google Scholar] [CrossRef]
- Sun, L.; Wu, J.; Du, F.; Chen, X.; Chen, Z.J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 2013, 339, 786–791. [Google Scholar] [CrossRef]
- Ishikawa, H.; Ma, Z.; Barber, G.N. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 2009, 461, 788–792. [Google Scholar] [CrossRef]
- West, A.P.; Khoury-Hanold, W.; Staron, M.; Tal, M.C.; Pineda, C.M.; Lang, S.M.; Bestwick, M.; Duguay, B.A.; Raimundo, N.; MacDuff, D.A.; et al. Mitochondrial DNA stress primes the antiviral innate immune response. Nature 2015, 520, 553–557. [Google Scholar] [CrossRef]
- Härtlova, A.; Erttmann, S.F.; Raffi, F.A.; Schmalz, A.M.; Resch, U.; Anugula, S.; Lienenklaus, S.; Nilsson, L.M.; Kröger, A.; Nilsson, J.A.; et al. DNA damage primes the type I interferon system via the cytosolic DNA sensor STING to promote anti-microbial innate immunity. Immunity 2015, 42, 332–343. [Google Scholar] [CrossRef] [PubMed]
- Visitchanakun, P.; Kaewduangduen, W.; Chareonsappakit, A.; Susantitaphong, P.; Pisitkun, P.; Ritprajak, P.; Townamchai, N.; Leelahavanichkul, A. Interference on Cytosolic DNA Activation Attenuates Sepsis Severity: Experiments on Cyclic GMP–AMP Synthase (cGAS) Deficient Mice. Int. J. Mol. Sci. 2021, 22, 11450. [Google Scholar] [CrossRef] [PubMed]
- Suksawad, N.; Udompornpitak, K.; Thawinpipat, N.; Korwattanamongkol, P.; Visitchanakun, P.; Phuengmaung, P.; Saisorn, W.; Kueanjinda, P.; Leelahavanichkul, A. Cyclic GMP–AMP Synthase (cGAS) Deletion Reduces Severity in Bilateral Nephrectomy Mice through Changes in Neutrophil Extracellular Traps and Mitochondrial Respiration. Biomedicines 2023, 11, 1208. [Google Scholar] [CrossRef] [PubMed]
- Marchi, S.; Guilbaud, E.; Tait, S.W.G.; Yamazaki, T.; Galluzzi, L. Mitochondrial control of inflammation. Nat. Rev. Immunol. 2023, 23, 159–173. [Google Scholar] [CrossRef]
- López-Armada, M.J.; Riveiro-Naveira, R.R.; Vaamonde-García, C.; Valcárcel-Ares, M.N. Mitochondrial dysfunction and the inflammatory response. Mitochondrion 2013, 13, 106–118. [Google Scholar] [CrossRef]
- Decout, A.; Katz, J.D.; Venkatraman, S.; Ablasser, A. The cGAS–STING pathway as a therapeutic target in inflammatory diseases. Nat. Rev. Immunol. 2021, 21, 548–569. [Google Scholar] [CrossRef]
- Huang, L.S.; Hong, Z.; Wu, W.; Xiong, S.; Zhong, M.; Gao, X.; Rehman, J.; Malik, A.B. mtDNA Activates cGAS Signaling and Suppresses the YAP-Mediated Endothelial Cell Proliferation Program to Promote Inflammatory Injury. Immunity 2020, 52, 475–486.e475. [Google Scholar] [CrossRef]
- Binmama, S.; Dang, C.P.; Visitchanakun, P.; Hiengrach, P.; Somboonna, N.; Cheibchalard, T.; Pisitkun, P.; Chindamporn, A.; Leelahavanichkul, A. Beta-Glucan from S. cerevisiae Protected AOM-Induced Colon Cancer in cGAS-Deficient Mice Partly through Dectin-1-Manipulated Macrophage Cell Energy. Int. J. Mol. Sci. 2022, 23, 10951. [Google Scholar] [CrossRef]
- Kumpunya, S.; Thim-uam, A.; Thumarat, C.; Leelahavanichkul, A.; Kalpongnukul, N.; Chantaravisoot, N.; Pisitkun, T.; Pisitkun, P. cGAS deficiency enhances inflammasome activation in macrophages and inflammatory pathology in pristane-induced lupus. Front. Immunol. 2022, 13, 1010764. [Google Scholar] [CrossRef]
- McQuiston, T.J.; Williamson, P.R. Paradoxical roles of alveolar macrophages in the host response to Cryptococcus neoformans. J. Infect. Chemother. 2012, 18, 1–9. [Google Scholar] [CrossRef][Green Version]
- Klein, I.; Cornejo, J.C.; Polakos, N.K.; John, B.; Wuensch, S.A.; Topham, D.J.; Pierce, R.H.; Crispe, I.N. Kupffer cell heterogeneity: Functional properties of bone marrow derived and sessile hepatic macrophages. Blood 2007, 110, 4077–4085. [Google Scholar] [CrossRef] [PubMed]
- Boulet, N.; Pensier, J.; Occean, B.V.; Peray, P.F.; Mimoz, O.; Rickard, C.M.; Buetti, N.; Lefrant, J.Y.; Muller, L.; Roger, C. Central venous catheter-related infections: A systematic review, meta-analysis, trial sequential analysis and meta-regression comparing ultrasound guidance and landmark technique for insertion. Crit. Care 2024, 28, 378. [Google Scholar] [CrossRef] [PubMed]
- Phuengmaung, P.; Panpetch, W.; Singkham-In, U.; Chatsuwan, T.; Chirathaworn, C.; Leelahavanichkul, A. Presence of Candida tropicalis on Staphylococcus epidermidis Biofilms Facilitated Biofilm Production and Candida Dissemination: An Impact of Fungi on Bacterial Biofilms. Front. Cell. Infect. Microbiol. 2021, 11, 763239. [Google Scholar] [CrossRef]
- Sherertz, R.J.; Forman, D.M.; Solomon, D.D. Efficacy of dicloxacillin-coated polyurethane catheters in preventing subcutaneous Staphylococcus aureus infection in mice. Antimicrob. Agents Chemother. 1989, 33, 1174–1178. [Google Scholar] [CrossRef] [PubMed]
- Song, K.H.; Ge, X.; Engelbach, J.A.; Thio, L.L.; Neil, J.J.; Ackerman, J.J.H.; Garbow, J.R. Subcutaneous deuterated substrate administration in mice: An alternative to tail vein infusion. Magn. Reson. Med. 2024, 91, 681–686. [Google Scholar] [CrossRef]
- Lew, S.Q.; Gruia, A. Ofloxacin solution for persistent exit-site and tunnel infection in peritoneal dialysis. Perit. Dial. Int. 2013, 33, 101–102. [Google Scholar] [CrossRef]
- Barraclough, K.; Hawley, C.M.; McDonald, S.P.; Brown, F.G.; Rosman, J.B.; Wiggins, K.J.; Bannister, K.M.; Johnson, D.W. Polymicrobial peritonitis in peritoneal dialysis patients in Australia: Predictors, treatment, and outcomes. Am. J. Kidney Dis. 2010, 55, 121–131. [Google Scholar] [CrossRef]
- Tang, A.; Shi, Y.; Dong, Q.; Wang, S.; Ge, Y.; Wang, C.; Gong, Z.; Zhang, W.; Chen, W. Prognostic differences in sepsis caused by gram-negative bacteria and gram-positive bacteria: A systematic review and meta-analysis. Crit. Care 2023, 27, 467. [Google Scholar] [CrossRef]
- Phuengmaung, P.; Mekjaroen, J.; Saisorn, W.; Chatsuwan, T.; Somparn, P.; Leelahavanichkul, A. Rapid Synergistic Biofilm Production of Pseudomonas and Candida on the Pulmonary Cell Surface and in Mice, a Possible Cause of Chronic Mixed Organismal Lung Lesions. Int. J. Mol. Sci. 2022, 23, 9202. [Google Scholar] [CrossRef]
- Tateda, K.; Matsumoto, T.; Miyazaki, S.; Yamaguchi, K. Lipopolysaccharide-induced lethality and cytokine production in aged mice. Infect. Immun. 1996, 64, 769–774. [Google Scholar] [CrossRef]
- Li, S.; Hu, Q.; Huang, J.; Wu, X.; Ren, J. Mitochondria-Derived Damage-Associated Molecular Patterns in Sepsis: From Bench to Bedside. Oxid. Med. Cell. Longev. 2019, 2019, 6914849. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.; Sheeja Prabhakaran, H.; Zhang, Y.-Y.; Luo, G.; He, W.; Liou, Y.-C. Mitochondrial dysfunction in sepsis: Mechanisms and therapeutic perspectives. Crit. Care 2024, 28, 292. [Google Scholar] [CrossRef]
- Yamamoto, K.; Tsujimura, Y.; Ato, M. Catheter-associated Mycobacterium intracellulare biofilm infection in C3HeB/FeJ mice. Sci. Rep. 2023, 13, 17148. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Qiu, L.; Wu, H.; Song, Z.; Ke, P.; Wu, X. Focus on the cGAS-STING Signaling Pathway in Sepsis and Its Inflammatory Regulatory Effects. J. Inflamm. Res. 2024, 17, 3629–3639. [Google Scholar] [CrossRef]
- Garcia-Alonso, L.; Holland, C.H.; Ibrahim, M.M.; Turei, D.; Saez-Rodriguez, J. Benchmark and integration of resources for the estimation of human transcription factor activities. Genome Res. 2019, 29, 1363–1375. [Google Scholar] [CrossRef]
- Schubert, M.; Klinger, B.; Klunemann, M.; Sieber, A.; Uhlitz, F.; Sauer, S.; Garnett, M.J.; Bluthgen, N.; Saez-Rodriguez, J. Perturbation-response genes reveal signaling footprints in cancer gene expression. Nat. Commun. 2018, 9, 20. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, T.; Fong, C. The resolution of inflammation: Anti-inflammatory roles for NF-kappaB. Int. J. Biochem. Cell Biol. 2010, 42, 519–523. [Google Scholar] [CrossRef]
- Kim, C.; Sano, Y.; Todorova, K.; Carlson, B.A.; Arpa, L.; Celada, A.; Lawrence, T.; Otsu, K.; Brissette, J.L.; Arthur, J.S.; et al. The kinase p38 alpha serves cell type-specific inflammatory functions in skin injury and coordinates pro- and anti-inflammatory gene expression. Nat. Immunol. 2008, 9, 1019–1027. [Google Scholar] [CrossRef]
- Rodriguez-Prados, J.C.; Traves, P.G.; Cuenca, J.; Rico, D.; Aragones, J.; Martin-Sanz, P.; Cascante, M.; Bosca, L. Substrate fate in activated macrophages: A comparison between innate, classic, and alternative activation. J. Immunol. 2010, 185, 605–614. [Google Scholar] [CrossRef]
- Jha, A.K.; Huang, S.C.; Sergushichev, A.; Lampropoulou, V.; Ivanova, Y.; Loginicheva, E.; Chmielewski, K.; Stewart, K.M.; Ashall, J.; Everts, B.; et al. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity 2015, 42, 419–430. [Google Scholar] [CrossRef]
- Zhan, X.; Cui, R.; Geng, X.; Li, J.; Zhou, Y.; He, L.; Cao, C.; Zhang, C.; Chen, Z.; Ying, S. LPS-induced mitochondrial DNA synthesis and release facilitate RAD50-dependent acute lung injury. Signal Transduct. Target. Ther. 2021, 6, 103. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Menoret, A.; Farragher, C.; Ouyang, Z.; Bonin, C.; Holvoet, P.; Vella, A.T.; Zhou, B. Single cell transcriptomics based-MacSpectrum reveals novel macrophage activation signatures in diseases. JCI Insight 2019, 5, e126453. [Google Scholar] [CrossRef]
- Doi, K.; Leelahavanichkul, A.; Yuen, P.S.; Star, R.A. Animal models of sepsis and sepsis-induced kidney injury. J. Clin. Investig. 2009, 119, 2868–2878. [Google Scholar] [CrossRef] [PubMed]
- Charoensappakit, A.; Sae-Khow, K.; Rattanaliam, P.; Vutthikraivit, N.; Pecheenbuvan, M.; Udomkarnjananun, S.; Leelahavanichkul, A. Cell-free DNA as diagnostic and prognostic biomarkers for adult sepsis: A systematic review and meta-analysis. Sci. Rep. 2023, 13, 19624. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, Z. The role of macrophages polarization in sepsis-induced acute lung injury. Front. Immunol. 2023, 14, 1209438. [Google Scholar] [CrossRef]
- Xu, W.; Hou, H.; Yang, W.; Tang, W.; Sun, L. Immunologic role of macrophages in sepsis-induced acute liver injury. Int. Immunopharmacol. 2024, 143, 113492. [Google Scholar] [CrossRef]
- Yu, L.; Liu, P. Cytosolic DNA sensing by cGAS: Regulation, function, and human diseases. Signal Transduct. Target. Ther. 2021, 6, 170. [Google Scholar] [CrossRef]
- Cao, D.J.; Schiattarella, G.G.; Villalobos, E.; Jiang, N.; May, H.I.; Li, T.; Chen, Z.J.; Gillette, T.G.; Hill, J.A. Cytosolic DNA Sensing Promotes Macrophage Transformation and Governs Myocardial Ischemic Injury. Circulation 2018, 137, 2613–2634. [Google Scholar] [CrossRef]
- Zong, Q.; Zhang, H.; Liu, F.; Li, J.; Liu, Q.; Duan, Z.; Duan, W.; Ruan, M.; Zhang, J.; Liu, Y.; et al. Activation of the cGAS-STING pathway by viral dsDNA leading to M1 polarization of macrophages mediates antiviral activity against hepatitis B virus. Immunobiology 2024, 229, 152810. [Google Scholar] [CrossRef]
- Kaewduangduen, W.; Visitchanakun, P.; Saisorn, W.; Phawadee, A.; Manonitnantawat, C.; Chutimaskul, C.; Susantitaphong, P.; Ritprajak, P.; Somboonna, N.; Cheibchalard, T.; et al. Blood Bacteria-Free DNA in Septic Mice Enhances LPS-Induced Inflammation in Mice through Macrophage Response. Int. J. Mol. Sci. 2022, 23, 1907. [Google Scholar] [CrossRef]
- Lama, L.; Adura, C.; Xie, W.; Tomita, D.; Kamei, T.; Kuryavyi, V.; Gogakos, T.; Steinberg, J.I.; Miller, M.; Ramos-Espiritu, L.; et al. Development of human cGAS-specific small-molecule inhibitors for repression of dsDNA-triggered interferon expression. Nat. Commun. 2019, 10, 2261. [Google Scholar] [CrossRef] [PubMed]
- Jaggi, U.; Matundan, H.H.; Yu, J.; Hirose, S.; Mueller, M.; Wormley, F.L., Jr.; Ghiasi, H. Essential role of M1 macrophages in blocking cytokine storm and pathology associated with murine HSV-1 infection. PLoS Pathog. 2021, 17, e1009999. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Saeed, A.; Liu, Q.; Jiang, Q.; Xu, H.; Xiao, G.G.; Rao, L.; Duo, Y. Macrophages in immunoregulation and therapeutics. Signal Transduct. Target. Ther. 2023, 8, 207. [Google Scholar] [CrossRef]
- Hannigan, B.M.; Barnett, Y.A.; Armstrong, D.B.; McKelvey-Martin, V.J.; McKenna, P.G. Thymidine kinases: The enzymes and their clinical usefulness. Cancer Biother. 1993, 8, 189–197. [Google Scholar] [CrossRef]
- Carmena, M.; Earnshaw, W.C. The cellular geography of aurora kinases. Nat. Rev. Mol. Cell Biol. 2003, 4, 842–854. [Google Scholar] [CrossRef]
- Saisorn, W.; Phuengmaung, P.; Issara-Amphorn, J.; Makjaroen, J.; Visitchanakun, P.; Sae-khow, K.; Boonmee, A.; Benjaskulluecha, S.; Nita-Lazar, A.; Palaga, T.; et al. Less Severe Lipopolysaccharide-Induced Inflammation in Conditional mgmt-Deleted Mice with LysM-Cre System: The Loss of DNA Repair in Macrophages. Int. J. Mol. Sci. 2023, 24, 10139. [Google Scholar] [CrossRef] [PubMed]
- Fekadu, J.; Modlich, U.; Bader, P.; Bakhtiar, S. Understanding the Role of LFA-1 in Leukocyte Adhesion Deficiency Type I (LAD I): Moving towards Inflammation? Int. J. Mol. Sci. 2022, 23, 3578. [Google Scholar] [CrossRef]
- Pan, J.; Zhou, L.; Zhang, C.; Xu, Q.; Sun, Y. Targeting protein phosphatases for the treatment of inflammation-related diseases: From signaling to therapy. Signal Transduct. Target. Ther. 2022, 7, 177. [Google Scholar] [CrossRef]
- Jing, J.; Yang, I.V.; Hui, L.; Patel, J.A.; Evans, C.M.; Prikeris, R.; Kobzik, L.; O’Connor, B.P.; Schwartz, D.A. Role of macrophage receptor with collagenous structure in innate immune tolerance. J. Immunol. 2013, 190, 6360–6367. [Google Scholar] [CrossRef]
- Bartucci, R.; Salvati, A.; Olinga, P.; Boersma, Y.L. Vanin 1: Its Physiological Function and Role in Diseases. Int. J. Mol. Sci. 2019, 20, 3891. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, L.; Jiang, C.; Pan, K.; Deng, J.; Wan, C. MMP9 protects against LPS-induced inflammation in osteoblasts. Innate Immun. 2020, 26, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Masuda, K.; Ripley, B.; Nishimura, R.; Mino, T.; Takeuchi, O.; Shioi, G.; Kiyonari, H.; Kishimoto, T. Arid5a controls IL-6 mRNA stability, which contributes to elevation of IL-6 level in vivo. Proc. Natl. Acad. Sci. USA 2013, 110, 9409–9414. [Google Scholar] [CrossRef] [PubMed]
- Sarode, P.; Zheng, X.; Giotopoulou, G.A.; Weigert, A.; Kuenne, C.; Gunther, S.; Friedrich, A.; Gattenlohner, S.; Stiewe, T.; Brune, B.; et al. Reprogramming of tumor-associated macrophages by targeting beta-catenin/FOSL2/ARID5A signaling: A potential treatment of lung cancer. Sci. Adv. 2020, 6, eaaz6105. [Google Scholar] [CrossRef]
- Koussounadis, A.; Langdon, S.P.; Um, I.H.; Harrison, D.J.; Smith, V.A. Relationship between differentially expressed mRNA and mRNA-protein correlations in a xenograft model system. Sci. Rep. 2015, 5, 10775. [Google Scholar] [CrossRef]
- Nie, L.; Wu, G.; Zhang, W. Correlation of mRNA expression and protein abundance affected by multiple sequence features related to translational efficiency in Desulfovibrio vulgaris: A quantitative analysis. Genetics 2006, 174, 2229–2243. [Google Scholar] [CrossRef] [PubMed]
- Cheneval, D.; Kastelic, T.; Fuerst, P.; Parker, C.N. A Review of Methods to Monitor the Modulation of mRNA Stability: A Novel Approach to Drug Discovery and Therapeutic Intervention. SLAS Discov. 2010, 15, 609–622. [Google Scholar] [CrossRef]
- Nowak, R.P.; Yue, H.; Park, E.Y.; Fischer, E.S. Methods for Quantitative Assessment of Protein Degradation. Methods Mol. Biol. 2021, 2365, 247–263. [Google Scholar] [CrossRef]
- Xiao, K.; Liu, C.; Tu, Z.; Xu, Q.; Chen, S.; Zhang, Y.; Wang, X.; Zhang, J.; Hu, C.A.; Liu, Y. Activation of the NF-κB and MAPK Signaling Pathways Contributes to the Inflammatory Responses, but Not Cell Injury, in IPEC-1 Cells Challenged with Hydrogen Peroxide. Oxid. Med. Cell. Longev. 2020, 2020, 5803639. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Guan, D.; Wang, S.; Chai, L.Y.A.; Xu, S.; Lam, K.P. Glycolysis and Oxidative Phosphorylation Play Critical Roles in Natural Killer Cell Receptor-Mediated Natural Killer Cell Functions. Front. Immunol. 2020, 11, 202. [Google Scholar] [CrossRef]
- Shen, X.; Sun, C.; Cheng, Y.; Ma, D.; Sun, Y.; Lin, Y.; Zhao, Y.; Yang, M.; Jing, W.; Cui, X.; et al. cGAS Mediates Inflammation by Polarizing Macrophages to M1 Phenotype via the mTORC1 Pathway. J. Immunol. 2023, 210, 1098–1107. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, R.; Gu, H.; Zhang, E.; Qu, J.; Cao, W.; Huang, X.; Yan, H.; He, J.; Cai, Z. Metabolic reprogramming in macrophage responses. Biomark. Res. 2021, 9, 1. [Google Scholar] [CrossRef]
- Keating, S.E.; Zaiatz-Bittencourt, V.; Loftus, R.M.; Keane, C.; Brennan, K.; Finlay, D.K.; Gardiner, C.M. Metabolic Reprogramming Supports IFN-gamma Production by CD56bright NK Cells. J. Immunol. 2016, 196, 2552–2560. [Google Scholar] [CrossRef]
- Yan, S.; Dong, J.; Qian, C.; Chen, S.; Xu, Q.; Lei, H.; Wang, X. The mTORC1 Signaling Support Cellular Metabolism to Dictate Decidual NK Cells Function in Early Pregnancy. Front. Immunol. 2022, 13, 771732. [Google Scholar] [CrossRef] [PubMed]
- Morita, M.; Gravel, S.P.; Chenard, V.; Sikstrom, K.; Zheng, L.; Alain, T.; Gandin, V.; Avizonis, D.; Arguello, M.; Zakaria, C.; et al. mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metab. 2013, 18, 698–711. [Google Scholar] [CrossRef]
- Wu, D.-H.; Zhao, Z.-L.; Yin, W.-T.; Liu, H.; Xiang, X.-Y.; Zhu, L.-J.; Li, J.-Q.; Yan, Z.-H.; Li, Y.-J.; Jian, Y.-P.; et al. STING exerts antiviral innate immune response by activating pentose phosphate pathway. Cell Commun. Signal. 2024, 22, 599. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Zhang, S.; Vuckovic, I.; Jeon, R.; Lerman, A.; Folmes, C.D.; Dzeja, P.P.; Herrmann, J. Glycolytic Stimulation Is Not a Requirement for M2 Macrophage Differentiation. Cell Metab. 2018, 28, 463–475.e464. [Google Scholar] [CrossRef] [PubMed]
- Byles, V.; Covarrubias, A.J.; Ben-Sahra, I.; Lamming, D.W.; Sabatini, D.M.; Manning, B.D.; Horng, T. The TSC-mTOR pathway regulates macrophage polarization. Nat. Commun. 2013, 4, 2834. [Google Scholar] [CrossRef]
- Zhang, K.M.; Zhao, D.C.; Li, Z.Y.; Wang, Y.; Liu, J.N.; Du, T.; Zhou, L.; Chen, Y.H.; Yu, Q.C.; Chen, Q.S.; et al. Inactivated cGAS-STING Signaling Facilitates Endocrine Resistance by Forming a Positive Feedback Loop with AKT Kinase in ER+HER2- Breast Cancer. Adv. Sci. 2024, 11, e2403592. [Google Scholar] [CrossRef]
- Stahl-Meyer, J.; Stahl-Meyer, K.; Jäättelä, M. Control of mitosis, inflammation, and cell motility by limited leakage of lysosomes. Curr. Opin. Cell Biol. 2021, 71, 29–37. [Google Scholar] [CrossRef]
- Baz, A.A.; Hao, H.; Lan, S.; Li, Z.; Liu, S.; Jin, X.; Chen, S.; Chu, Y. Emerging insights into macrophage extracellular traps in bacterial infections. FASEB J. 2024, 38, e23767. [Google Scholar] [CrossRef]
- Weng, W.; Liu, Y.; Hu, Z.; Li, Z.; Peng, X.; Wang, M.; Dong, B.; Zhong, S.; Jiang, Y.; Pan, Y. Macrophage extracellular traps promote tumor-like biologic behaviors of fibroblast-like synoviocytes through cGAS-mediated PI3K/Akt signaling pathway in patients with rheumatoid arthritis. J. Leukoc. Biol. 2024, 115, 116–129. [Google Scholar] [CrossRef] [PubMed]
- Doster, R.S.; Rogers, L.M.; Gaddy, J.A.; Aronoff, D.M. Macrophage Extracellular Traps: A Scoping Review. J. Innate Immun. 2018, 10, 3–13. [Google Scholar] [CrossRef]
- Song, J.-X.; Villagomes, D.; Zhao, H.; Zhu, M. cGAS in nucleus: The link between immune response and DNA damage repair. Front. Immunol. 2022, 13, 1076784. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Chen, Q.; Chen, Z.; Sun, Z.; Chen, W.; Wei, R. Mitochondrial DNA-Activated cGAS-STING Signaling in Environmental Dry Eye. Investig. Ophthalmol. Vis. Sci. 2024, 65, 33. [Google Scholar] [CrossRef]
- Gordon, S.; Plüddemann, A. Tissue macrophages: Heterogeneity and functions. BMC Biol. 2017, 15, 53. [Google Scholar] [CrossRef]
- Phuengmaung, P.; Somparn, P.; Panpetch, W.; Singkham-In, U.; Wannigama, D.L.; Chatsuwan, T.; Leelahavanichkul, A. Coexistence of Pseudomonas aeruginosa With Candida albicans Enhances Biofilm Thickness Through Alginate-Related Extracellular Matrix but Is Attenuated by N-acetyl-l-cysteine. Front. Cell. Infect. Microbiol. 2020, 10, 594336. [Google Scholar] [CrossRef]
- Saisorn, W.; Santiworakul, C.; Phuengmaung, P.; Siripen, N.; Rianthavorn, P.; Leelahavanichkul, A. Extracellular traps in peripheral blood mononuclear cell fraction in childhood-onset systemic lupus erythematosus. Sci. Rep. 2024, 14, 23177. [Google Scholar] [CrossRef] [PubMed]
- Kueanjinda, P.; Roytrakul, S.; Palaga, T. A Novel Role of Numb as A Regulator of Pro-inflammatory Cytokine Production in Macrophages in Response to Toll-like Receptor 4. Sci. Rep. 2015, 5, 12784. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef]
- Galaxy, C. The Galaxy platform for accessible, reproducible, and collaborative data analyses: 2024 update. Nucleic Acids Res. 2024, 52, W83–W94. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdottir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Jaroonwitchawan, T.; Visitchanakun, P.; Dang, P.C.; Ritprajak, P.; Palaga, T.; Leelahavanichkul, A. Dysregulation of Lipid Metabolism in Macrophages Is Responsible for Severe Endotoxin Tolerance in FcgRIIB-Deficient Lupus Mice. Front. Immunol. 2020, 11, 959. [Google Scholar] [CrossRef]
- Sae-Khow, K.; Charoensappakit, A.; Udompornpitak, K.; Saisorn, W.; Issara-Amphorn, J.; Palaga, T.; Leelahavanichkul, A. Syk inhibitor attenuates lupus in FcγRIIb(-)(/-) mice through the Inhibition of DNA extracellular traps from macrophages and neutrophils via p38MAPK-dependent pathway. Cell Death Discov. 2025, 11, 63. [Google Scholar] [CrossRef]
| Primers | Sequences | Accession Number | |
|---|---|---|---|
| Cyclic GMP–AMP synthase (cGAS) | Forward | 5′-ATGTGAAGATTTCGCTCCTAATGA-3′ | KC294567.1 |
| Reverse | 5′-GAAATGACTCAGCGGATTTCCT-3′ | ||
| Nuclear factor kappa B (NF-κB) | Forward | 5′-CTTCCTCAGCCATGGTACCTCT-3′ | M61909.1 |
| Reverse | 5′-CAAGTCTTCATCAGCATCAAACTG-3′ | ||
| Interleukin-1β (IL-1β) | Forward | 5′-GAAATGCCACCTTTTGACAGTG-3′ | NM_008361.4 |
| Reverse | 5′-TGGATGCTCTCATCAGGACAG-3′ | ||
| Inducible nitric oxide synthase (iNOS) | Forward | 5′-ACCCACATCTGGCAGAATGAG-3′ | AF427516.1 |
| Reverse | 5′-AGCCATGACCTTTCGCATTAG-3′ | ||
| Resistin-like molecule-α (Fizz-1) | Forward | 5′-GCCAGGTCCTGGAACCTTTC- 3′ | AF323082.1 |
| Reverse | 5′-GGAGCAGGGAGATGCAGATGA-3′ | ||
| Arginase-1 (Arg-1) | Forward | 5′-CTTGGCTTGCTTCGGAACTC-3′ | NM_007482.3 |
| Reverse | 5′-GGAGAAGGCGTTTGCTTAGTTC-3′ | ||
| Transforming growth factor (TGF-β) | Forward | 5′-CAGAGCTGCGCTTGCAGAG-3′ | AH003562.3 |
| Reverse | 5′-GTCAGCAGCCGGTTACCAAG-3′ | ||
| Beta-actin (β-actin) | Forward | 5′-CGGTTCCGATGCCCTGAGGCTCTT-3′ | NM_007393.5 |
| Reverse | 5′-CGTCACACTTCATGATGGAATTGA-3′ | ||
| Mitochondrial DNA (mtDNA) | Forward | 5′-CGTACACCCTCTAACCTAGAGAAGG-3′ | PV231059.1 |
| Reverse | 5′-GGTTTTAAGTCTTACGCAATTTCC-3′ | ||
| β2-microglobulin (β2M) | Forward | 5′-TTCTGGTGCTTGTCTCACTGA-3′ | NM_009735.3 |
| Reverse | 5′-CAGTATGTTCGGCTTCCCATTC-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suksamai, C.; Kaewduangduen, W.; Phuengmaung, P.; Sae-Khow, K.; Charoensappakit, A.; Udomkarnjananun, S.; Lotinun, S.; Kueanjinda, P.; Leelahavanichkul, A. Cyclic GMP-AMP Synthase (cGAS) Deletion Promotes Less Prominent Inflammatory Macrophages and Sepsis Severity in Catheter-Induced Infection and LPS Injection Models. Int. J. Mol. Sci. 2025, 26, 5069. https://doi.org/10.3390/ijms26115069
Suksamai C, Kaewduangduen W, Phuengmaung P, Sae-Khow K, Charoensappakit A, Udomkarnjananun S, Lotinun S, Kueanjinda P, Leelahavanichkul A. Cyclic GMP-AMP Synthase (cGAS) Deletion Promotes Less Prominent Inflammatory Macrophages and Sepsis Severity in Catheter-Induced Infection and LPS Injection Models. International Journal of Molecular Sciences. 2025; 26(11):5069. https://doi.org/10.3390/ijms26115069
Chicago/Turabian StyleSuksamai, Chatsuree, Warerat Kaewduangduen, Pornpimol Phuengmaung, Kritsanawan Sae-Khow, Awirut Charoensappakit, Suwasin Udomkarnjananun, Sutada Lotinun, Patipark Kueanjinda, and Asada Leelahavanichkul. 2025. "Cyclic GMP-AMP Synthase (cGAS) Deletion Promotes Less Prominent Inflammatory Macrophages and Sepsis Severity in Catheter-Induced Infection and LPS Injection Models" International Journal of Molecular Sciences 26, no. 11: 5069. https://doi.org/10.3390/ijms26115069
APA StyleSuksamai, C., Kaewduangduen, W., Phuengmaung, P., Sae-Khow, K., Charoensappakit, A., Udomkarnjananun, S., Lotinun, S., Kueanjinda, P., & Leelahavanichkul, A. (2025). Cyclic GMP-AMP Synthase (cGAS) Deletion Promotes Less Prominent Inflammatory Macrophages and Sepsis Severity in Catheter-Induced Infection and LPS Injection Models. International Journal of Molecular Sciences, 26(11), 5069. https://doi.org/10.3390/ijms26115069







