Low Efficiency of Homology-Independent Targeted Integration for CRISPR/Cas9 Correction in the Vicinity of the SLC26A4 c.919-2A>G Variant
Abstract
1. Introduction
2. Results
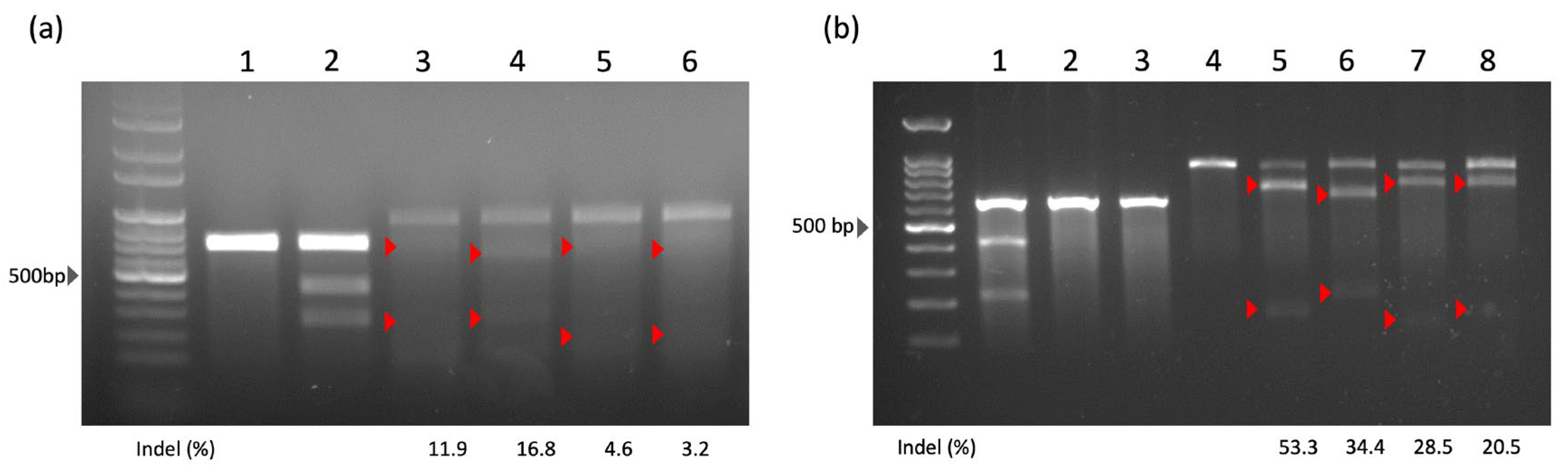
2.1. The Surveyor Assay Results
2.2. Statistics of HITI Editing Efficiency
3. Discussion
4. Materials and Methods
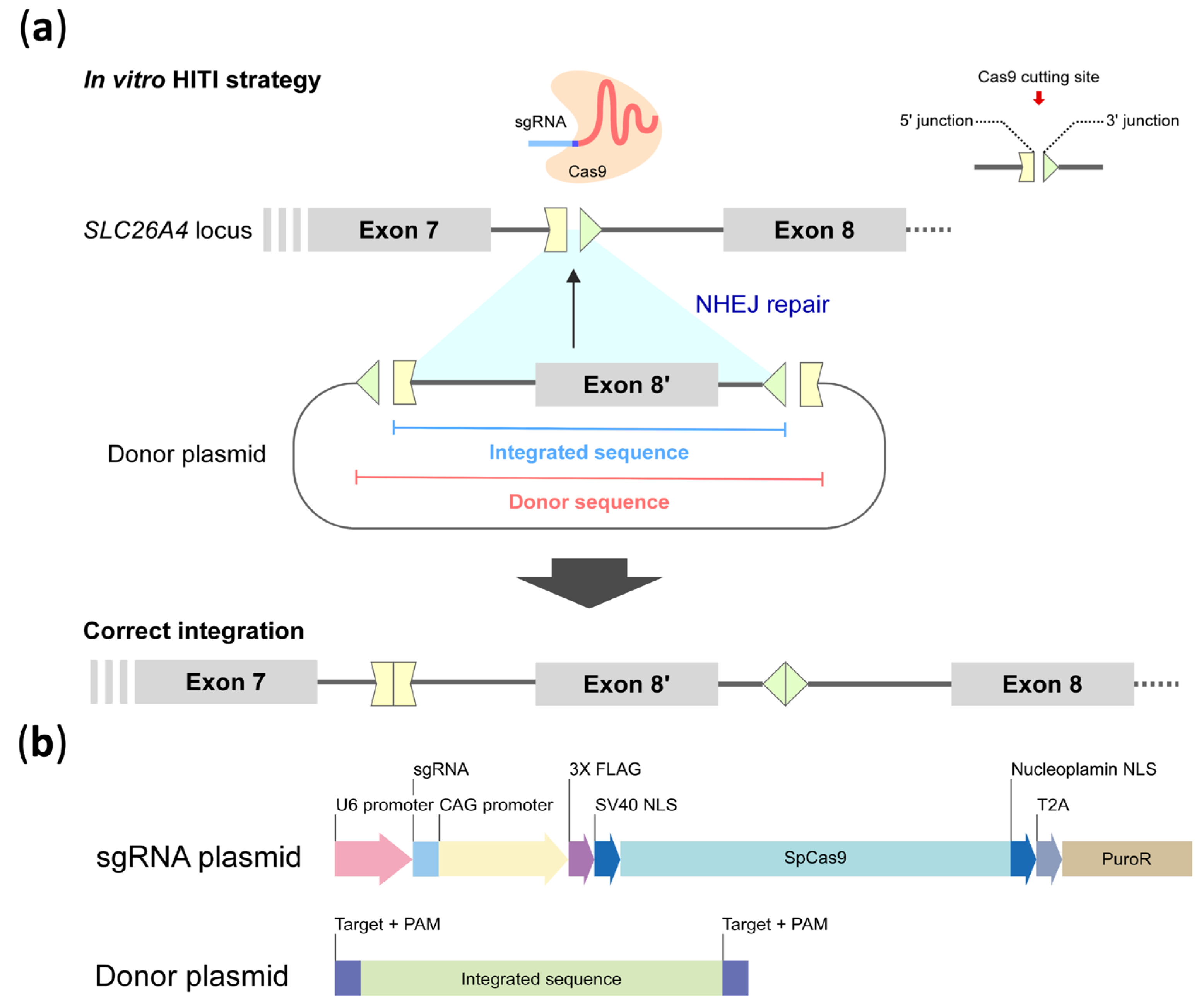
4.1. HITI Plasmid System
4.2. Cell Culture and Transfection
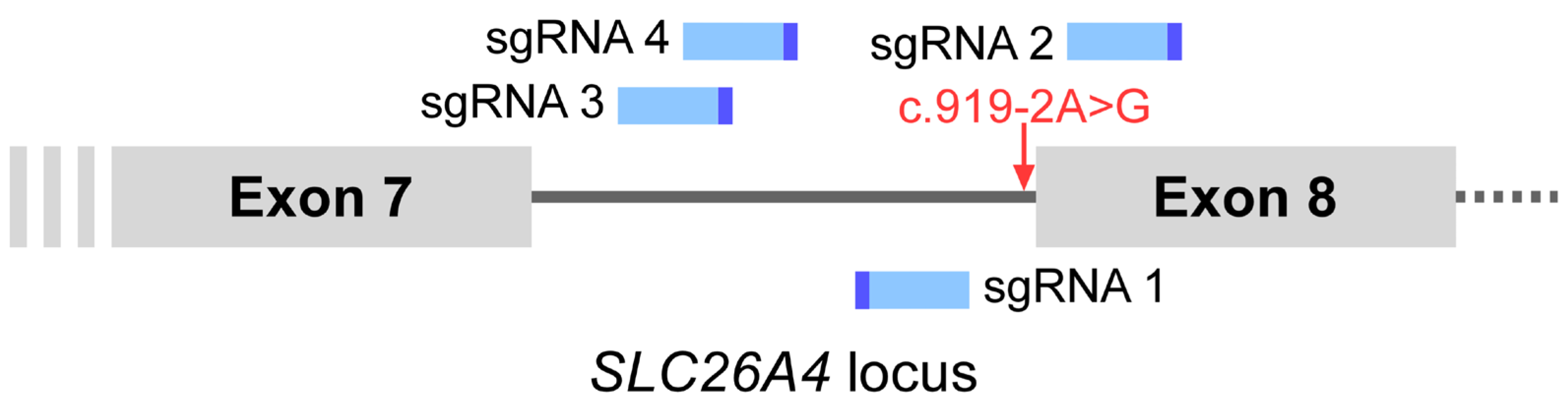
4.3. Targeting Efficiency of sgRNAs and Selection
4.4. HITI
4.5. Amplicon-Based NGS and Data Processing of Reads
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A

References
- Hilgert, N.; Smith, R.J.H.; Van Camp, G. Forty-six genes causing nonsyndromic hearing impairment: Which ones should be analyzed in DNA diagnostics? Mutat. Res. 2009, 681, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Li, Z.; Wang, W.; Huang, S.; Lu, Y.; Gao, Z.; Wang, L.; Kang, D.; Li, L.; Liu, Y.; et al. A quantitative cSMART assay for noninvasive prenatal screening of autosomal recessive nonsyndromic hearing loss caused by GJB2 and SLC26A4 mutations. Genet. Med. 2017, 19, 1309–1316. [Google Scholar] [CrossRef] [PubMed]
- Li, X.C.; Everett, L.A.; Lalwani, A.K.; Desmukh, D.; Friedman, T.B.; Green, E.D.; Wilcox, E.R. A mutation in PDS causes non-syndromic recessive deafness. Nat. Genet. 1998, 18, 215–217. [Google Scholar] [CrossRef] [PubMed]
- Mori, T.; Westerberg, B.D.; Atashband, S.; Kozak, F.K. Natural history of hearing loss in children with enlarged vestibular aqueduct syndrome. J. Otolaryngol. Head. Neck Surg. 2008, 37, 112–118. [Google Scholar]
- Jiang, L.; Wang, D.; He, Y.; Shu, Y. Advances in gene therapy hold promise for treating hereditary hearing loss. Mol. Ther. 2023, 31, 934–950. [Google Scholar] [CrossRef]
- Royaux, I.E.; Wall, S.M.; Karniski, L.P.; Everett, L.A.; Suzuki, K.; Knepper, M.A.; Green, E.D. Pendrin, encoded by the Pendred syndrome gene, resides in the apical region of renal intercalated cells and mediates bicarbonate secretion. Proc. Natl. Acad. Sci. USA 2001, 98, 4221–4226. [Google Scholar] [CrossRef]
- Hosoya, M.; Fujioka, M.; Sone, T.; Okamoto, S.; Akamatsu, W.; Ukai, H.; Ueda, H.R.; Ogawa, K.; Matsunaga, T.; Okano, H. Cochlear Cell Modeling Using Disease-Specific iPSCs Unveils a Degenerative Phenotype and Suggests Treatments for Congenital Progressive Hearing Loss. Cell Rep. 2017, 18, 68–81. [Google Scholar] [CrossRef]
- Royaux, I.E.; Belyantseva, I.A.; Wu, T.; Kachar, B.; Everett, L.A.; Marcus, D.C.; Green, E.D. Localization and functional studies of pendrin in the mouse inner ear provide insight about the etiology of deafness in pendred syndrome. J. Assoc. Res. Otolaryngol. 2003, 4, 394–404. [Google Scholar] [CrossRef]
- Bidart, J.M.; Mian, C.; Lazar, V.; Russo, D.; Filetti, S.; Caillou, B.; Schlumberger, M. Expression of pendrin and the Pendred syndrome (PDS) gene in human thyroid tissues. J. Clin. Endocrinol. Metab. 2000, 85, 2028–2033. [Google Scholar] [CrossRef][Green Version]
- Everett, L.A.; Morsli, H.; Wu, D.K.; Green, E.D. Expression pattern of the mouse ortholog of the Pendred’s syndrome gene (Pds) suggests a key role for pendrin in the inner ear. Proc. Natl. Acad. Sci. USA 1999, 96, 9727–9732. [Google Scholar] [CrossRef]
- Shcheynikov, N.; Yang, D.; Wang, Y.; Zeng, W.; Karniski, L.P.; So, I.; Wall, S.M.; Muallem, S. The Slc26a4 transporter functions as an electroneutral Cl-/I-/HCO3- exchanger: Role of Slc26a4 and Slc26a6 in I- and HCO3- secretion and in regulation of CFTR in the parotid duct. J. Physiol. 2008, 586, 3813–3824. [Google Scholar] [CrossRef] [PubMed]
- Nakao, I.; Kanaji, S.; Ohta, S.; Matsushita, H.; Arima, K.; Yuyama, N.; Yamaya, M.; Nakayama, K.; Kubo, H.; Watanabe, M.; et al. Identification of pendrin as a common mediator for mucus production in bronchial asthma and chronic obstructive pulmonary disease. J. Immunol. 2008, 180, 6262–6269. [Google Scholar] [CrossRef]
- Azaiez, H.; Booth, K.T.; Ephraim, S.S.; Crone, B.; Black-Ziegelbein, E.A.; Marini, R.J.; Shearer, A.E.; Sloan-Heggen, C.M.; Kolbe, D.; Casavant, T. Genomic landscape and mutational signatures of deafness-associated genes. Am. J. Hum. Genet. 2018, 103, 484–497. [Google Scholar] [CrossRef]
- Tsukada, K.; Nishio, S.-y.; Hattori, M.; Usami, S.-i. Ethnic-specific spectrum of GJB2 and SLC26A4 mutations: Their origin and a literature review. Ann. Otol. Rhinol. Laryngol. 2015, 124, 61S–76S. [Google Scholar] [CrossRef]
- Dai, P.; Li, Q.; Huang, D.; Yuan, Y.; Kang, D.; Miller, D.T.; Shao, H.; Zhu, Q.; He, J.; Yu, F. SLC26A4 c.919-2A>G varies among Chinese ethnic groups as a cause of hearing loss. Genet. Med. 2008, 10, 586–592. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.C.; Yeh, T.H.; Chen, P.J.; Hsu, C.J. Prevalent SLC26A4 mutations in patients with enlarged vestibular aqueduct and/or Mondini dysplasia: A unique spectrum of mutations in Taiwan, including a frequent founder mutation. Laryngoscope 2005, 115, 1060–1064. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-C.; Tsai, C.-Y.; Lin, Y.-H.; Chen, P.-Y.; Lin, P.-H.; Cheng, Y.-F.; Wu, C.-M.; Lin, Y.-H.; Lee, C.-Y.; Erdenechuluun, J. Genetic epidemiology and clinical features of hereditary hearing impairment in the Taiwanese population. Genes 2019, 10, 772. [Google Scholar] [CrossRef]
- Tsukamoto, K.; Suzuki, H.; Harada, D.; Namba, A.; Abe, S.; Usami, S.-i. Distribution and frequencies of PDS (SLC26A4) mutations in Pendred syndrome and nonsyndromic hearing loss associated with enlarged vestibular aqueduct: A unique spectrum of mutations in Japanese. Eur. J. Hum. Genet. 2003, 11, 916–922. [Google Scholar] [CrossRef]
- Park, H.J.; Shaukat, S.; Liu, X.Z.; Hahn, S.H.; Naz, S.; Ghosh, M.; Kim, H.N.; Moon, S.K.; Abe, S.; Tukamoto, K.; et al. Origins and frequencies of SLC26A4 (PDS) mutations in east and south Asians: Global implications for the epidemiology of deafness. J. Med. Genet. 2003, 40, 242–248. [Google Scholar] [CrossRef]
- Erdenechuluun, J.; Lin, Y.-H.; Ganbat, K.; Bataakhuu, D.; Makhbal, Z.; Tsai, C.-Y.; Lin, Y.-H.; Chan, Y.-H.; Hsu, C.-J.; Hsu, W.-C. Unique spectra of deafness-associated mutations in Mongolians provide insights into the genetic relationships among Eurasian populations. PLoS ONE 2018, 13, e0209797. [Google Scholar] [CrossRef]
- Cheng, Y.F.; Chan, Y.H.; Hu, C.J.; Lu, Y.C.; Saeki, T.; Hosoya, M.; Saegusa, C.; Fujioka, M.; Okano, H.; Weng, S.M.; et al. Generation of a human iPS cell line (CGMH.SLC26A4919-2) from a Pendred syndrome patient carrying SLC26A4 c.919-2A>G splice-site mutation. Stem Cell Res. 2019, 40, 101524. [Google Scholar] [CrossRef]
- Li, G.; Li, X.; Zhuang, S.; Wang, L.; Zhu, Y.; Chen, Y.; Sun, W.; Wu, Z.; Zhou, Z.; Chen, J.; et al. Gene editing and its applications in biomedicine. Sci. China Life Sci. 2022, 65, 660–700. [Google Scholar] [CrossRef] [PubMed]
- Rodgers, K.; McVey, M. Error-Prone Repair of DNA Double-Strand Breaks. J. Cell. Physiol. 2016, 231, 15–24. [Google Scholar] [CrossRef]
- Orthwein, A.; Noordermeer, S.M.; Wilson, M.D.; Landry, S.; Enchev, R.I.; Sherker, A.; Munro, M.; Pinder, J.; Salsman, J.; Dellaire, G.; et al. A mechanism for the suppression of homologous recombination in G1 cells. Nature 2015, 528, 422–426. [Google Scholar] [CrossRef]
- Saha, J.; Wang, S.-Y.; Davis, A.J. Chapter Five—Examining DNA Double-Strand Break Repair in a Cell Cycle-Dependent Manner. In Methods in Enzymology; Eichman, B.F., Ed.; Academic Press: Cambridge, MA, USA, 2017; Volume 591, pp. 97–118. [Google Scholar]
- Lieber, M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010, 79, 181–211. [Google Scholar] [CrossRef] [PubMed]
- Merkle, F.T.; Neuhausser, W.M.; Santos, D.; Valen, E.; Gagnon, J.A.; Maas, K.; Sandoe, J.; Schier, A.F.; Eggan, K. Efficient CRISPR-Cas9-Mediated Generation of Knockin Human Pluripotent Stem Cells Lacking Undesired Mutations at the Targeted Locus. Cell Rep. 2015, 11, 875–883. [Google Scholar] [CrossRef] [PubMed]
- Orlando, S.J.; Santiago, Y.; DeKelver, R.C.; Freyvert, Y.; Boydston, E.A.; Moehle, E.A.; Choi, V.M.; Gopalan, S.M.; Lou, J.F.; Li, J.; et al. Zinc-finger nuclease-driven targeted integration into mammalian genomes using donors with limited chromosomal homology. Nucleic Acids Res. 2010, 38, e152. [Google Scholar] [CrossRef]
- Suzuki, K.; Tsunekawa, Y.; Hernandez-Benitez, R.; Wu, J.; Zhu, J.; Kim, E.J.; Hatanaka, F.; Yamamoto, M.; Araoka, T.; Li, Z.; et al. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature 2016, 540, 144–149. [Google Scholar] [CrossRef]
- Coolidge, C.J.; Seely, R.J.; Patton, J.G. Functional analysis of the polypyrimidine tract in pre-mRNA splicing. Nucleic Acids Res. 1997, 25, 888–896. [Google Scholar] [CrossRef]
- Anna, A.; Monika, G. Splicing mutations in human genetic disorders: Examples, detection, and confirmation. J. Appl. Genet. 2018, 59, 253–268. [Google Scholar] [CrossRef]
- Watakabe, A.; Tanaka, K.; Shimura, Y. The role of exon sequences in splice site selection. Genes Dev. 1993, 7, 407–418. [Google Scholar] [CrossRef]
- Kim, M.A.; Kim, S.H.; Ryu, N.; Ma, J.H.; Kim, Y.R.; Jung, J.; Hsu, C.J.; Choi, J.Y.; Lee, K.Y.; Wangemann, P.; et al. Gene therapy for hereditary hearing loss by SLC26A4 mutations in mice reveals distinct functional roles of pendrin in normal hearing. Theranostics 2019, 9, 7184–7199. [Google Scholar] [CrossRef] [PubMed]
- Takeda, H.; Miwa, T.; Kim, M.Y.; Choi, B.Y.; Orita, Y.; Minoda, R. Prenatal electroporation-mediated gene transfer restores Slc26a4 knock-out mouse hearing and vestibular function. Sci. Rep. 2019, 9, 17979. [Google Scholar] [CrossRef] [PubMed]
- Newby, G.A.; Liu, D.R. In vivo somatic cell base editing and prime editing. Mol. Ther. 2021, 29, 3107–3124. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef]
- Yeh, W.-H.; Shubina-Oleinik, O.; Levy, J.M.; Pan, B.; Newby, G.A.; Wornow, M.; Burt, R.; Chen, J.C.; Holt, J.R.; Liu, D.R. In vivo base editing restores sensory transduction and transiently improves auditory function in a mouse model of recessive deafness. Sci. Transl. Med. 2020, 12, eaay9101. [Google Scholar] [CrossRef] [PubMed]
- Nami, F.; Basiri, M.; Satarian, L.; Curtiss, C.; Baharvand, H.; Verfaillie, C. Strategies for In Vivo Genome Editing in Nondividing Cells. Trends Biotechnol. 2018, 36, 770–786. [Google Scholar] [CrossRef]
- Genovese, P.; Schiroli, G.; Escobar, G.; Tomaso, T.D.; Firrito, C.; Calabria, A.; Moi, D.; Mazzieri, R.; Bonini, C.; Holmes, M.C.; et al. Targeted genome editing in human repopulating haematopoietic stem cells. Nature 2014, 510, 235–240. [Google Scholar] [CrossRef]
- Nishiyama, J.; Mikuni, T.; Yasuda, R. Virus-Mediated Genome Editing via Homology-Directed Repair in Mitotic and Postmitotic Cells in Mammalian Brain. Neuron 2017, 96, 755–768.e5. [Google Scholar] [CrossRef]
- Tornabene, P.; Ferla, R.; Llado-Santaeularia, M.; Centrulo, M.; Dell’Anno, M.; Esposito, F.; Marrocco, E.; Pone, E.; Minopoli, R.; Iodice, C.; et al. Therapeutic homology-independent targeted integration in retina and liver. Nat. Commun. 2022, 13, 1963. [Google Scholar] [CrossRef]
- Bandara, R.A.; Chen, Z.R.; Hu, J. Potential of helper-dependent Adenoviral vectors in CRISPR-cas9-mediated lung gene therapy. Cell Biosci. 2021, 11, 145. [Google Scholar] [CrossRef] [PubMed]
- Bloomer, H.; Smith, R.H.; Hakami, W.; Larochelle, A. Genome editing in human hematopoietic stem and progenitor cells via CRISPR-Cas9-mediated homology-independent targeted integration. Mol. Ther. 2021, 29, 1611–1624. [Google Scholar] [CrossRef] [PubMed]
- Onishi, A.; Tsunekawa, Y.; Mandai, M.; Ishimaru, A.; Ohigashi, Y.; Sho, J.; Yasuda, K.; Suzuki, K.; Izpisua Belmonte, J.C.; Matsuzaki, F.; et al. Optimization of HITI-Mediated Gene Insertion for Rhodopsin and Peripherin-2 in Mouse Rod Photoreceptors: Targeting Dominant Retinitis Pigmentosa. Investig. Ophthalmol. Vis. Sci. 2024, 65, 38. [Google Scholar] [CrossRef]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Thorvaldsdóttir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Masella, A.P.; Lalansingh, C.M.; Sivasundaram, P.; Fraser, M.; Bristow, R.G.; Boutros, P.C. BAMQL: A query language for extracting reads from BAM files. BMC Bioinform. 2016, 17, 305. [Google Scholar] [CrossRef]

| No. | sgRNA and PAM Sequence | Out-of-Frame Scores |
|---|---|---|
| sgRNA1 | AAAGATGTTAAAAACTCCAT TGG | 53.9 |
| sgRNA2 | ATTGCTACTGCCATTTCATA TGG | 69.8 |
| sgRNA3 | TTAGAAAGTTCAGCATTATT TGG | 68.4 |
| sgRNA4 | CATTATTTGGTTGACAAACA AGG | 70.5 |
| Name | SLC-1 * | SLC-2 * | SLC-3 * |
|---|---|---|---|
| Raw reads | 17,435 | 18,381 | 22,062 |
| Refined reads | 17,184 | 18,181 | 21,755 |
| Refined reads with QueryWT | 16,466 (95.82%) | 16,561 (91.09%) | 20,688 (95.10%) |
| Refined reads with QueryCI | 0 | 27 (0.15%) | 0 |
| Refined reads with deletion | 14 (0.08%) | 193 (1.06%) | 19 (0.09%) |
| Refined reads with insertion | 0 | 434 (2.39%) | 0 |
| Refined reads with indel | 0 | 3 (0.02%) | 1 (<0.01%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ho, C.-H.; Tsai, C.-Y.; Chang, C.-C.; Hu, C.-J.; Huang, C.-Y.; Lu, Y.-C.; Lin, P.-H.; Lin, C.-H.; Lin, H.-I.; OuYang, C.-H.; et al. Low Efficiency of Homology-Independent Targeted Integration for CRISPR/Cas9 Correction in the Vicinity of the SLC26A4 c.919-2A>G Variant. Int. J. Mol. Sci. 2025, 26, 4980. https://doi.org/10.3390/ijms26114980
Ho C-H, Tsai C-Y, Chang C-C, Hu C-J, Huang C-Y, Lu Y-C, Lin P-H, Lin C-H, Lin H-I, OuYang C-H, et al. Low Efficiency of Homology-Independent Targeted Integration for CRISPR/Cas9 Correction in the Vicinity of the SLC26A4 c.919-2A>G Variant. International Journal of Molecular Sciences. 2025; 26(11):4980. https://doi.org/10.3390/ijms26114980
Chicago/Turabian StyleHo, Chang-Han, Cheng-Yu Tsai, Chi-Chieh Chang, Chin-Ju Hu, Cheng-Yen Huang, Ying-Chang Lu, Pei-Hsuan Lin, Chin-Hsien Lin, Han-I Lin, Chih-Hsin OuYang, and et al. 2025. "Low Efficiency of Homology-Independent Targeted Integration for CRISPR/Cas9 Correction in the Vicinity of the SLC26A4 c.919-2A>G Variant" International Journal of Molecular Sciences 26, no. 11: 4980. https://doi.org/10.3390/ijms26114980
APA StyleHo, C.-H., Tsai, C.-Y., Chang, C.-C., Hu, C.-J., Huang, C.-Y., Lu, Y.-C., Lin, P.-H., Lin, C.-H., Lin, H.-I., OuYang, C.-H., Hsu, C.-J., Liu, T.-C., Chen, Y.-T., Chan, Y.-H., Cheng, Y.-F., & Wu, C.-C. (2025). Low Efficiency of Homology-Independent Targeted Integration for CRISPR/Cas9 Correction in the Vicinity of the SLC26A4 c.919-2A>G Variant. International Journal of Molecular Sciences, 26(11), 4980. https://doi.org/10.3390/ijms26114980








