1. Introduction
Cytochrome P450s (CYPs) are a large family of enzymes comprising 57 genes in humans. CYPs catalyze the oxidation of numerous hydrophobic chemicals, including endogenous compounds, therapeutic drugs, and environmental toxins [
1,
2]. Individual CYPs are classified into families and sub-families according to their sequence similarities. CYPs from families 1, 2, and 3 are responsible for the metabolism of 70–80% of clinically used drugs, including CYP1A2, 2A6, 2B6, 2C8, 2C9, 2C19, 2D6, 2E1, and 3A4/5 [
1,
3]. Drug-metabolizing CYPs are highly expressed in the liver and are also present in other tissues, such as small intestine, lung, kidney, and adipose tissues [
4]. In the liver and small intestine, these enzymes are primarily responsible for phase I reactions. Phase I drug metabolism covers a variety of reactions which include oxidation, reduction, and hydrolysis [
1]. These reactions introduce polar groups onto the parent compound, converting hydrophobic drugs into more water-soluble compounds facilitating their elimination from the body. CYP gene expression is often regulated by xenobiotic receptors. For example, the aryl hydrocarbon receptor upregulates the expression of CYP1A1 and 1A2 mRNA in response to a range of aromatic hydrocarbon agonists [
5]. Furthermore, the constitutive androstane receptor and pregnane X receptor upregulate several xenobiotic-metabolizing enzymes upon activation, including CYP2B6, 2C8, 2C9, 2C19, and 3A4 [
6,
7]. These CYPs are responsible for metabolizing multiple drugs, leading to altered drug levels and effectiveness. Identifying CYP inhibitors is a critical part of drug development to minimize the potential for drug–drug interactions as required in regulatory guidance from US Food and Drug Administration (FDA) [
8] and European Medicines Agency (EMA) [
9].
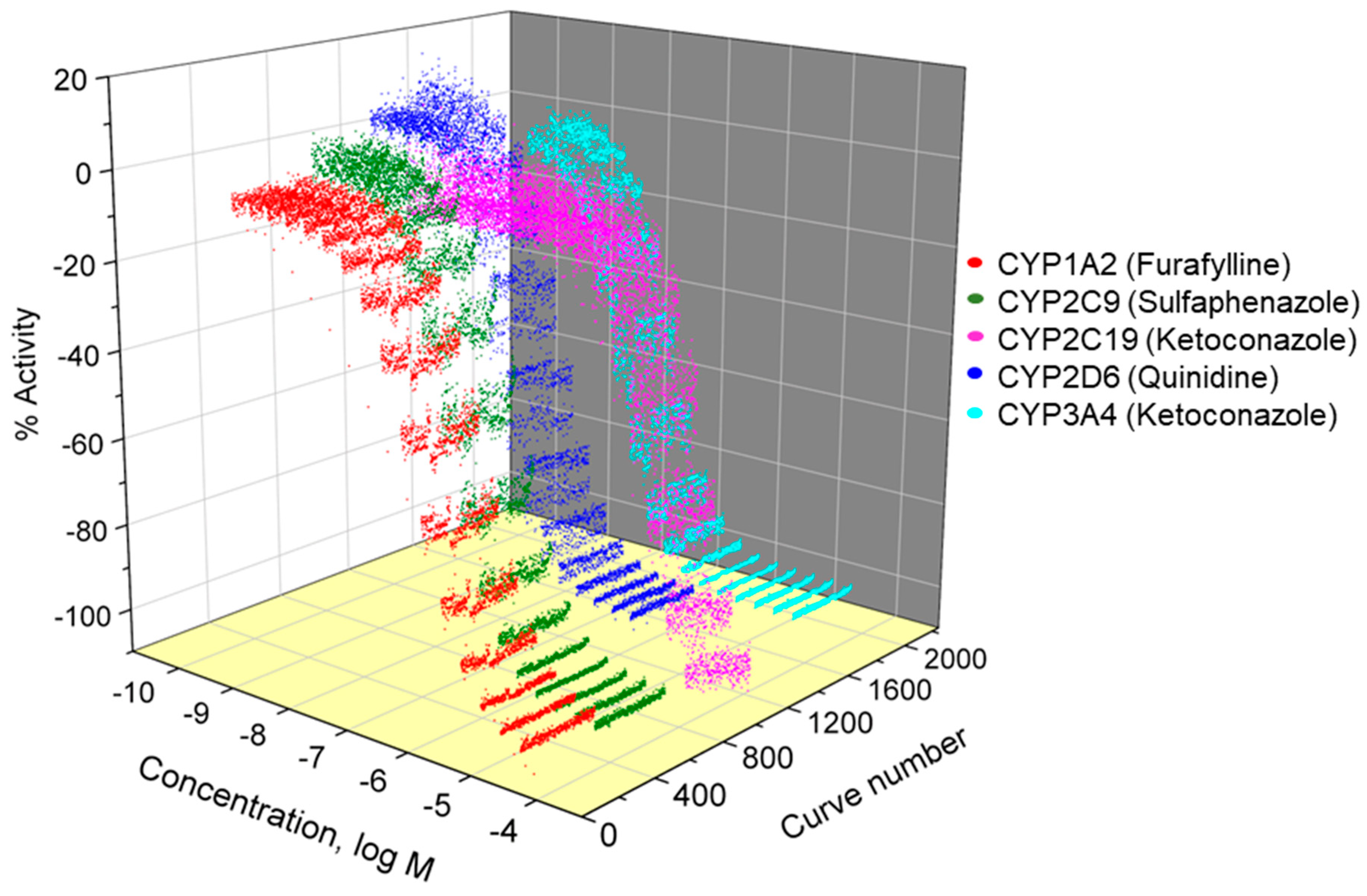
We identified CYP inhibitors from large compound libraries by employing quantitative high-throughput screening (qHTS) using P450-Glo™ assays as a part of the Tox21 program. The Tox21 program aims to assess the potential toxicological effects of a large collection of chemicals (Tox21 10K compound library) using qHTS [
10,
11]. To represent a more physiologically relevant model in humans, the Tox2110K library has recently been screened using assays that incorporated human liver microsomes to identify chemicals that are either bioactivated or detoxified by metabolism [
12,
13]. In the present study, five CYP inhibition screens were performed in a qHTS format to obtain datasets consisting of concentration–response curves, potencies, and efficacies for each test compound against human CYP1A2, 2C9, 2C19, 2D6, and 3A4. The compounds are clustered based on their structural similarity and each cluster was evaluated for their enrichment of active (i.e., CYP inhibiting) compounds. We identified several structural classes that inhibit the activity of all five tested CYP enzymes with over 90% of the class members as actives, including azole fungicides, phenylenediamines, and proton pump inhibitors (PPIs). In addition to the pan-active clusters, several CYP-selective clusters were identified, such as polycyclic aromatic hydrocarbons (PAHs) in the CYP1A2 assay. Identifying CYP inhibitors can provide valuable tools for understanding the mechanisms of drug and environmental chemical toxicity.
3. Discussion
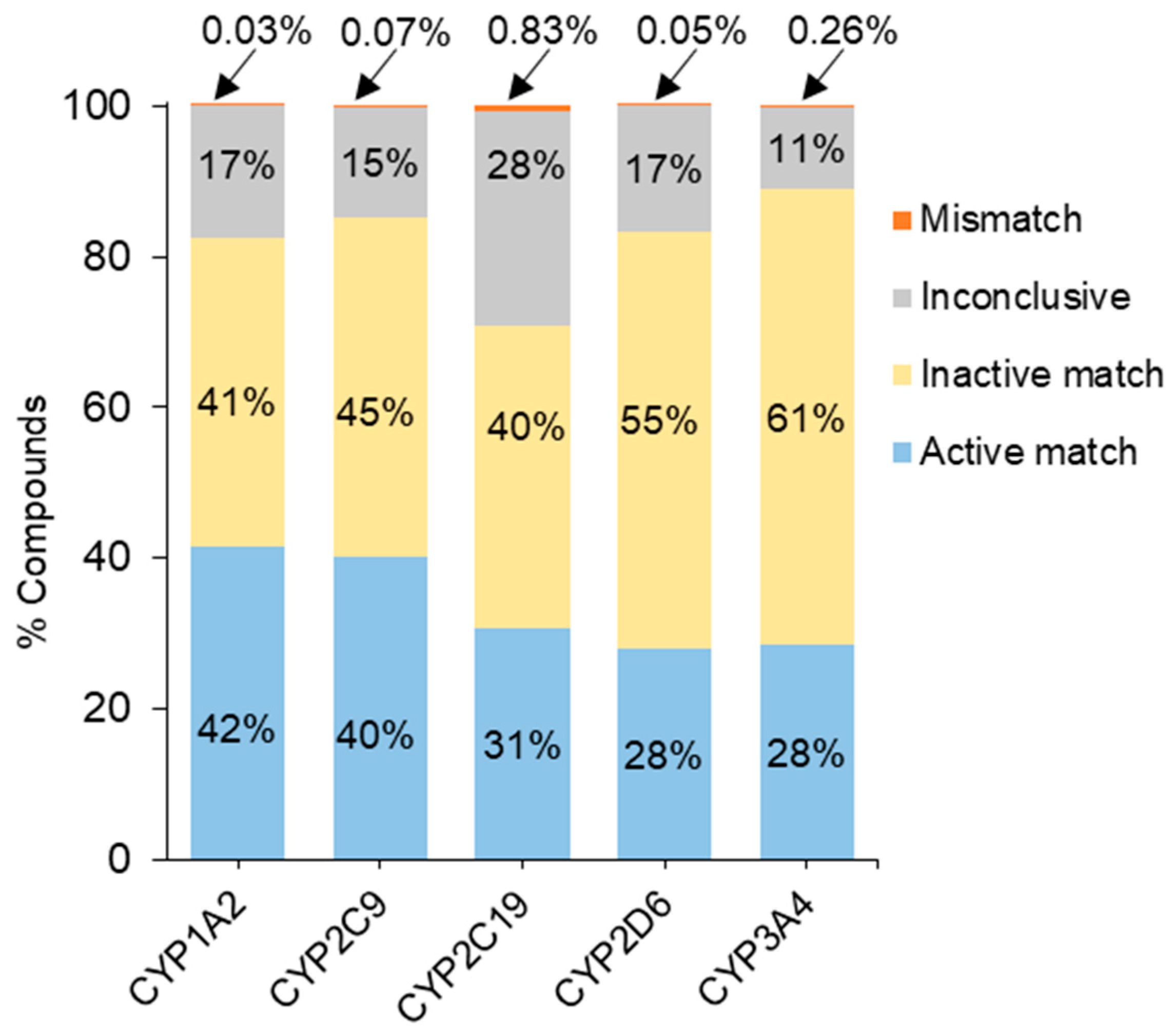
The Tox21 10K compound library, consisting of 7316 unique compounds, was screened in five CYP assays for human CYP1A2, 2C9, 2C19, 2D6, and 3A4 using P450-Glo™ assay technology [
14,
24]. Different luciferin substrates were used for each P450-Glo™ assay, as these substrates are specific to each CYP enzyme [
25]. The five CYP screenings performed in three independent runs exhibited high reproducibility, with minimal activity mismatch rates. Of the 7316 compounds tested, 68% were active inhibitors in at least one CYP assay and 7.6% demonstrated inhibitory activity against all five CYP enzymes. Notably, CYP2C9 and 2C19 have the most overlapping active compounds, likely related to their high structural and functional similarity among the five CYPs included. The Tox21 10K compounds were clustered based on structural similarity, and each structural cluster was evaluated for its enrichment of active inhibitors in five CYP assays. Structural analysis has identified both pan-CYP and/or CYP-selective clusters that are enriched with actives, such as azole fungicides, proton pump inhibitors, and/or PAHs, respectively [
5,
16,
17] that were confirmed in the current study. Previously unknown potent compounds (IC
50 values less than 1 µM) were identified from the arylamine, indanedione, organophosphate, indole alkaloid, and corticosteroid classes in CYP1A2, 2C9, 2C19, 2D6, and 3A4 assays, respectively. These potentially novel compounds were either inactive or less potent (IC
50 > 20 µM) in the luciferase counter-screen assay, indicating minimal interference with the luciferase signal. Although the inhibitory potential of these compounds has yet to be validated in vivo, several publicly or commercially available tools, such as ADMET-AI [
26] or ADMET Predictor
® from Stimulation Plus, can be used to rapidly and accurately predict their ADMET properties, a critical step for identifying drug-like candidates. Some therapeutic compounds from the aforementioned chemical classes, such as yohimbine and loteprednol etaboanate, have Cmax (maximum plasma concentration) values of 75 ng/mL and 139 pg/mL, respectively [
27,
28,
29]. The IC
50 values for these drugs in the current study are 0.3 µM and 0.68 µM, respectively. For yohimbine, the ratio of IC
50 and Cmax value is less than 10, suggesting it may be an effective inhibitor within the therapeutic range [
30]. In contrast, the IC
50 value for loteprednol in this study may not fall within the comparable therapeutic range, particularly for drugs primarily administered topically or intranasally [
28,
29].
Surprisingly, activators for CYP2C9, 2C19, or 3A4 were also identified during this study. However, the apparent activators identified in biochemical-based in vitro assays are not true endpoints of gene induction, which entails the activation of CYP gene expression. For example, common CYP inducers, including carbamazepine, phenobarbital, phenytoin, and rifampicin [
31], were all shown to be either inactive or inhibitors in the current study. This apparent activation of CYPs indicates positive cooperativity, in which the binding of a substrate to the active site increases the affinity of other binding sites [
32]. This phenomenon, particularly associated with the CYP3A4 enzyme, has been reported as heterotropic cooperativity [
33], which involves the simultaneous binding of two different substrates to the spatially distinct substrate-binding domains within the enzyme active site or effector site [
34] and hence results in the formation of distinct enzyme–substrate complexes. Although the detailed mechanism underlying allosteric or heterotropic cooperativity in CYP3A4 activation remains unclear, in silico approaches may offer valuable tools for evaluating these interactions. Previous studies have identified potential ligand-binding sites in CYP3A4 and highlighted Phe123 as a key residue mediating heterotropic allosteric interactions [
35,
36].
The five CYP enzymes included in the current study, representing the CYP 1, 2, and 3 families, are responsible for the biotransformation of 70–80% of clinically approved drugs [
1,
3]. Several drugs and environmental chemicals act as both substrates and inducers or inhibitors of CYPs. This is due to non-selectivity, i.e., the same drug can act as a substrate for several CYP isoforms, leading to the formation of reactive or secondary metabolites that can inhibit or induce CYPs [
37]. Moreover, the assay technology used in the current study (P450-Glo™) is unable to distinguish between inhibitors and substrates, and additional studies such as metabolic stability assays are needed to make those distinctions. As previously reported [
38], the substrate depletion method can be used to determine the intrinsic clearance of the compound and evaluate its metabolic stability. Drug classes such as beta-blockers and corticosteroids, identified as inhibitors in this study, are known to be extensively metabolized by CYP2D6 and 3A4, respectively [
21,
39]. However, the structural classes included in the current study are populated with selective inhibitors identified in the corresponding CYP assays. The CYP2D6 and 3A4 assays demonstrated the highest activity across several classes of therapeutic drugs, whereas CYP2C19 had the lowest activity rate within these therapeutic drug classes, which is not surprising given that CYP2D6 and 3A4 contribute to over 50% of CYP-related drug metabolism [
1].
Among the five CYPs, CYP3A4 is notable for exhibiting substrate-dependent inhibition due to its multiple substrate’s sites. The three distinct substrate recognition sites are typically grouped based on representative substrates, testosterone, midazolam, and nifedipine, with most other substrates falling within one of these categories [
40]. The probe substrate used in the current study, luciferin-PPXE [
25], is known to be inhibited by midazolam and nifedipine. In the current study, both compounds demonstrated CYP3A4 inhibition, with IC
50 values of 11.4 µM for midazolam and 13.4 µM for nifedipine. Therefore, any compound that inhibits a CYP3A4 assay using either of those substrates is also expected to inhibit the luciferin-PPXE assay. In contrast, testosterone has been shown to stimulate the luciferin-PPXE reaction. In the current study, testosterone activated CYP3A4 with an AC
50 of 4.2 µM, suggesting that compounds binding in a similar manner may also enhance the assay signal. Thus, the luciferin-PPXE assay can detect effects from compounds interacting with any of the three known substrate-binding configurations. Importantly, luciferin-PPXE is the only DMSO-tolerant probe substrate for CYP3A4, which is critical for screening purposes, as all test compounds in this study were dissolved in DMSO, resulting in a final concentration of 0.5% DMSO in the assay.
The screening data revealed that most structural classes of compounds exhibited non-selective CYP inhibition due to the high homology in the binding pocket regions of the CYPs, all of which contain the heme iron motif. The majority of the potent inhibitors identified across all CYP assays are azole compounds, suggesting that their potency arises from the presence of heme-binding groups in their chemical structures [
41]. Several chemical classes, including those from the PAHs and quaternary ammonium compounds (QACs) found throughout the library, were enriched with CYP1A2 and 2D6 inhibitors, respectively, as most of these chemicals inhibit CYPs through a mechanism-based action. This process leads to the formation of reactive metabolites that covalently bind to the enzymes resulting in their inactivation [
42]. However, CYP-selective inhibitors pose drug interaction challenges, particularly when targeting enzymes with high homology. In addition to enzyme inactivation, reactive metabolites can covalently bind to other cellular macromolecules leading to adverse effects such as hepatotoxicity and carcinogenicity. This also includes the metabolic activation of specific natural products from dietary supplements that can result in toxicities [
43,
44]. Studies have shown that a polyphenol-rich diet from plant-based foods, along with parabens and phthalates used in consumer products, can inhibit expression of some CYP isoforms [
45,
46]. Therefore, evaluating the potential of a drug candidate or an environmental chemical to inhibit CYP activity is a crucial step in therapeutic drug development or chemical synthesis.
The current study identified potential CYP inhibitors from the Tox21 10K compound library and assessed their CYP inhibitory activities by clustering them based on structural similarity, resulting in the identification of several pan-CYP and CYP-selective chemical classes. These screening datasets offer valuable insights into the chemical classes associated with CYP inhibition, which could significantly aid early-stage drug development and chemical synthesis. Additionally, given that the Tox21 10K library contains more than two-thirds environmental chemicals, these data may also be instrumental in predicting toxicity from the biotransformation of these chemicals.