Exploring Inflammatory Asthma Phenotypes: Proteomic Signatures in Serum and Induced Sputum
Abstract
1. Introduction
2. Results
2.1. Clinical Characteristics
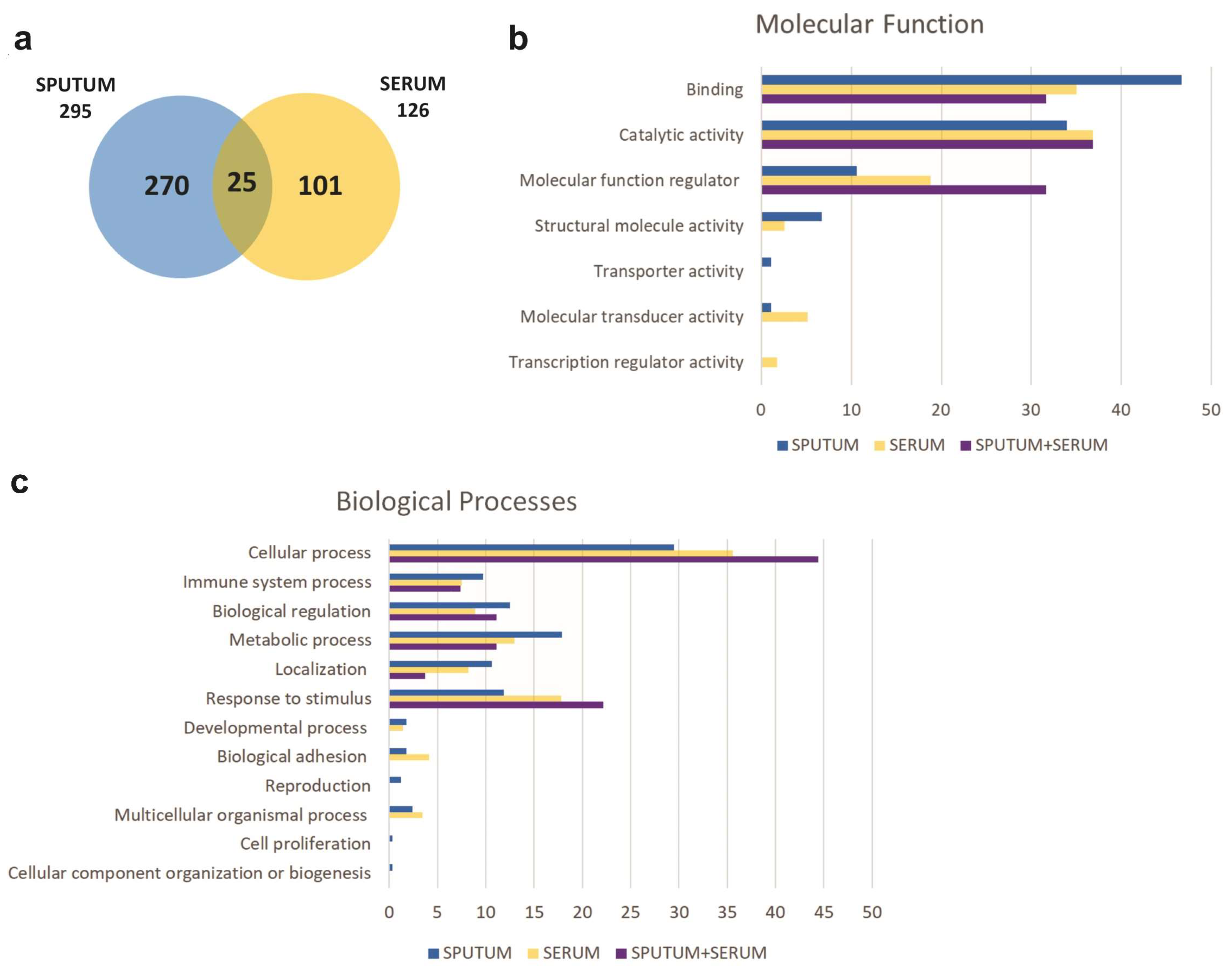
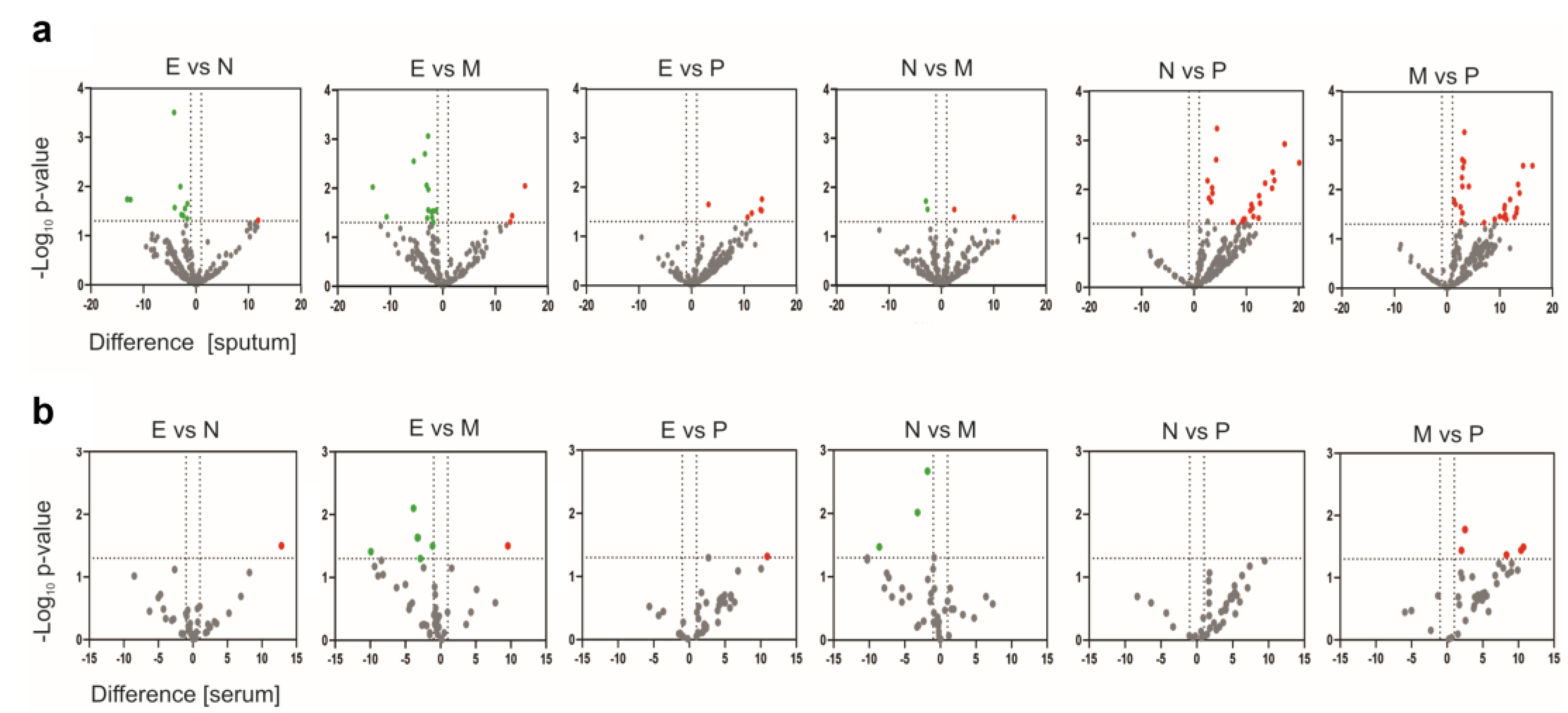
2.2. Proteomic Profiling
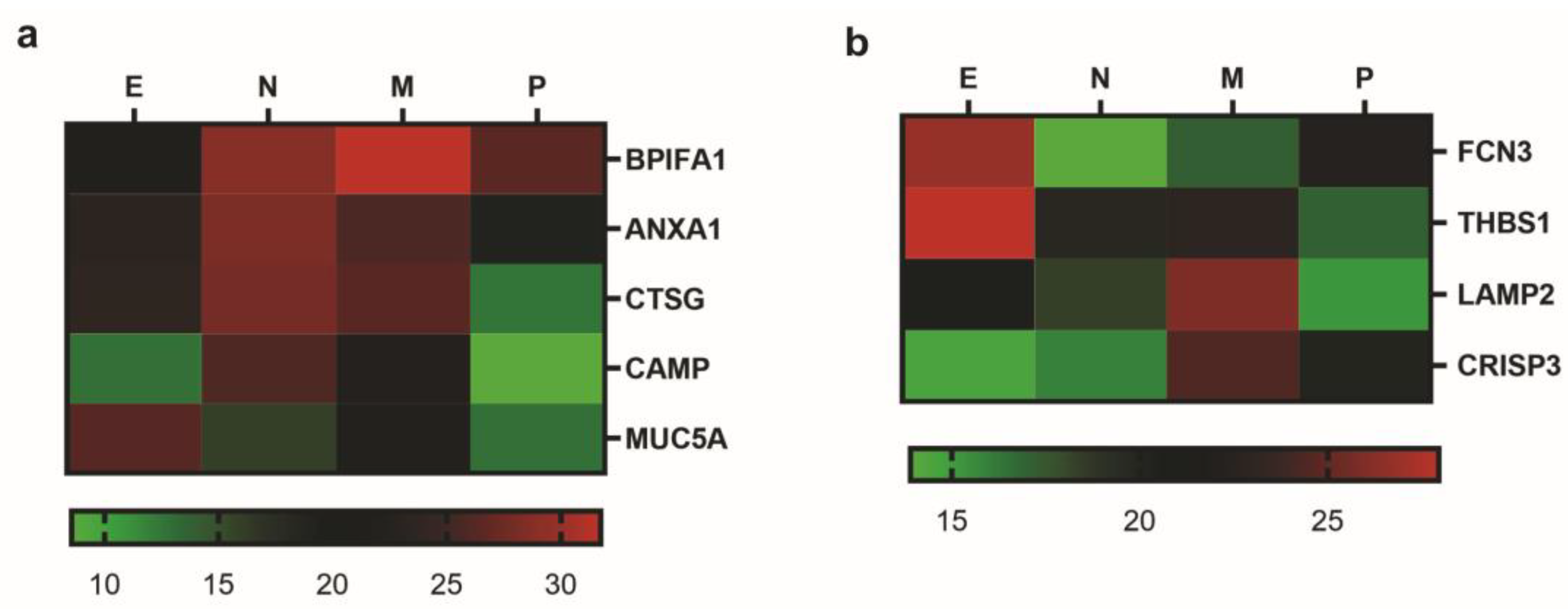
2.3. Identification of Biomarkers
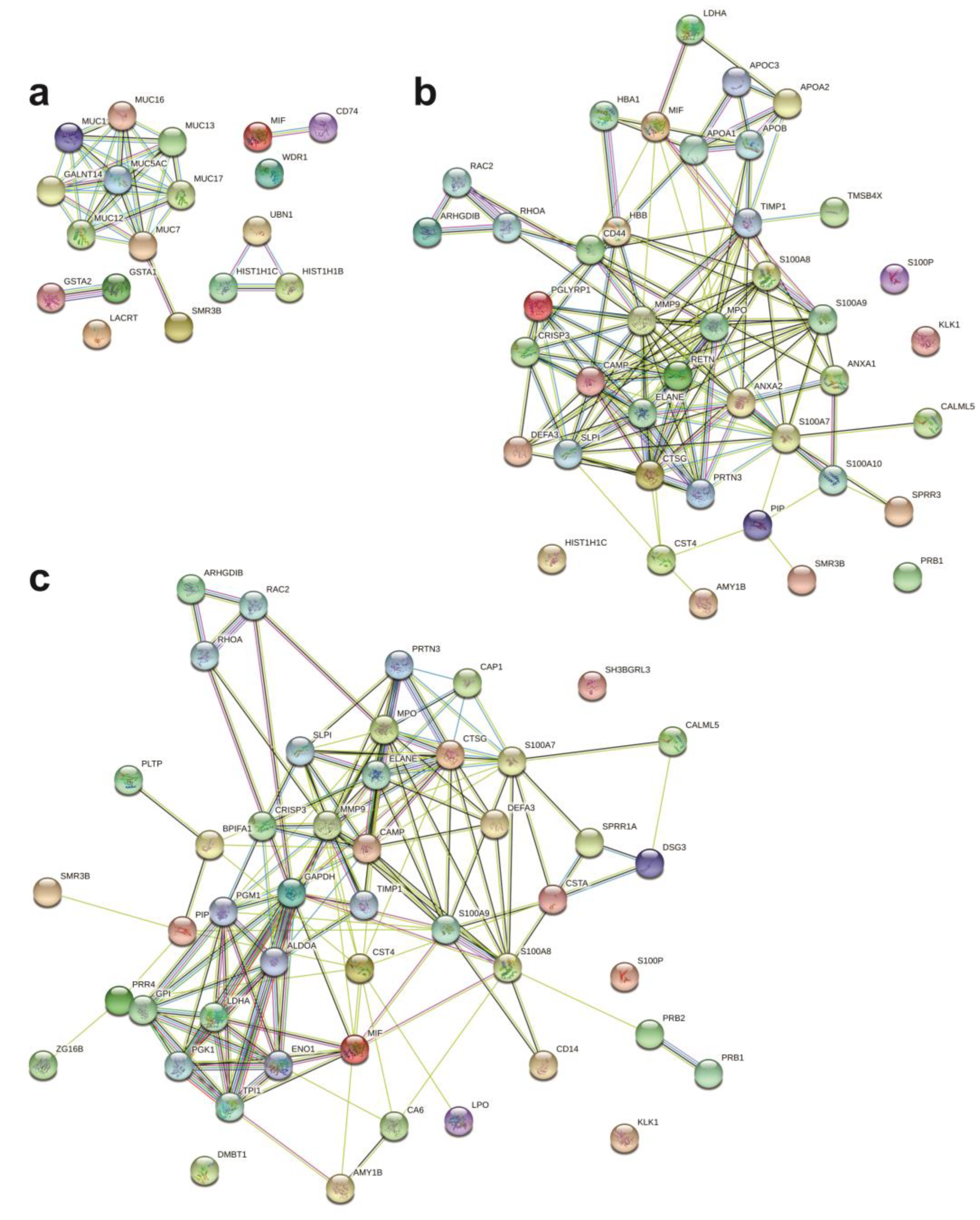
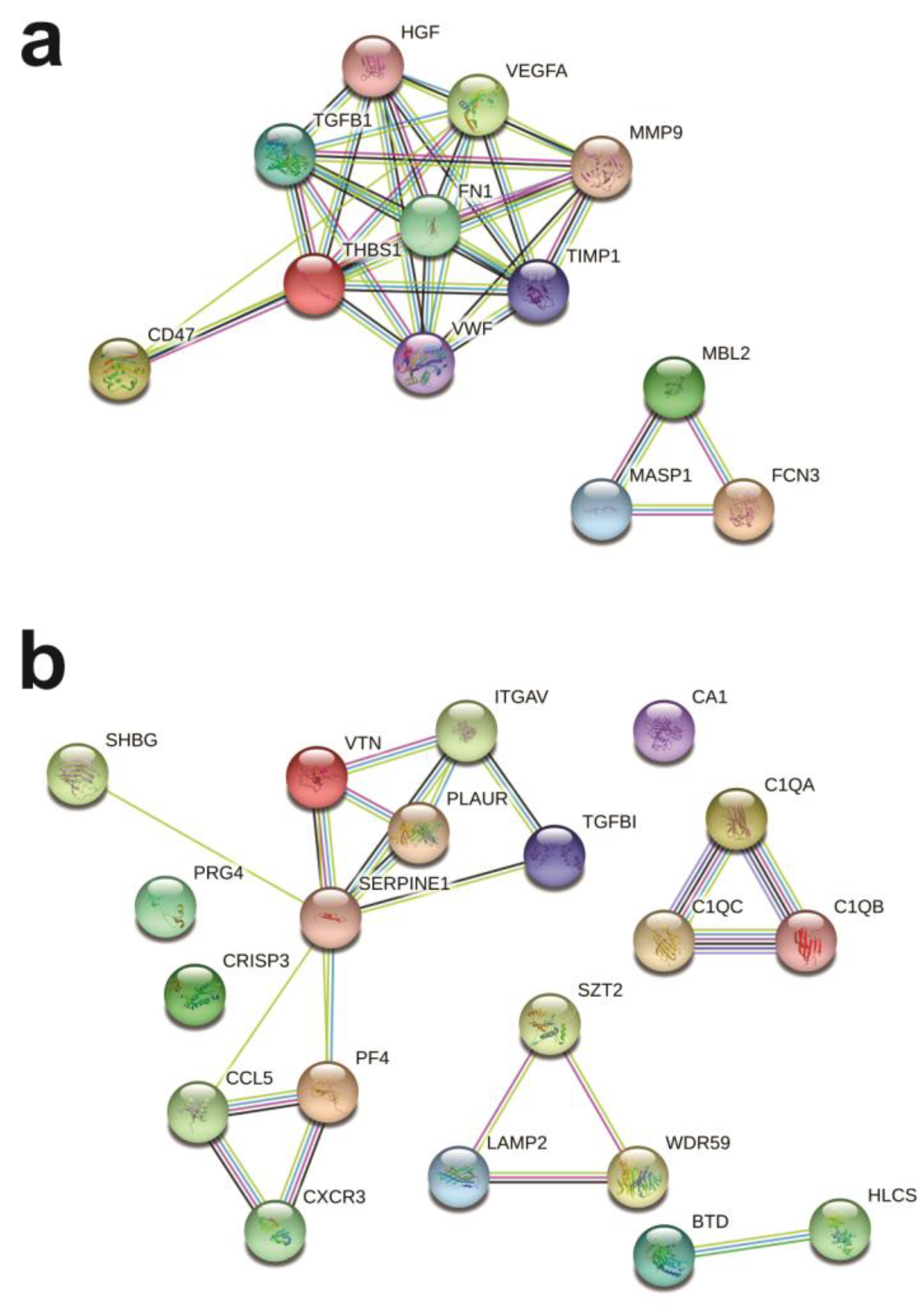
2.4. Protein Networks
3. Discussion
4. Materials and Methods
4.1. Subject Recruitment
4.2. Sample Collection and Phenotyping
4.3. Sample Preparation
4.4. Proteomic Analysis
4.5. Functional and Network Analysis
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Global Initiative for Asthma. Global Strategy for Asthma Management and Prevention. 2022. Available online: www.ginasthma.org (accessed on 5 September 2023).
- Hamilton, D.; Lehman, H. Asthma Phenotypes as a Guide for Current and Future Biologic Therapies. Clin. Rev. Allergy Immunol. 2019, 59, 160–174. [Google Scholar] [CrossRef] [PubMed]
- Taunk, S.T.; Cardet, J.C.; Ledford, D.K. Clinical Implications of Asthma Endotypes and Phenotypes. Allergy Asthma Proc. 2022, 45, 375–382. [Google Scholar] [CrossRef]
- Woodruff, P.G.; Modrek, B.; Choy, D.F.; Jia, G.; Abbas, A.R.; Ellwanger, A.; Koth, L.L.; Arron, J.R.; Fahy, J. V T-Helper Type 2-Driven Inflammation Defines Major Subphenotypes of Asthma. Am. J. Respir. Crit. Care Med. 2009, 180, 388–395. [Google Scholar] [CrossRef] [PubMed]
- Papi, A.; Brightling, C.; Pedersen, S.E.; Reddel, H.K. Asthma. Lancet 2018, 391, 783–800. [Google Scholar] [CrossRef] [PubMed]
- Nelson, R.K.; Bush, A.; Stokes, J.; Nair, P.; Akuthota, P. Eosinophilic Asthma. J. Allergy Clin. Immunol. Pract. 2020, 8, 465–473. [Google Scholar] [CrossRef]
- Woodruff, P.G.; Khashayar, R.; Lazarus, S.C.; Janson, S.; Avila, P.; Boushey, H.A.; Segal, M.; Fahy, J.V. Relationship between Airway Inflammation, Hyperresponsiveness, and Obstruction in Asthma. J. Allergy Clin. Immunol. 2001, 108, 753–758. [Google Scholar] [CrossRef]
- Gao, H.; Ying, S.; Dai, Y. Pathological Roles of Neutrophil-Mediated Inflammation in Asthma and Its Potential for Therapy as a Target. J. Immunol. Res. 2017, 2017, 3743048. [Google Scholar] [CrossRef]
- Choy, D.F.; Hart, K.M.; Borthwick, L.A.; Shikotra, A.; Nagarkar, D.R.; Siddiqui, S.; Jia, G.; Ohri, C.M.; Doran, E.; Vannella, K.M.; et al. TH2 and TH17 Inflammatory Pathways Are Reciprocally Regulated in Asthma. Sci. Transl. Med. 2015, 7, 301ra129. [Google Scholar] [CrossRef]
- Simpson, J.L.; Scott, R.; Boyle, M.J.; Gibson, P.G. Inflammatory Subtypes in Asthma: Assessment and Identification Using Induced Sputum. Respirology 2006, 11, 54–61. [Google Scholar] [CrossRef]
- Pembrey, L.; Barreto, M.L.; Douwes, J.; Cooper, P.; Henderson, J.; Mpairwe, H.; Ardura-Garcia, C.; Chico, M.; Brooks, C.; Cruz, A.A.; et al. Understanding Asthma Phenotypes: The World Asthma Phenotypes (WASP) International Collaboration. ERJ Open Res. 2018, 4, 1–11. [Google Scholar] [CrossRef]
- Eguiluz-Gracia, I.; Tay, T.R.; Hew, M.; Escribese, M.M.; Barber, D.; O’Hehir, R.E.; Torres, M.J. Recent Developments and Highlights in Biomarkers in Allergic Diseases and Asthma. Allergy Eur. J. Allergy Clin. Immunol. 2018, 73, 2290–2305. [Google Scholar] [CrossRef]
- Schofield, J.P.R.; Burg, D.; Nicholas, B.; Strazzeri, F.; Brandsma, J.; Staykova, D.; Folisi, C.; Bansal, A.T.; Xian, Y.; Guo, Y.; et al. Stratification of Asthma Phenotypes by Airway Proteomic Signatures. J. Allergy Clin. Immunol. 2019, 144, 70–82. [Google Scholar] [CrossRef]
- Kostikas, K.; Brindicci, C.; Patalano, F. Blood Eosinophils as Biomarkers to Drive Treatment Choices in Asthma and COPD. Curr. Drug Targets 2018, 19, 1882–1896. [Google Scholar] [CrossRef]
- Akar-Ghibril, N.; Casale, T.; Custovic, A.; Phipatanakul, W. Allergic Endotypes and Phenotypes of Asthma. J. Allergy Clin. Immunol. Pract. 2020, 8, 429–440. [Google Scholar] [CrossRef]
- Porpodis, K.; Tsiouprou, I.; Apostolopoulos, A.; Ntontsi, P.; Fouka, E.; Papakosta, D.; Vliagoftis, H.; Domvri, K. Eosinophilic Asthma, Phenotypes-Endotypes and Current Biomarkers of Choice. J. Pers. Med. 2022, 12, 1093. [Google Scholar] [CrossRef] [PubMed]
- Izuhara, K.; Ohta, S.; Ono, J. Using Periostin as a Biomarker in the Treatment of Asthma. Allergy Asthma Immunol. Res. 2016, 8, 491–498. [Google Scholar] [CrossRef]
- Tiotiu, A. Biomarkers in Asthma: State of the Art. Asthma Res. Pract. 2018, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Kuruvilla, M.E.; Lee, F.E.H.; Lee, G.B. Understanding Asthma Phenotypes, Endotypes, and Mechanisms of Disease. Clin. Rev. Allergy Immunol. 2019, 56, 219–233. [Google Scholar] [CrossRef] [PubMed]
- Popović-Grle, S.; Štajduhar, A.; Lampalo, M.; Rnjak, D. Biomarkers in Different Asthma Phenotypes. Genes 2021, 12, 801. [Google Scholar] [CrossRef] [PubMed]
- McGrath, K.W.; Icitovic, N.; Boushey, H.A.; Lazarus, S.C.; Sutherland, E.R.; Chinchilli, V.M.; Fahy, J.V. A Large Subgroup of Mild-to-Moderate Asthma Is Persistently Noneosinophilic. Am. J. Respir. Crit. Care Med. 2012, 185, 612–619. [Google Scholar] [CrossRef] [PubMed]
- Schleich, F.N.; Manise, M.; Sele, J.; Henket, M.; Seidel, L.; Louis, R. Distribution of Sputum Cellular Phenotype in a Large Asthma Cohort: Predicting Factors for Eosinophilic vs Neutrophilic Inflammation. BMC Pulm. Med. 2013, 13, 11. [Google Scholar] [CrossRef]
- Coumou, H.; Westerhof, G.A.; de Nijs, S.B.; Amelink, M.; Bel, E.H. Diagnosing Persistent Blood Eosinophilia in Asthma with Single Blood Eosinophil or Exhaled Nitric Oxide Level. Respir. Med. 2018, 141, 81–86. [Google Scholar] [CrossRef]
- Wagener, A.H.; De Nijs, S.B.; Lutter, R.; Sousa, A.R.; Weersink, E.J.M.; Bel, E.H.; Sterk, P.J.; Ahw, S. External Validation of Blood Eosinophils, FE NO and Serum Periostin as Surrogates for Sputum Eosinophils in Asthma. Thorax 2015, 70, 115–120. [Google Scholar] [CrossRef]
- Gharib, S.A.; Nguyen, E.V.; Lai, Y.; Plampin, J.D.; Goodlett, D.R.; Hallstrand, T.S. Induced Sputum Proteome in Healthy Subjects and Asthmatic Patients. J. Allergy Clin. Immunol. 2011, 128, 1176–1184. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and Integrative Analysis of Large Gene Lists Using DAVID Bioinformatics Resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Hiemstra, P.S.; Amatngalim, G.D.; Van Der Does, A.M.; Taube, C. Antimicrobial Peptides and Innate Lung Defenses: Role in Infectious and Noninfectious Lung Diseases and Therapeutic Applications. Chest 2016, 149, 545–551. [Google Scholar] [CrossRef] [PubMed]
- Jiao, D.; Wong, C.K.; Tsang, M.S.M.; Chu, I.M.T.; Liu, D.; Zhu, J.; Chu, M.; Lam, C.W.K. Activation of Eosinophils Interacting with Bronchial Epithelial Cells by Antimicrobial Peptide LL-37: Implications in Allergic Asthma. Sci. Rep. 2017, 7, 1848. [Google Scholar] [CrossRef]
- Roider, E.; Ruzicka, T.; Schauber, J. Vitamin D, the Cutaneous Barrier, Antimicrobial Peptides and Allergies: Is There a Link? Allergy, Asthma Immunol. Res. 2013, 5, 119–128. [Google Scholar] [CrossRef]
- Xiao, W.; Hsu, Y.P.; Ishizaka, A.; Kirikae, T.; Moss, R.B. Sputum Cathelicidin, Urokinase Plasminogen Activation System Components, and Cytokines Discriminate Cystic Fibrosis, COPD, and Asthma Inflammation. Chest 2005, 128, 2316–2326. [Google Scholar] [CrossRef] [PubMed]
- Ronchetti, S.; Ricci, E.; Migliorati, G.; Gentili, M.; Riccardi, C. How Glucocorticoids Affect the Neutrophil Life. Int. J. Mol. Sci. 2018, 19, 4090. [Google Scholar] [CrossRef]
- Ibrahim, A.A.; Ramadan, A.; Wahby, A.A.; Draz, I.H.; El Baroudy, N.R.; Abdel Hamid, T.A. Evaluation of MiR-196a2 Expression and Annexin A1 Level in Children with Bronchial AsthmaEvaluation of MiR-196a2 Expression and Annexin A1 Level in Children. Allergol. Immunopathol. 2020, 48, 458–464. [Google Scholar] [CrossRef]
- Ng, F.S.P.; Wong, K.Y.; Guan, S.P.; Mustafa, F.B.; Kajiji, T.S.; Bist, P.; Biswas, S.K.; Wong, W.S.F.; Lim, L.H.K. Annexin-1-Deficient Mice Exhibit Spontaneous Airway Hyperresponsiveness and Exacerbated Allergen-Specific Antibody Responses in a Mouse Model of Asthma. Clin. Exp. Allergy 2011, 41, 1793–1803. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Lee, P.H.; Kim, B.G.; Seo, H.J.; Baek, A.R.; Park, J.S.; Lee, J.H.; Park, S.W.; Kim, D.J.; Park, C.S.; et al. Annexin A1 in Plasma from Patients with Bronchial Asthma: Its Association with Lung Function. BMC Pulm. Med. 2018, 18, 1. [Google Scholar] [CrossRef] [PubMed]
- Tsou, Y.A.; Tung, M.C.; Alexander, K.A.; Chang, W.D.; Tsai, M.H.; Chen, H.L.; Chen, C.M. The Role of BPIFA1 in Upper Airway Microbial Infections and Correlated Diseases. Biomed. Res. Int. 2018, 2018, 2021890. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Huang, J.; Moore, P.J.; Little, M.S.; Walton, W.G.; Fellner, R.C.; Alexis, N.E.; Peter Di, Y.; Redinbo, M.R.; Tilley, S.L.; et al. Identification of BPIFA1/SPLUNC1 as an Epithelium-Derived Smooth Muscle Relaxing Factor. Nat. Commun. 2017, 8, 14118. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, N.; Li, X.; Seibold, M.A.; Jarjour, N.N.; Denlinger, L.C.; Castro, M.; Coverstone, A.M.; Teague, W.G.; Boomer, J.; Bleecker, E.R.; et al. The Effect of BPIFA1/SPLUNC1 Genetic Variation on Its Expression and Function in Asthmatic Airway Epithelium. JCI Insight 2019, 4, e127237. [Google Scholar] [CrossRef]
- Zhang, R.; Trower, J.; Wu, T. Degradation of Bacterial Permeability Family Member A1 (BPIFA1) by House Dust Mite (HDM) Cysteine Protease Der p 1 Abrogates Immune Modulator Function. Int. J. Biol. Macromol. 2020, 164, 4022–4031. [Google Scholar] [CrossRef]
- Dasgupta, A.; Chakraborty, R.; Saha, B.; Suri, H.; Singh, P.; Raj, A.; Taneja, B.; Dash, D.; Sengupta, S.; Agrawal, A. Sputum Protein Biomarkers in Airway Diseases: A Pilot Study. Int. J. Chron. Obstruct. Pulmon. Dis. 2021, 16, 2203–2215. [Google Scholar] [CrossRef]
- Evans, C.M.; Raclawska, D.S.; Ttofali, F.; Liptzin, D.R.; Fletcher, A.A.; Harper, D.N.; McGing, M.A.; McElwee, M.M.; Williams, O.W.; Sanchez, E.; et al. The Polymeric Mucin Muc5ac Is Required for Allergic Airway Hyperreactivity. Nat. Commun. 2015, 6, 6281. [Google Scholar] [CrossRef]
- Lachowicz-Scroggins, M.E.; Yuan, S.; Kerr, S.C.; Dunican, E.M.; Yu, M.; Carrington, S.D.; Fahy, J.V. Abnormalities in MUC5AC and MUC5B Protein in Airway Mucus in Asthma. Am. J. Respir. Crit. Care Med. 2016, 194, 1296–1299. [Google Scholar] [CrossRef]
- Bonser, L.; Erle, D. Airway Mucus and Asthma: The Role of MUC5AC and MUC5B. J. Clin. Med. 2017, 6, 112. [Google Scholar] [CrossRef]
- Matsushita, M.; Fujita, T. Ficolins and the Lectin Complement Pathway. Immunol. Rev. 2001, 180, 78–85. [Google Scholar] [CrossRef]
- Alam, R.; Good, J.; Rollins, D.; Verma, M.; Chu, H.W.; Pham, T.H.; Martin, R.J. Airway and Serum Biochemical Correlates of Refractory Neutrophilic Asthma. J. Allergy Clin. Immunol. 2017, 140, 1004–1014.e13. [Google Scholar] [CrossRef]
- Freishtat, R.J.; Benton, A.S.; Watson, A.M.; Wang, Z.; Rose, M.C.; Hoffman, E.P. Delineation of a Gene Network Underlying the Pulmonary Response to Oxidative Stress in Asthma. J. Investig. Med. 2009, 57, 756–764. [Google Scholar] [CrossRef]
- Kılıç, A.; Ameli, A.; Park, J.A.; Kho, A.T.; Tantisira, K.; Santolini, M.; Cheng, F.; Mitchel, J.A.; McGill, M.; O’Sullivan, M.J.; et al. Mechanical Forces Induce an Asthma Gene Signature in Healthy Airway Epithelial Cells. Sci. Rep. 2020, 10, 966. [Google Scholar] [CrossRef]
- Chen, L.; Xu, J.; Chu, X.; Ju, C. MicroRNA-98 Interferes with Thrombospondin 1 Expression in Peripheral B Cells of Patients with Asthma. Biosci. Rep. 2017, 37, BSR20170149. [Google Scholar] [CrossRef]
- Udby, L.; Sørensen, O.E.; Borregaard, N.; Kjeldsen, L.; Calafat, J. Identification of Human Cysteine-Rich Secretory Protein 3 (CRISP-3) as a Matrix Protein in a Subset of Peroxidase-Negative Granules of Neutrophils and in the Granules of Eosinophils. J. Leukoc. Biol. 2002, 72, 462–469. [Google Scholar] [CrossRef]
- Udby, L.; Cowland, J.B.; Johnsen, A.H.; Sørensen, O.E.; Borregaard, N.; Kjeldsen, L. An ELISA for SGP28/CRISP-3, a Cysteine-Rich Secretory Protein in Human Neutrophils, Plasma, and Exocrine Secretions. J. Immunol. Methods 2002, 263, 43–55. [Google Scholar] [CrossRef] [PubMed]
- Bigler, J.; Boedigheimer, M.; Schofield, J.P.R.; Skipp, P.J.; Corfield, J.; Rowe, A.; Sousa, A.R.; Timour, M.; Twehues, L.; Hu, X.; et al. A Severe Asthma Disease Signature from Gene Expression Profiling of Peripheral Blood from U-BIOPRED Cohorts. Am. J. Respir. Crit. Care Med. 2017, 195, 1311–1320. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Huang, W.; Chen, N.; Shang, Y.; Zhang, H. SMC3 May Play an Important Role in Atopic Asthma Development. Clin. Respir. J. 2016, 10, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Thaysen-Andersen, M.; Venkatakrishnan, V.; Loke, I.; Laurini, C.; Diestel, S.; Parker, B.L.; Packer, N.H. Human Neutrophils Secrete Bioactive Paucimannosidic Proteins from Azurophilic Granules into Pathogen-Infected Sputum. J. Biol. Chem. 2015, 290, 8789–8802. [Google Scholar] [CrossRef] [PubMed]
- Gukovskaya, A.S.; Gukovsky, I.; Algül, H.; Habtezion, A. Autophagy, Inflammation, and Immune Dysfunction in the Pathogenesis of Pancreatitis. Gastroenterology 2017, 153, 1212–1226. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Pavlidis, S.; Ng Kee Kwong, F.; Hoda, U.; Rossios, C.; Sun, K.; Loza, M.; Baribaud, F.; Chanez, P.; Fowler, S.J.; et al. Sputum Proteomics and Airway Cell Transcripts of Current and Ex-Smokers with Severe Asthma in U-BIOPRED: An Exploratory Analysis. Eur. Respir. J. 2018, 51, 1702173. [Google Scholar] [CrossRef] [PubMed]
- Katayama, S.; Hammar, K.S.; Krjutškov, K.; Einarsdottir, E.; Hedlin, G.; Kere, J.; Söderhäll, C. Acute Wheeze-Specific Gene Module Shows Correlation with Vitamin D and Asthma Medication. Eur. Respir. J. 2020, 55, 1901330. [Google Scholar] [CrossRef]
- Pavord, I.D.; Pizzichini, M.M.; Pizzichini, E.; Hargreave, F.E. The Use of Induced Sputum to Investigate Airway Inflammation. Pneumologie 1997, 52, 498. [Google Scholar] [CrossRef]
- Pizzichini, E.; Pizzichini, M.M.M.; Kidney, J.C.; Efthimiadis, A.; Hussack, P.; Popov, T.; Cox, G.; Dolovich, J.; O’Byrne, P.; Hargreave, F.E. Induced Sputum, Bronchoalveolar Lavage and Blood from Mild Asthmatics: Inflammatory Cells, Lymphocyte Subsets and Soluble Markers Compared. Eur. Respir. J. 1998, 11, 828–834. [Google Scholar] [CrossRef]
| E | N | M | P | p Value | |
|---|---|---|---|---|---|
| (n = 8) | (n = 8) | (n = 8) | (n = 8) | ||
| Age, years | 48.6 ± 21.2 | 56.1 ± 13.7 | 55.5 ±10.0 | 48.9 ± 14.8 | 0.43 |
| Female sex, n (%) | 4 (50) | 4 (50) | 6 (75) | 5 (62.5) | 0.71 |
| BMI, kg/m2 | 25.1 ± 5.9 | 25.9 ± 2.3 | 28.9 ± 5.2 | 26.2 ± 5.3 | 0.57 |
| Age at onset, ≥12 years | 5 (62.5) | 3 (37.5) | 6 (75) | 5 (62.5) | 0.50 |
| SPT positivity, n (%) | 6 (75) | 8 (100) | 5 (62.5) | 5 (62.5) | 0.27 |
| FEV1, % predicted | 73.9 ± 12.5 | 71.5 ± 21.4 | 74.3 ± 15.6 | 78.5 ± 10.9 | 0.93 |
| Sputum eosinophils, % | 25 (12.5–36.7) | 0.5 (0–1.2) | 10 (4.9–13.2) | 0.5 (0–1) | <0.0001 a,b,c |
| Sputum neutrophils, % | 47 (40.5–60.2) | 94.5 (89.7–95.7) | 80.5 (76–84.5) | 43 (20.2–60.7) | <0.0001 a,c,d |
| Blood eosinophils/µL | 303.5 (171.2–511.2) | 144 (102.7–246.5) | 137.5 (115–207.7) | 286.5 (102.2–450) | 0.25 |
| Blood neutrophils/µL | 4232 (2948.7–7220.7) | 4250 (3827.5–4596.5) | 3831.5 (3303.5–4541.5) | 3612 (2513.5–4513.7) | 0.79 |
| GINA Step | |||||
| 3, n (%) | 0 (0) | 0 (0) | 0 (0) | 1 (12.5) | 0.53 |
| 4, n (%) | 4 (50) | 4 (50) | 4 (50) | 5 (62.5) | |
| 5, n (%) | 4 (50) | 4 (50) | 4 (50) | 2 (25) | |
| Inhaled corticosteroid, mcg | 1000 (800–1200) | 1000 (800–1200) | 1200 (950–1200) | 800 (700–900) | 0.79 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maia, L.P.; Cunha, T.M.; Santos, P.S.; Martins, M.M.; Briza, P.; Ferreira, F.; Amorim, M.M.; Caetano, L.B.; Farias, C.F.; Santoro, I.L.; et al. Exploring Inflammatory Asthma Phenotypes: Proteomic Signatures in Serum and Induced Sputum. Int. J. Mol. Sci. 2024, 25, 3501. https://doi.org/10.3390/ijms25063501
Maia LP, Cunha TM, Santos PS, Martins MM, Briza P, Ferreira F, Amorim MM, Caetano LB, Farias CF, Santoro IL, et al. Exploring Inflammatory Asthma Phenotypes: Proteomic Signatures in Serum and Induced Sputum. International Journal of Molecular Sciences. 2024; 25(6):3501. https://doi.org/10.3390/ijms25063501
Chicago/Turabian StyleMaia, Larissa Prado, Thulio Marquez Cunha, Paula Souza Santos, Mario Machado Martins, Peter Briza, Fatima Ferreira, Maria Marta Amorim, Lilian Ballini Caetano, Camyla Fernandes Farias, Ilka Lopes Santoro, and et al. 2024. "Exploring Inflammatory Asthma Phenotypes: Proteomic Signatures in Serum and Induced Sputum" International Journal of Molecular Sciences 25, no. 6: 3501. https://doi.org/10.3390/ijms25063501
APA StyleMaia, L. P., Cunha, T. M., Santos, P. S., Martins, M. M., Briza, P., Ferreira, F., Amorim, M. M., Caetano, L. B., Farias, C. F., Santoro, I. L., Godoy Fernandes, A. L., & Goulart, L. R. (2024). Exploring Inflammatory Asthma Phenotypes: Proteomic Signatures in Serum and Induced Sputum. International Journal of Molecular Sciences, 25(6), 3501. https://doi.org/10.3390/ijms25063501






