Blood Transcriptomics Identifies Multiple Gene Expression Pathways Associated with the Clinical Efficacy of Hymenoptera Venom Immunotherapy
Abstract
1. Introduction
2. Results
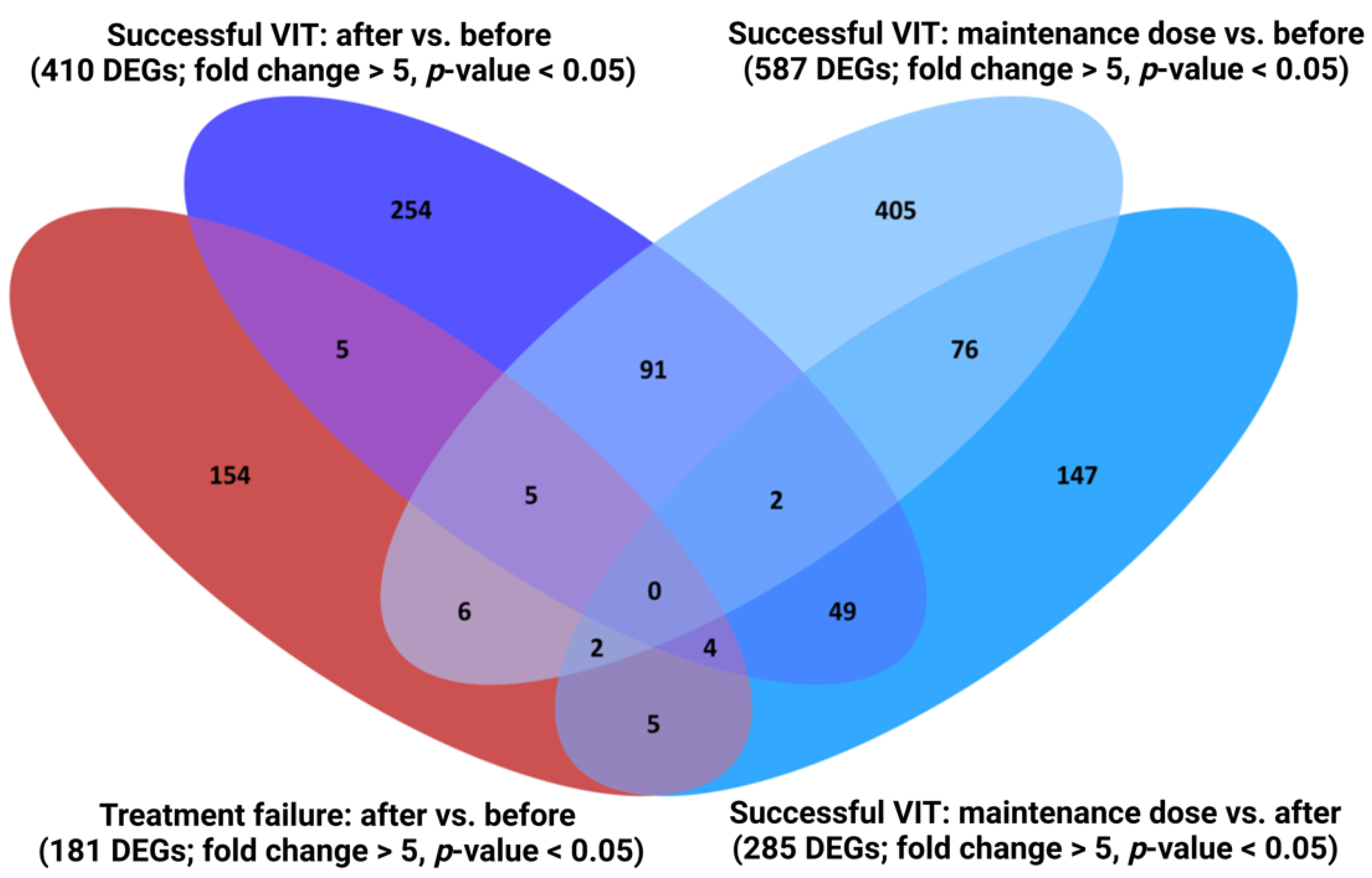
2.1. Characterisation of Transcriptome Fingerprint Characteristic of Successful VIT
2.2. The Immune System Seems to Be Activated after Reaching the Maintenance Dose and, Conversely, Significantly Suppressed after Finishing VIT
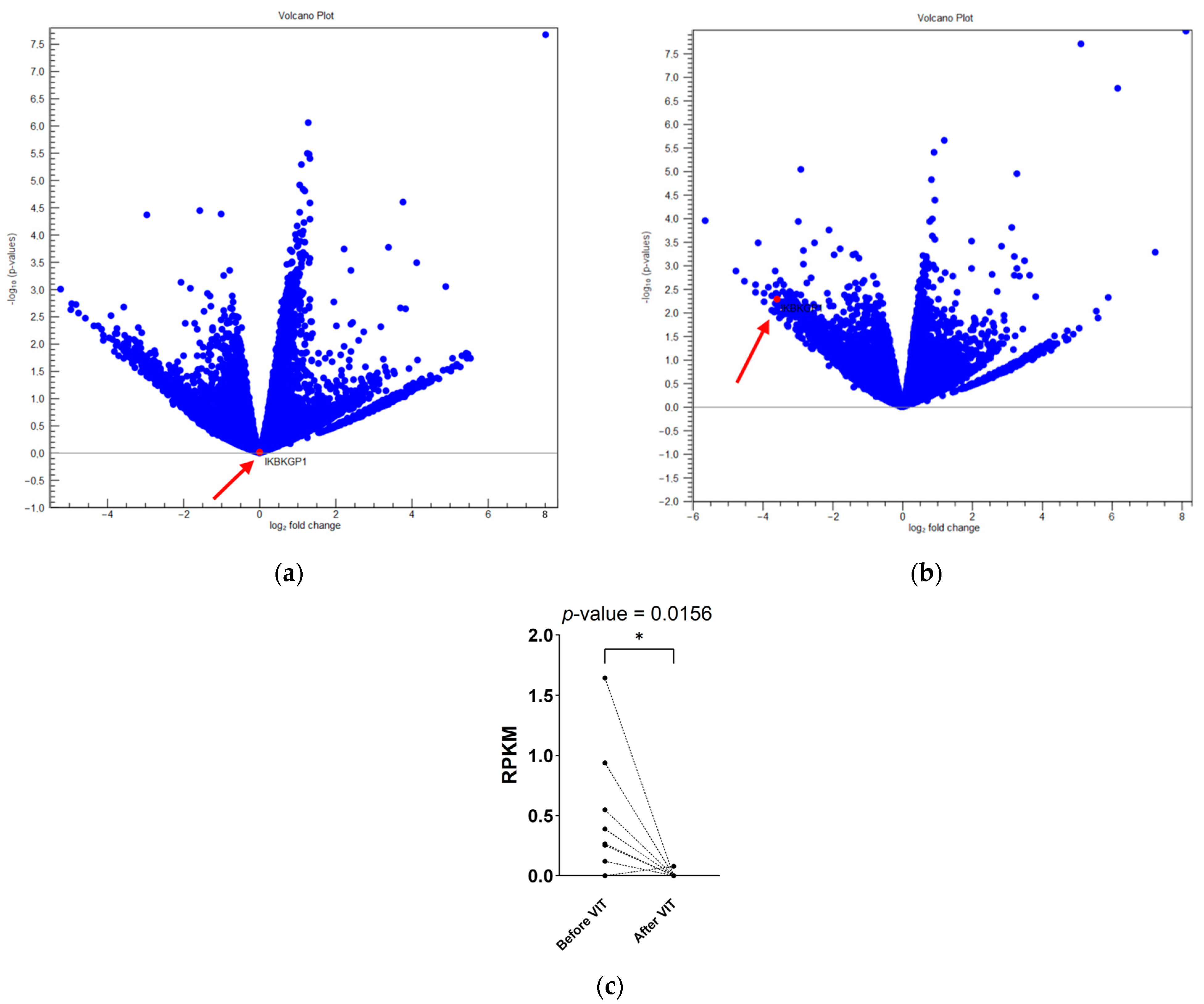
2.3. NFκB Pathway Inhibition and Downregulation of DUX4 Transcripts Are Important for Early Protection and Tolerance Induction
2.4. Th2- and Th17-Related DEGs Are Significantly Downregulated after Successful Immunotherapy
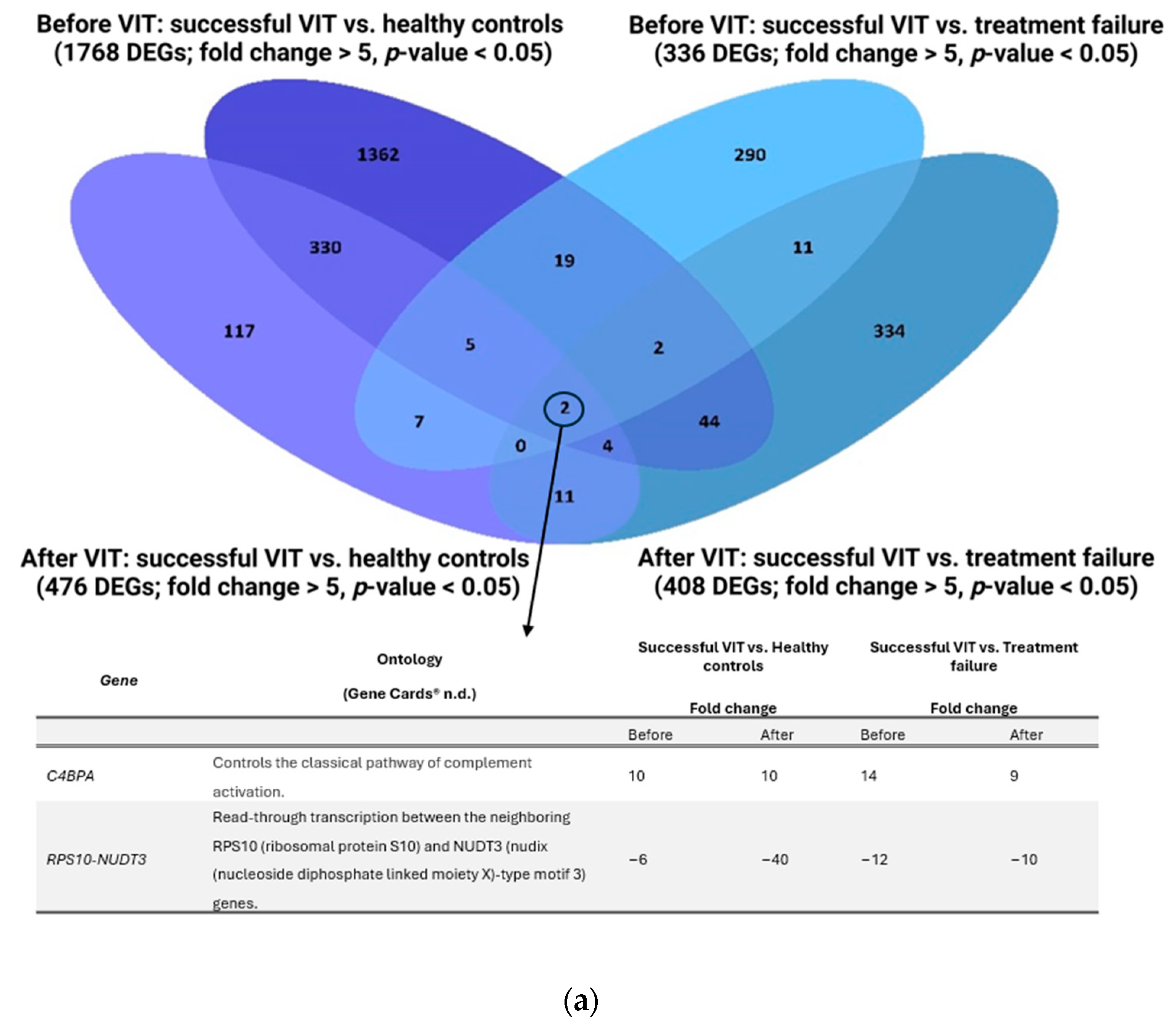
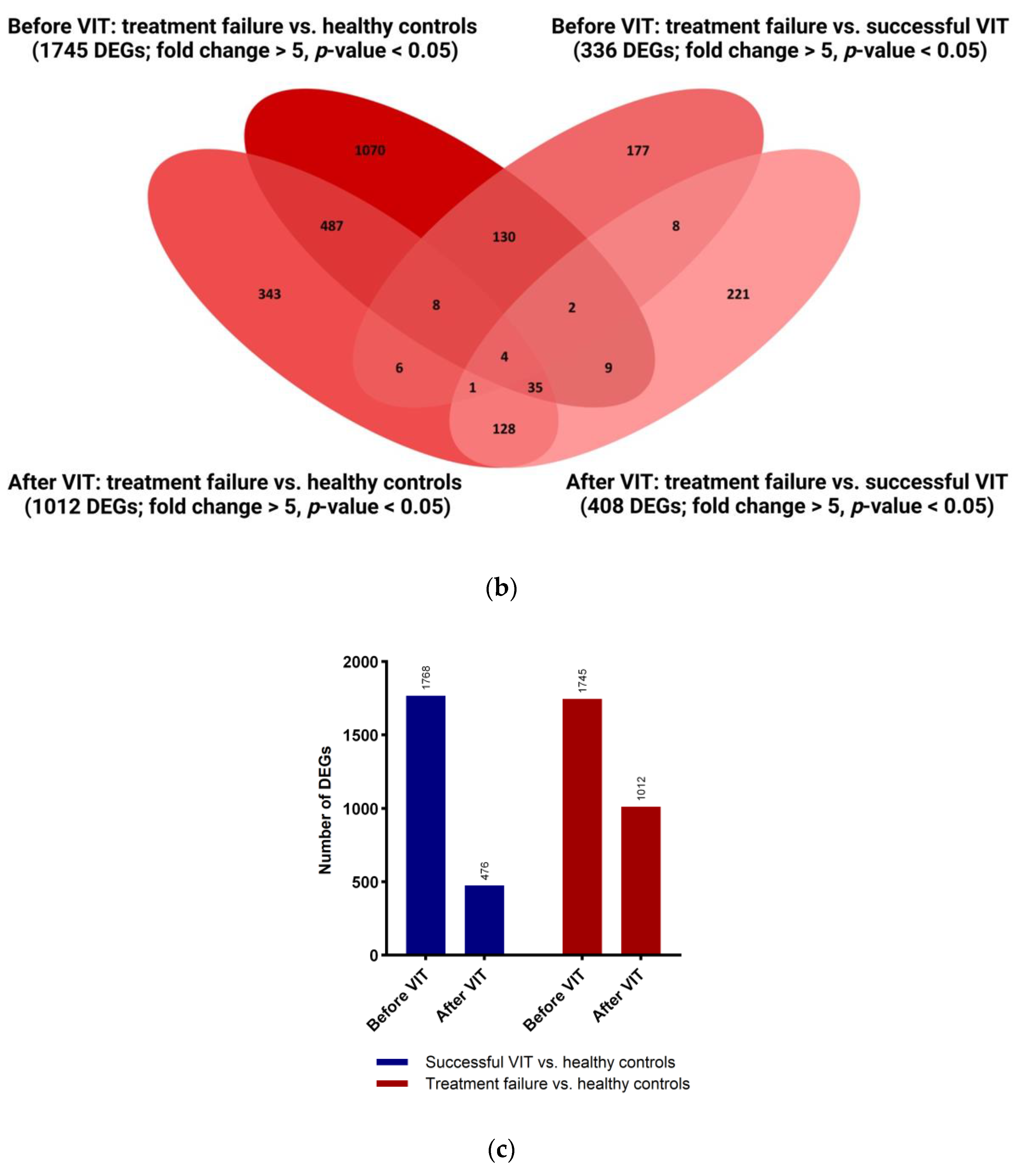
2.5. Successful Venom Immunotherapy Seems to Restore the Balance of the Immune System to a State Similar to That of Healthy Individuals
2.6. Two Biomarkers Specific for Successful VIT, Regardless of the Time of Sampling, Are C4BPA and RPS10-NUDT3
2.7. Inhibition of Macrophage Alternative Signalling Pathway in Synergy with Inhibition of PPAR Pathway Results in Silencing of Th2 Immune Response Characteristic of Successful VIT
2.8. Protein–Protein Interaction before and after Venom Immunotherapy Identifies Different Master Regulators
2.9. Different Mechanisms after Sting Challenge in Groups with Successful VIT and Treatment Failure
3. Discussion
4. Materials and Methods
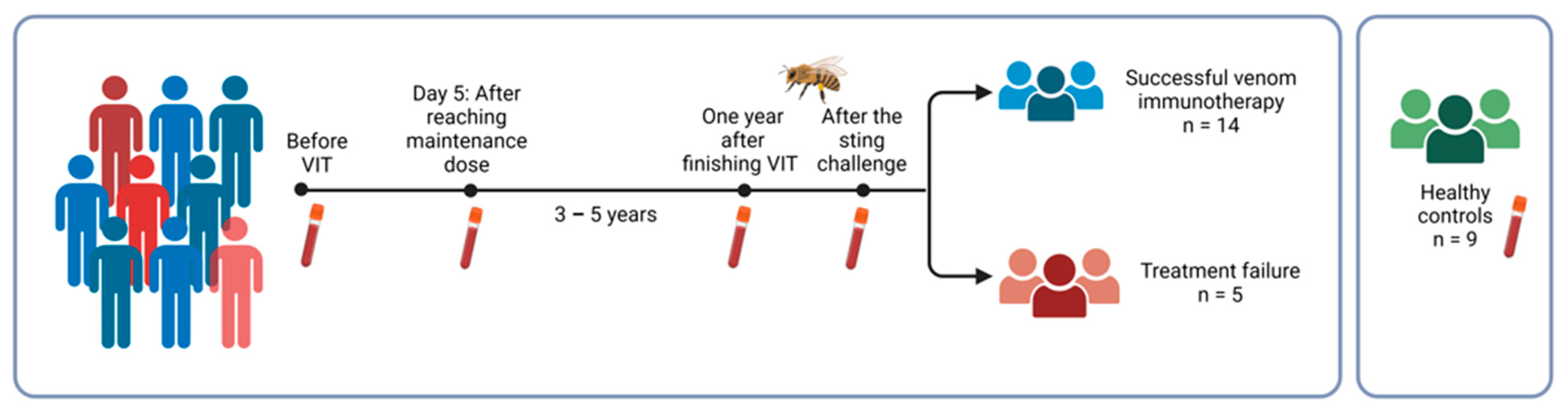
4.1. Study Group
4.2. Allergy Diagnostic Tests
4.3. RNA Samples
4.4. RNA Sequencing and Bioinformatics Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lydyard, P.; Whelan, A.; Fanger, M.W. Immunology, 2nd ed.; Garland Science/Bios Scientific Publishers: New York, NY, USA, 2004. [Google Scholar]
- Sturm, G.J.; Varga, E.; Roberts, G.; Mosbech, H.; Bilò, M.B.; Akdis, C.A.; Antolín-Amérigo, D.; Cichocka-Jarosz, E.; Gawlik, R.; Jakob, T.; et al. EAACI guidelines on allergen immunotherapy: Hymenoptera venom allergy. Allergy Eur. J. Allergy Clin. Immunol. 2018, 73, 744–764. [Google Scholar] [CrossRef]
- De Graaf, D.C.; Aerts, M.; Danneels, E.; Devreese, B. Bee, wasp and ant venomics pave the way for a component-resolved diagnosis of sting allergy. J. Proteom. 2009, 72, 145–154. [Google Scholar] [CrossRef]
- Biló, B.M.; Rueff, F.; Mosbech, H.; Bonifazi, F.; Oude-Elberink, J.N.G.; The EAACI Interest Group on Insect Venom Hypersensitivity. Diagnosis of Hymenoptera venom allergy. Allergy Eur. J. Allergy Clin. Immunol. 2005, 60, 1339–1349. [Google Scholar] [CrossRef]
- Bilo, B.; Bonifazi, F. Epidemiology of insect-venom anaphylaxis. Curr. Opin. Allergy Clin. Immunol. 2008, 8, 330–337. [Google Scholar] [CrossRef] [PubMed]
- Bilò, M.B. Anaphylaxis caused by Hymenoptera stings: From epidemiology to treatment. Allergy 2011, 66, 35–37. [Google Scholar] [CrossRef]
- Pühringer, V.; Jilma, B.; Herkner, H. Population-based incidence of all-cause anaphylaxis and its development over time: A systematic review and meta-analysis. Front. Allergy 2023, 4, 1249280. [Google Scholar] [CrossRef] [PubMed]
- Worm, M.; Monaret-Vautrin, A.; Scherer, K.; Lang, R. First European data from the network of severe allergic reactions (NORA). Eur. J. Allergy Clin. Immunol. 2014, 69, 1397–1404. [Google Scholar] [CrossRef] [PubMed]
- Rijavec, M.; Inkret, J.; Bidovec-stojkovi, U. Fatal Hymenoptera Venom—Triggered Anaphylaxis in Patients with Unrecognized Clonal Mast Cell Disorder—Is Mastocytosis to Blame? Int. J. Mol. Sci. 2023, 24, 16368. [Google Scholar] [CrossRef] [PubMed]
- Ruëff, F.; Vos, B.; Elberink, J.O.; Bender, A.; Chatelain, R.; Dugas-Breit, S.; Horny, H.; Küchenhoff, H.; Linhardt, A.; Mastnik, S.; et al. Predictors of clinical effectiveness of Hymenoptera venom immunotherapy. Clin. Exp. Allergy 2014, 44, 736–746. [Google Scholar] [CrossRef]
- Lang, R.; Hawranek, T. Hymenoptera Venom Immunotherapy and Field Stings. J. Investig. Allergol. Clin. Immunol. 2006, 16, 224. [Google Scholar]
- Jarkvist, J.; Salehi, C.; Akin, C.; Gülen, T. Venom immunotherapy in patients with clonal mast cell disorders: IgG4 correlates with protection. Allergy Eur. J. Allergy Clin. Immunol. 2020, 75, 169–177. [Google Scholar] [CrossRef]
- Kačar, M.; Rijavec, M.; Šelb, J.; Korošec, P. Clonal mast cell disorders and hereditary α-tryptasemia as risk factors for anaphylaxis. Clin. Exp. Allergy 2023, 53, 392–404. [Google Scholar] [CrossRef]
- Bonadonna, P.; Zanotti, R.; Pagani, M.; Bonifacio, M.; Scaffidi, L.; Olivieri, E.; Franchini, M.; Reccardini, F.; Costantino, M.T.; Roncallo, C.; et al. Anaphylactic Reactions After Discontinuation of Hymenoptera Venom Immunotherapy: A Clonal Mast Cell Disorder Should Be Suspected. J. Allergy Clin. Immunol. Pract. 2018, 6, 1368–1372. [Google Scholar] [CrossRef] [PubMed]
- Shamji, M.H.; Durham, S.R. Mechanisms of allergen immunotherapy for inhaled allergens and predictive biomarkers. J. Allergy Clin. Immunol. 2017, 140, 1485–1498. [Google Scholar] [CrossRef] [PubMed]
- Möbs, C.; Müller, J.; Rudzio, A.; Pickert, J.; Blank, S.; Jakob, T.; Spillner, E.P.W. Decline of Ves v 5-specific blocking capacity in wasp venom-allergic patients after stopping allergen immunotherapy. Allergy 2015, 70, 715–719. [Google Scholar] [CrossRef] [PubMed]
- Eržen, R.; Košnik, M.; Silar, M.K.P. Basophil response and the induction of a tolerance in venom immunotherapy: A long-term sting challenge study. Allergy 2012, 67, 822–830. [Google Scholar] [CrossRef] [PubMed]
- Žitnik, S.E.; Vesel, T.; Avčin, T.; Šilar, M.; Košnik, M.; Korošec, P. Monitoring honeybee venom immunotherapy in children with the basophil activation test. Pediatr. Allergy Immunol. 2012, 23, 166–172. [Google Scholar] [CrossRef]
- Čelesnik, N.; Vesel, T.; Rijavec, M.; Šilar, M.; Eržen, R.; Košnik, M.; Žitnik, S.E.K.; Avčin, T.; Korošec, P. Short-term venom immunotherapy induces desensitization of FcεRI-mediated basophil response. Allergy Eur. J. Allergy Clin. Immunol. 2012, 67, 1594–1600. [Google Scholar] [CrossRef]
- Foschi, F.G.; Emiliani, F.; Savini, S.; Quercia, O.; Stefanini, G.F. CD30 serum levels and response to Hymenoptera venom immunotherapy. J. Investig. Allergol. Clin. Immunol. 2008, 18, 279–283. [Google Scholar]
- Packi, K.; Matysiak, J.; Matuszewska, E.; Bręborowicz, A.; Kycler, Z.; Matysiak, J. New biomarkers of hymenoptera venom allergy in a group of inflammation factors. Int. J. Environ. Res. Public Health 2021, 18, 4011. [Google Scholar] [CrossRef]
- Frick, M.; Fischer, J.; Helbling, A.; Ruëff, F.; Wieczorek, D.; Ollert, M.; Pfützner, W.; Müller, S.; Huss-Marp, J.; Dorn, B.; et al. Predominant Api m 10 sensitization as risk factor for treatment failure in honey bee venom immunotherapy. J. Allergy Clin. Immunol. 2016, 138, 1663–1671.e9. [Google Scholar] [CrossRef]
- Jakob, T.; Rauber, M.M.; Perez-Riverol, A.; Spillner, E.; Blank, S. The Honeybee Venom Major Allergen Api m 10 (Icarapin) and Its Role in Diagnostics and Treatment of Hymenoptera Venom Allergy. Curr. Allergy Asthma Rep. 2020, 20, 48. [Google Scholar] [CrossRef] [PubMed]
- Blank, S.; Jakwerth, C.A.; Zissler, U.M.; Schmidt-Weber, C.B. Molecular determination of insect venom allergies. Expert Rev. Mol. Diagn. 2022, 22, 983–986. [Google Scholar] [CrossRef] [PubMed]
- Demšar Luzar, A.; Korošec, P.; Košnik, M.; Zidarn, M.; Rijavec, M. Hymenoptera venom immunotherapy: Immune mechanisms of induced protection and tolerance. Cells 2021, 10, 1575. [Google Scholar] [CrossRef] [PubMed]
- Tsicopoulos, A. Venom immunotherapy modulates interleukin-4 and interferon-γ messenger RNA expression of peripheral T lymphocytes. Immunology 1996, 87, 593–598. [Google Scholar] [CrossRef]
- Konno, S.; Hizawa, N.; Nishimura, M.; Huang, S.K. Osteopontin: A potential biomarker for successful bee venom immunotherapy and a potential molecule for inhibiting IgE-mediated allergic responses. Allergol. Int. 2006, 55, 355–359. [Google Scholar] [CrossRef][Green Version]
- Konno, S.; Golden, D.B.K.; Schroeder, J.; Hamilton, R.G.; Lichtenstein, L.M.; Huang, S.K. Increased expression of osteopontin is associated with long-term bee venom immunotherapy. J. Allergy Clin. Immunol. 2005, 115, 1063–1067. [Google Scholar] [CrossRef]
- Niedoszytko, M.; Bruinenberg, M.; de Monchy, J.; Wijmenga, C.; Platteel, M.; Jassem, E.; Elberink, J.N.O. Gene expression analysis in predicting the effectiveness of insect venom immunotherapy. J. Allergy Clin. Immunol. 2010, 125, 1092–1097. [Google Scholar] [CrossRef]
- Niedoszytko, M.; Bruinenberg, M.; de Monchy, J.; Weersma, R.K.; Wijmenga, C.; Jassem, E.; Elberink, J.N.O. Changes in gene expression caused by insect venom immunotherapy responsible for the long-term protection of insect venom-allergic patients. Ann. Allergy Asthma Immunol. 2011, 106, 502–510. [Google Scholar] [CrossRef] [PubMed]
- Karpinski, P.; Skiba, P.; Kosinska, M.; Rosiek-Biegus, M.; Królewicz, E.; Blin, N.; Meese, E.; Panaszek, B.; Nittner-Marszalska, M.; Sasiadek, M.M. Genome-wide analysis of gene expression after one year of venom immunotherapy. Immunol. Lett. 2018, 204, 23–28. [Google Scholar] [CrossRef]
- McKenzie, C.I.; Varese, N.; Aui, P.M.; Reinwald, S.; Wines, B.D.; Hogarth, P.M.; Thien, F.; Hew, M.; Rolland, J.M.; O’Hehir, R.E.; et al. RNA sequencing of single allergen-specific memory B cells after grass pollen immunotherapy: Two unique cell fates and CD29 as a biomarker for treatment effect. Allergy Eur. J. Allergy Clin. Immunol. 2023, 78, 822–835. [Google Scholar] [CrossRef]
- Chao, W.; Liao, W.; Wang, J.; Sung, H.; Chen, H.; Lin, F.; Huang, J.; Liu, K.; Ko, T. Single-cell profiling reveals novel cellular heterogeneity of monocytes during Hymenoptera venom allergy. Clin. Transl. Allergy 2022, 12, e12151. [Google Scholar] [CrossRef]
- Layhadi, J.A.; Moya, R.; Tan, T.J.; Lenormand, M.M.; Sharif, H.; Parkin, R.V.; Vila-Nadal, G.; Fedina, O.; Zhu, R.; Laisuan, W.; et al. Single-cell RNA sequencing identifies precise tolerogenic cellular and molecular pathways induced by depigmented-polymerized grass pollen allergen extract. J. Allergy Clin. Immunol. 2023, 151, 1357–1370.e9. [Google Scholar] [CrossRef] [PubMed]
- Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. Table 22-1 Blood Cells. In Molecular Biology of the Cell, 4th ed.; Garland Science: New York, NY, USA, 2002. [Google Scholar]
- Sivaprasad, U.; Kinker, K.G.; Ericksen, M.B.; Lindsey, M.; Gibson, A.M.; Bass, S.A.; Hershey, N.S.; Deng, J.; Medvedovic, M.; Hershey, G.K.K. SERPINB3/B4 Contributes to Early Inflammation and Barrier Dysfunction in an Experimental Murine Model of Atopic Dermatitis. J. Investig. Dermatol. 2015, 135, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Howell, W.M.; Carter, V.; Clark, B. The HLA system: Immunobiology, HLA typing, antibody screening and crossmatching techniques. J. Clin. Pathol. 2010, 63, 387–390. [Google Scholar] [CrossRef]
- Gene Cards®. The Human Gene Database n.d. Available online: https://www.genecards.org/ (accessed on 23 January 2024).
- Mocciaro, E.; Runfola, V.; Ghezzi, P.; Pannese, M.; Gabellini, D. DUX4 role in normal physiology and in FSHD muscular dystrophy. Cells 2021, 10, 3322. [Google Scholar] [CrossRef] [PubMed]
- Dmitriev, P.; Kiseleva, E.; Kharchenko, O.; Ivashkin, E.; Pichugin, A.; Dessen, P.; Robert, T.; Coppée, F.; Belayew, A.; Carnac, G.; et al. Dux4 controls migration of mesenchymal stem cells through the Cxcr4-Sdf1 axis. Oncotarget 2016, 7, 65090–65108. [Google Scholar] [CrossRef][Green Version]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.C. NF-κB signaling in inflammation. Signal Transduct. Target. Ther. 2017, 2, 17023. [Google Scholar] [CrossRef]
- Hayden, M.S.; Ghosh, S. NF-κB, the first quarter-century: Remarkable progress and outstanding questions. Genes Dev. 2012, 26, 203–234. [Google Scholar] [CrossRef]
- Gustavsson, E.K.; Sethi, S.; Gao, Y.; Brenton, J.W.; García-Ruiz, S.; Zhang, D.; Garza, R.; Reynolds, R.H.; Evans, J.R.; Chen, Z.; et al. The annotation and function of the Parkinson’s and Gaucher disease-linked gene GBA1 has been concealed by its protein-coding pseudogene GBAP1. BioRxiv 2023. [Google Scholar] [CrossRef]
- Pink, R.C.; Wicks, K.; Caley, D.P.; Punch, E.K.; Jacobs, L.; Carter, D.R.F. Pseudogenes: Pseudo-functional or key regulators in health and disease? RNA 2011, 17, 792–798. [Google Scholar] [CrossRef]
- Tan, L.; Qin, Y.; Xie, R.; Xia, T.; Duan, X.; Peng, L.; You, R.; Liu, Y.; Zou, X.; Zhang, M.; et al. Theranostics N6-methyladenosine-associated prognostic pseudogenes contribute to predicting immunotherapy benefits and therapeutic agents in head and neck squamous cell carcinoma. Theranostics 2022, 12, 7267–7288. [Google Scholar] [CrossRef] [PubMed]
- Kempiński, K.; Romantowski, J.; Maciejewska, A.; Pawłowski, R.; Chełmińska, M.; Jassem, E.; Niedoszytko, M. COMMD8 changes expression during initial phase of wasp venom immunotherapy. J. Gene Med. 2020, 22, e3243. [Google Scholar] [CrossRef] [PubMed]
- Starokadomskyy, P.; Gluck, N.; Li, H.; Chen, B.; Wallis, M.; Maine, G.N.; Mao, X.; Zaidi, I.W.; Hein, M.Y.; McDonald, F.J.; et al. CCDC22 deficiency in humans blunts activation of proinfammatory NF-κB signaling. J. Clin. Investig. 2013, 123, 2244–2256. [Google Scholar] [CrossRef] [PubMed]
- Ande, S.R.; Xu, Y.X.Z.; Mishra, S. Prohibitin: A potential therapeutic target in tyrosine kinase signaling. Signal Transduct. Target. Ther. 2017, 2, 17059. [Google Scholar] [CrossRef] [PubMed]
- Werner, L.M.; Criss, A.K. Diverse Functions of C4b-Binding Protein in Health and Disease. J. Immunol. 2023, 211, 1443–1449. [Google Scholar] [CrossRef] [PubMed]
- Sjef Verbeek, J.; Hirose, S.; Nishimura, H. The complex association of fcγriib with autoimmune susceptibility. Front. Immunol. 2019, 10, 2061. [Google Scholar] [CrossRef]
- Phillips, N.E.; Parker, D.C. Cross-linking of B lymphocyte Fc gamma receptors and membrane immunoglobulin inhibits anti-immunoglobulin-induced blastogenesis. J. Immunol. 1984, 132, 627–632. [Google Scholar] [CrossRef]
- Varma, R.; Puri, N. Dampening of mast cell secondary responses to allergen involves specific signalling and epigenetic changes. Cell Immunol. 2019, 344, 103944. [Google Scholar] [CrossRef]
- Odegaard, J.I.; Chawla, A. Alternative Macrophage Activation and Metabolism. Annu. Rev. Pathol. 2011, 6, 275–297. [Google Scholar] [CrossRef]
- Nobs, S.P.; Natali, S.; Pohlmeier, L.; Okreglicka, K.; Schneider, C.; Kurrer, M.; Sallusto, F.; Kopf, M. PPARγ in dendritic cells and T cells drives pathogenic type-2 effector responses in lung inflammation. J. Exp. Med. 2017, 214, 3015–3035. [Google Scholar] [CrossRef]
- Chen, T.; Tibbitt, C.A.; Feng, X.; Stark, J.M.; Rohrbeck, L.; Rausch, L.; Sedimbi, S.K.; Karlsson, M.C.I.; Lambrecht, B.N.; Hedestam, G.B.K.; et al. PPAR-promotes type 2 immune responses in allergy and nematode infection. Sci. Immunol. 2017, 2, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Micossé, C.; von Meyenn, L.; Steck, O.; Kipfer, E.; Adam, C.; Simillion, C.; Jafari, S.M.S.; Olah, P.; Yawlkar, N.; Simon, D.; et al. Human “TH9” cells are a subpopulation of PPAR-+ TH2 cells. Sci. Immunol. 2019, 4, 1–14. [Google Scholar] [CrossRef]
- McHugh, S.M.; Deighton, J.; Stewart, A.G.; Lachmann, P.J.; Ewan, P.W. Bee venom immunotherapy induces a shift in cytokine responses from a TH-2 to a TH-1 dominant pattern: Comparison of rush and conventional immunotherapy. Clin. Exp. Allergy 1995, 25, 828–838. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Zhang, W.; Xiang, R.; Tan, L.; Liu, P.; Tao, Z.; Deng, Y.; Tong, H.; Xu, Y. The early-phase transcriptome and the clinical efficacy analysis in three modes of subcutaneous immunotherapy for allergic rhinitis. World Allergy Organ. J. 2023, 16, 100811. [Google Scholar] [CrossRef] [PubMed]
- Karisola, P.; Palosuo, K.; Hinkkanen, V.; Wisgrill, L.; Savinko, T.; Fyhrquist, N.; Alenius, H.; Mäkelä, M.J. Integrative Transcriptomics Reveals Activation of Innate Immune Responses and Inhibition of Inflammation during Oral Immunotherapy for Egg Allergy in Children. Front. Immunol. 2021, 12, 704633. [Google Scholar] [CrossRef]
- Gruzelle, V.; Mailhol, C.; Waters, D.W.; Guilleminault, L. Clinical utility of rush venom immunotherapy: Current status. J. Asthma Allergy 2020, 13, 1–10. [Google Scholar] [CrossRef]
- Pospischil, I.M.; Kagerer, M.; Cozzio, A.; Angelova-Fischer, I.; Guenova, E.; Ballmer-Weber, B.; Hoetzenecker, W. Comparison of the Safety Profiles of 3 Different Hymenoptera Venom Immunotherapy Protocols: A Retrospective 2-Center Study of 143 Patients. Int. Arch. Allergy Immunol. 2020, 181, 783–789. [Google Scholar] [CrossRef]
- CLC Genomics Workbench 23.0.2 Online Manual. Available online: https://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/2302/index.php?manual=Introduction_CLC_Genomics_Workbench.html (accessed on 14 December 2023).
- Aken, B.L.; Ayling, S.; Barrell, D.; Clarke, L.; Curwen, V.; Fairley, S.; Banet, J.F.; Billis, K.; Girón, C.G.; Hourlier, T.; et al. The Ensembl gene annotation system. Database 2016, 2016, baw093. [Google Scholar] [CrossRef]
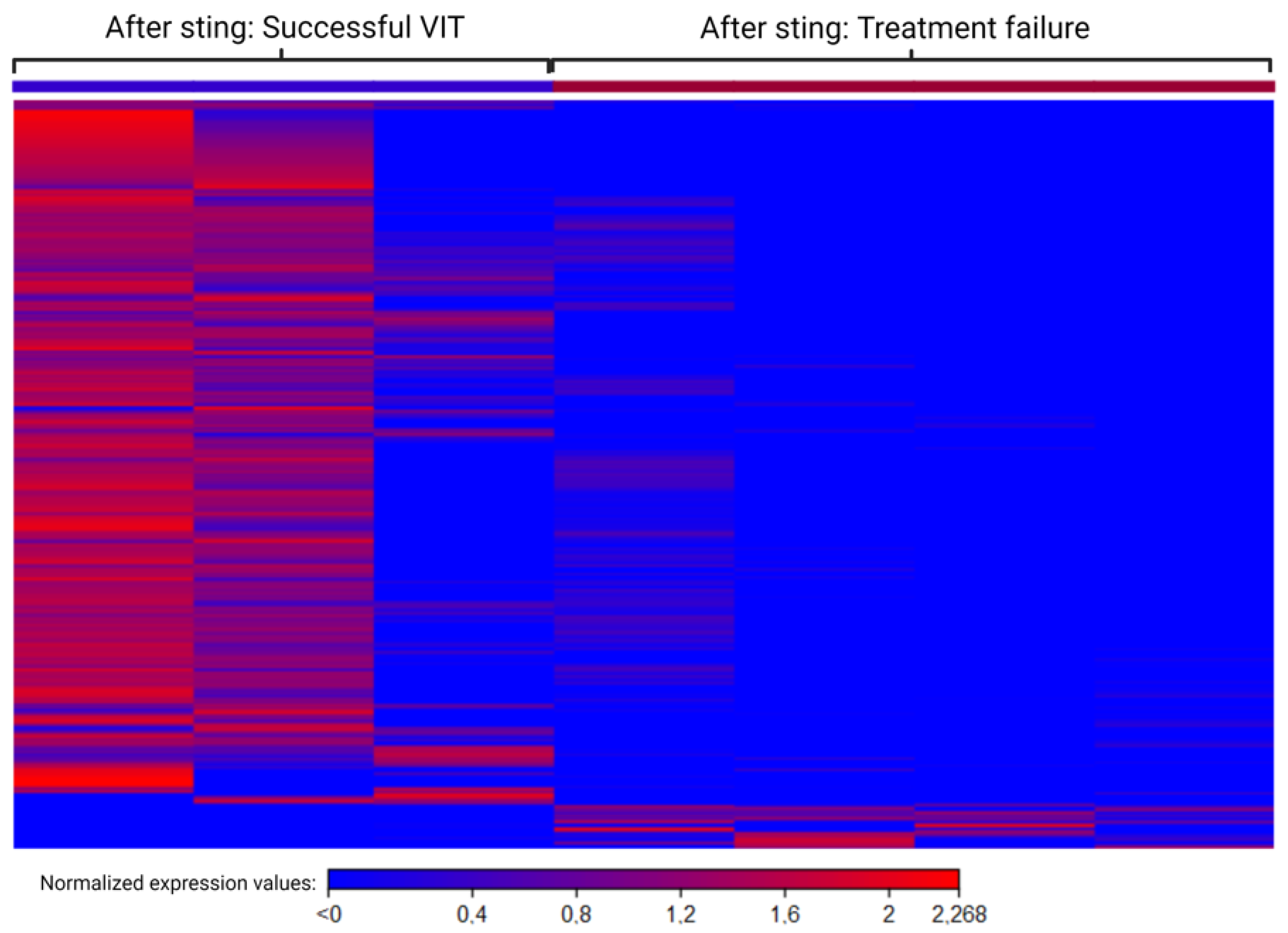
| Gene | Ontology [38] | Maintenance Dose vs. Before | Maintenance Dose vs. After | ||
|---|---|---|---|---|---|
| Fold Change | p-Value | Fold Change | p-Value | ||
| IGLV2–5 | Immunoglobulin Lambda Variable 2–5 | 6.4 | 0.04 | 6.5 | 0.03 |
| HLA-U | Major Histocompatibility Complex, Class I, U | 6.5 | 0.04 | 6.6 | 0.03 |
| SERPINB4 | May act as a protease inhibitor to modulate the host immune response against tumour cells | 7.2 | 0.02 | 7.7 | 0.01 |
| Gene | Ontology [38] | Maintenance Dose vs. Before | After vs. Before | ||
|---|---|---|---|---|---|
| Fold Change | p-Value | Fold Change | p-Value | ||
| DUX4L10 | Involvement in inflammation, cellular migration, and chemotaxis | −240.1 | 6.46 × 10−7 | −7.3 | 0.0009 |
| DUX4L12 | Involvement in inflammation, cellular migration, and chemotaxis | −90.1 | 5.48 × 10−5 | −6.5 | 0.002 |
| CFL1P2 | Required for the upregulation of the atypical chemokine receptor ACKR2 | −9.9 | 0.02 | −8.3 | 0.02 |
| HTN3 | They function as antimicrobial peptides and are important components of the innate immune system | −7.6 | 0.009 | −11.7 | 0.002 |
| CDH9 | Cadherins are calcium-dependent cell adhesion proteins | −7.0 | 0.008 | −5.6 | 0.01 |
| PCDHB8 | Calcium-dependent cell adhesion protein involved in cell self-recognition and non-self discrimination | −6.9 | 0.01 | −8.8 | 0.002 |
| USP17L22 | Regulates different cellular processes, which may include cell proliferation, progression through the cell cycle, apoptosis, cell migration, and the cellular response to viral infection | −6.5 | 0.01 | −13 | 0.004 |
| IKBKGP1 | The NF-kappa-B essential modulator (also known as NEMO) regulating the NFкB pathway, which is involved in many immune and inflammatory responses | −5.5 | 0.01 | −24.9 | 0.00001 |
| MAPK15 | Regulates several processes, such as autophagy, protein trafficking, and genome integrity, in a kinase activity-dependent manner | −5 | 0.03 | −5.6 | 0.01 |
| Gene | Ontology [38] | Fold Change | p-Value |
|---|---|---|---|
| PHBP6 | Interacts with STAT3 to affect IL17 secretion in T-helper Th17 cells | −24 | 0.001 |
| DUX4 | Involvement in inflammation, cellular migration, and chemotaxis | −18.5 | 0.0007 |
| ZG16 | Predicted to act upstream of or in defence response to Gram-positive bacteria | −12 | 0.007 |
| CACNG5 | Involved in TCR signalling | −10.4 | 0.01 |
| CPN1 | Among its related pathways are the complement cascade and innate immune system | −10 | 0.011 |
| TMEM196 | Predicted to be an integral component of the membrane. Diseases associated with TMEM196 include Adult-Onset Severe Asthma | −9 | 0.002 |
| IGSF21 | Proteins in this superfamily are usually found on or in cell membranes and act as receptors in immune response pathways | −9 | 0.007 |
| DUX4L14 | Involvement in inflammation, cellular migration, and chemotaxis | −8.3 | 0.03 |
| TDO2 | This gene encodes a heme enzyme that plays a critical role in tryptophan metabolism | −8 | 0.004 |
| MMD2 | Monocyte To Macrophage Differentiation Associated 2; annotations related to this gene include protein kinase activity | −8 | 0.026 |
| TWIST1 | Represses expression of pro-inflammatory cytokines such as TNFA and IL1B | −7 | 0.014 |
| DUX4L2 | Involvement in inflammation, cellular migration, and chemotaxis | −6.5 | 0.01 |
| UMOD | May serve as a receptor for the binding and endocytosis of cytokines (IL-1, IL-2) and TNF | −6 | 0.043 |
| DPT | Dermatopontin is postulated to modify the behaviour of TGF-beta through interaction with decorin. Enhances TGFB1 activity. | −6 | 0.014 |
| SLC17A2 | Predicted to be located in the membrane. Predicted to be active in the lysosome. | −6 | 0.018 |
| GNB3 | Related to chronic rhinosinusitis. | −6 | 0.006 |
| PLA2G10 | This gene encodes a member of the phospholipase A2 family of proteins. This enzyme plays a role in the production of various inflammatory lipid mediators, such as prostaglandins. | −5 | 0.016 |
| NLRP8 | NLRP genes play roles in the mammalian innate immune system through inflammasome formation and the activation of caspases | −5 | 0.042 |
| EBI3 | It encodes a secreted glycoprotein and forms interleukin 27 (IL-27). IL-27 partially regulates T-cell and inflammatory responses by activating the Jak/STAT pathway of CD4+ T cells. Among its related pathways are IL27-mediated signalling events and interleukin-12 family signalling | −5 | 0.040 |
| PCDHA9 | Potential calcium-dependent cell adhesion protein. It may be involved in establishing and maintaining specific neuronal connections in the brain | −5 | 0.013 |
| IGKV2D-28 | The V region of the variable domain of immunoglobulin light chains that participate in the antigen recognition | 6 | 0.002 |
| E2F4P1 | It plays a vital role in suppressing proliferation-associated genes, and its gene mutation and increased expression may be associated with human cancer | 7 | 0.042 |
| Patients on VIT | Healthy Controls | p-Value | ||
|---|---|---|---|---|
| Successful VIT | Unsuccessful VIT | |||
| No. of patients | 14 | 5 | 9 | / |
| Age [years], median (range) | 50 (27–63) | 49 (26–59) | 47 (24–58) | ns 1 |
| Sex, no. of patients (%) Female Male | 3 (21%) 11 (79%) | 0 5 (100%) | 2 (22%) 7 (78%) | ns |
| Specific honeybee IgE [kIU/L], median (range) 2 | 5.48 (0.45–52.40) | 3.46 (0.82–11.4) | / | ns |
| Double sensitisation; honeybee and wasp sIgE, no. of patients (%) 2 | 10 (71%) | 1 (20%) | / | ns |
| Total IgE [IU/mL], median (range) 2 | 113 (3–661) | 32 (8–297) | / | ns |
| Years of VIT, median (range) | 6 (6–7) | 7 (6–7) | / | ns |
| Complications during VIT, n (%) | 1 (7%) | 5 (100%) | / | 0.0005 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Demšar Luzar, A.; Korošec, P.; Košnik, M.; Zidarn, M.; Rijavec, M. Blood Transcriptomics Identifies Multiple Gene Expression Pathways Associated with the Clinical Efficacy of Hymenoptera Venom Immunotherapy. Int. J. Mol. Sci. 2024, 25, 3499. https://doi.org/10.3390/ijms25063499
Demšar Luzar A, Korošec P, Košnik M, Zidarn M, Rijavec M. Blood Transcriptomics Identifies Multiple Gene Expression Pathways Associated with the Clinical Efficacy of Hymenoptera Venom Immunotherapy. International Journal of Molecular Sciences. 2024; 25(6):3499. https://doi.org/10.3390/ijms25063499
Chicago/Turabian StyleDemšar Luzar, Ajda, Peter Korošec, Mitja Košnik, Mihaela Zidarn, and Matija Rijavec. 2024. "Blood Transcriptomics Identifies Multiple Gene Expression Pathways Associated with the Clinical Efficacy of Hymenoptera Venom Immunotherapy" International Journal of Molecular Sciences 25, no. 6: 3499. https://doi.org/10.3390/ijms25063499
APA StyleDemšar Luzar, A., Korošec, P., Košnik, M., Zidarn, M., & Rijavec, M. (2024). Blood Transcriptomics Identifies Multiple Gene Expression Pathways Associated with the Clinical Efficacy of Hymenoptera Venom Immunotherapy. International Journal of Molecular Sciences, 25(6), 3499. https://doi.org/10.3390/ijms25063499








