Reference Genes Selection and Validation for Cinnamomum burmanni by Real-Time Quantitative Polymerase Chain Reaction
Abstract
1. Introduction
2. Results
2.1. Primer Specificity and Amplification Efficiency of Candidate Reference Genes
2.2. Expression Analysis of Candidate Reference Genes of C. burmannii
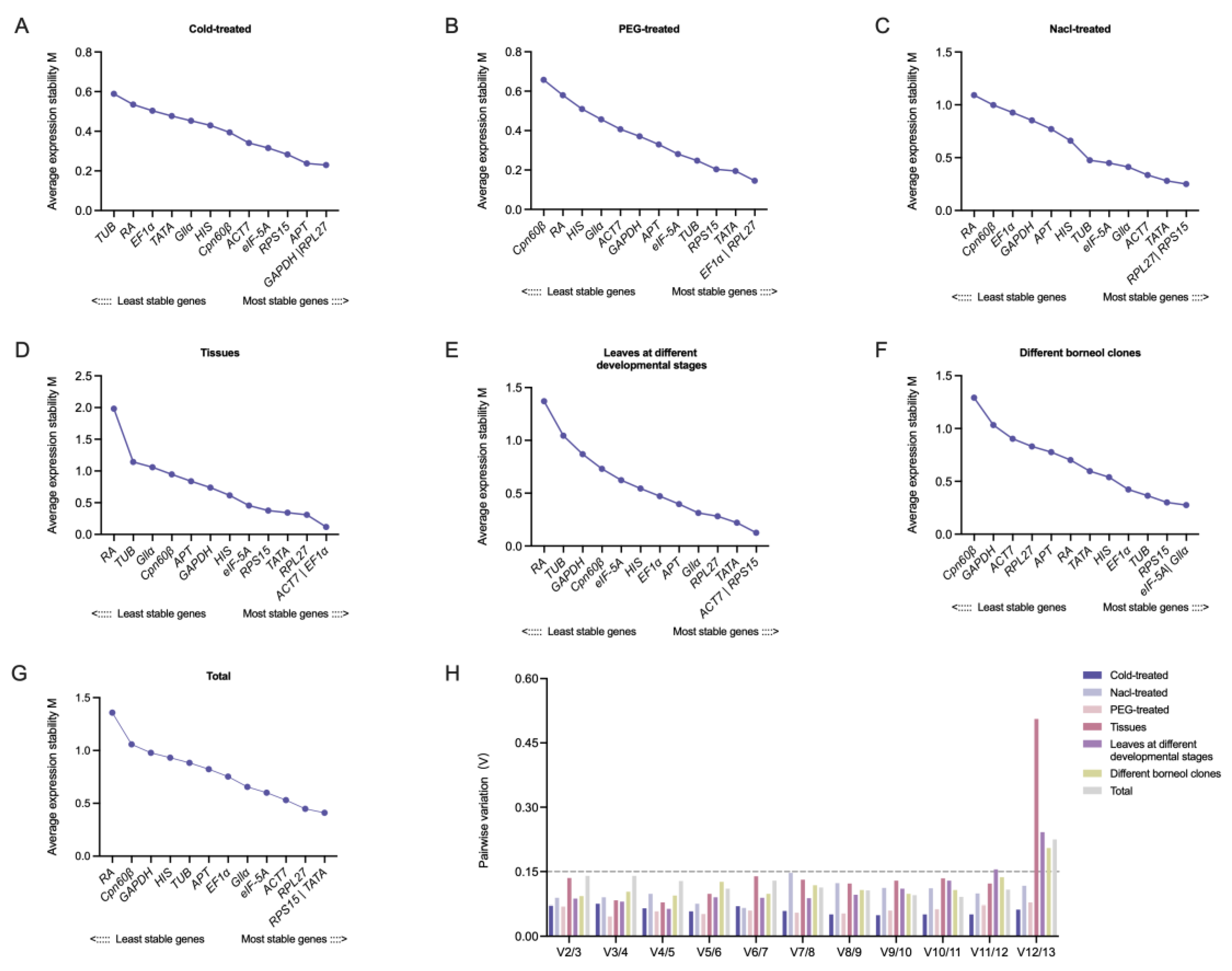
2.3. Gene Expression Stability Analysis
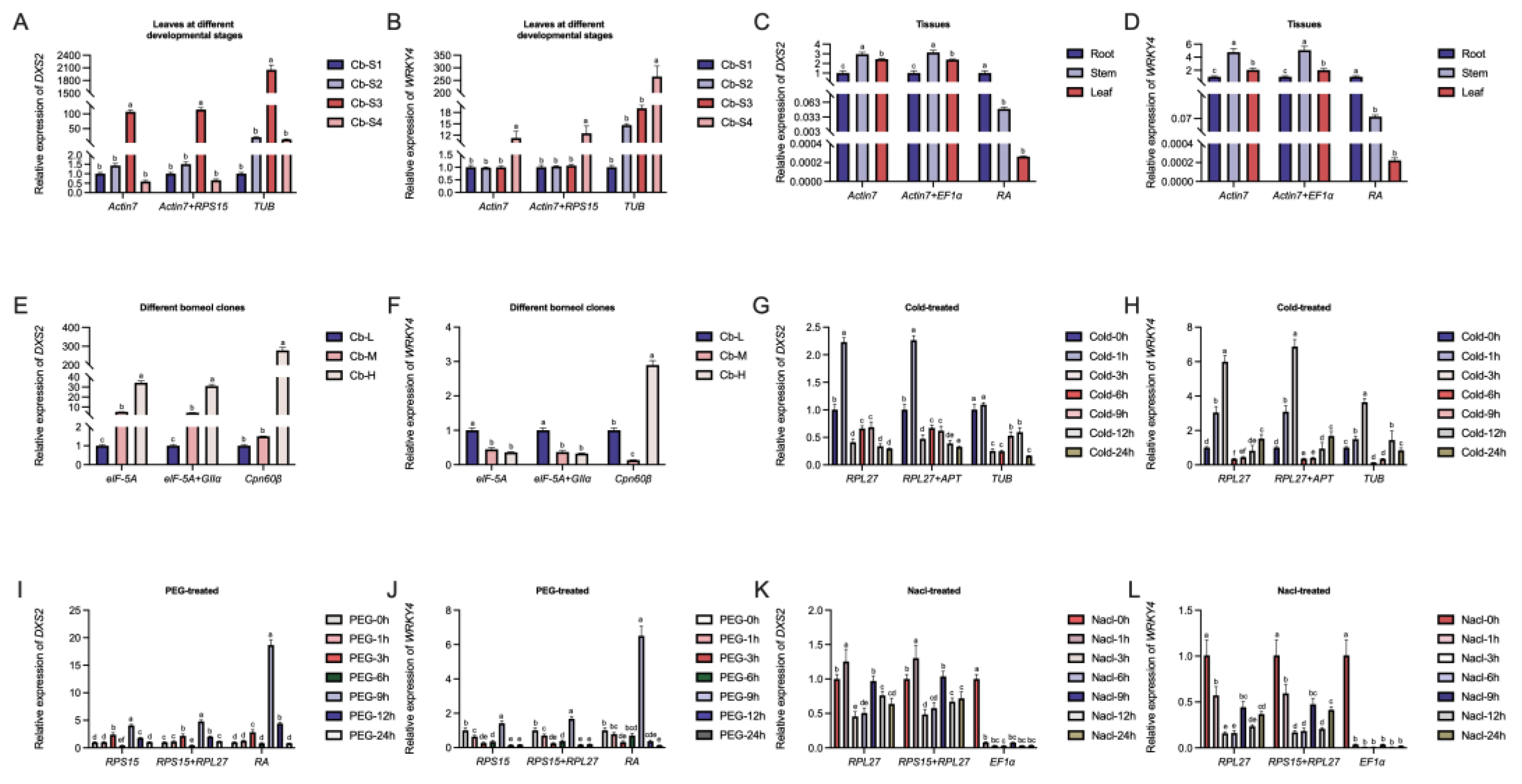
2.4. Reference Gene Validation
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. RNA Extraction and cDNA Synthesis
4.3. Candidate Reference Genes Selection and Primer Design
4.4. q-PCR Amplification
4.5. Data Analysis and Validation of Selected Reference Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ding, G.; Gao, Q.; Chen, J.; Zhao, J.; Zhang, G.; Liu, W. Validation of Potential Reference Genes for Real-Time qPCR Analysis in Pharaoh Ant, Monomorium pharaonis (Hymenoptera: Formicidae). Front. Physiol. 2022, 13, 852357. [Google Scholar] [CrossRef] [PubMed]
- Valifard, M.; Hir, R.L.; Müller, J.; Scheuring, D.; Neuhaus, H.E.; Pommerrenig, B. Vacuolar fructose transporter SWEET17 is critical for root development and drought tolerance. Plant Physiol. 2021, 187, 2716–2730. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Mao, Y.; Guo, Y.; Gao, J.; Liu, X.; Li, S.; Lin, Y.; Chen, H.; Wang, J.P.; Chiang, V.L. MYB transcription factor 161 mediates feedback regulation of Secondary wall-associated NAC-Domain 1 family genes for wood formation. Plant Physiol. 2020, 184, 1389–1406. [Google Scholar] [CrossRef] [PubMed]
- Gachon, C.; Mingam, A.; Charrier, B. Real-time PCR: What relevance to plant studies? J. Exp. Bot. 2004, 55, 1445–1454. [Google Scholar] [CrossRef] [PubMed]
- Udvardi, M.K.; Czechowski, T.; Scheible, W.R. Eleven golden rules of quantitative RT-PCR. Plant Cell 2008, 20, 1736–1737. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-Excel-Based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Zhang, K.; Li, M.; Cao, S.; Sun, Y.; Long, R.; Kang, J.; Yan, L.; Cui, H. Selection and validation of reference genes for target gene analysis with quantitative real-time PCR in the leaves and roots of Carex rigescens under abiotic stress. Ecotoxicol. Environ. Saf. 2019, 168, 127–137. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, S. Selection of the Reference Gene for Expression Normalization in Salsola ferganica under Abiotic Stress. Genes 2022, 13, 571. [Google Scholar] [CrossRef]
- Li, Z.; Lu, H.; He, Z.; Wang, C.; Wang, Y.; Ji, X. Selection of appropriate reference genes for quantitative real-time reverse transcription PCR in Betula platyphylla under salt and osmotic stress conditions. PLoS ONE 2019, 14, e0225926. [Google Scholar] [CrossRef]
- Belén, F.; Germán, L.; Lorenzo, L.; Raúl, C. Selection and optimization of reference genes for RT-qPCR normalization: A case study in Solanum lycopersicum exposed to UV-B. Plant Physiol. Biochem. 2021, 160, 269–280. [Google Scholar]
- Mo, Z.; Chen, Y.; Lou, W.; Jia, X.; Zhai, M.; Xuan, J.; Guo, Z.; Li, Y. Identification of suitable reference genes for normalization of real-time quantitative PCR data in pecan (Carya illinoinensis). Trees 2020, 34, 1233–1241. [Google Scholar] [CrossRef]
- Liu, X.; Liu, S.; Zhang, J.; Wu, Y.; Jia, X. Optimization of reference genes for qRT-PCR analysis of microRNA expression under abiotic stress conditions in sweetpotato. Plant Physiol. Biochem. 2020, 154, 379–386. [Google Scholar] [CrossRef]
- Jun, N.; Baoqing, Z.; Jian, C.; Peixue, L.; Libing, W.; Huitang, D.; Lin, Q.; Haiyan, Y.; Denglong, H.; Haiyan, Z. Selection of Reference Genes for Gene Expression Studies in Siberian Apricot (Prunus sibirica L.) Germplasm Using Quantitative Real-Time PCR. PLoS ONE 2014, 9, e103900. [Google Scholar]
- Palakolanu, S.R.; Dumbala, S.R.; Kaliamoorthy, S.; Pooja, B.M.; Vincent, V.; Sharma, K.K. Evaluation of Sorghum [Sorghum bicolor (L.)] Reference Genes in Various Tissues and under Abiotic Stress Conditions for Quantitative Real-Time PCR Data Normalization. Front. Plant Sci. 2016, 7, 529. [Google Scholar]
- Wang, J.J.; Han, S.; Yin, W.; Xia, X.; Liu, C. Comparison of Reliable Reference Genes Following Different Hormone Treatments by Various Algorithms for qRT-PCR Analysis of Metasequoia. Int. J. Mol. Sci. 2019, 20, 34. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, C.; Yang, H.; Lyu, L.; Li, W.; Wu, W. Selection and Validation of Candidate Reference Genes for Gene Expression Analysis by RT-qPCR inRubus. Int. J. Mol. Sci. 2021, 22, 10533. [Google Scholar] [CrossRef]
- Yang, Z.; Zhang, R.; Zhou, Z. Identification and Validation of Reference Genes for Gene Expression Analysis in Schima superba. Genes 2021, 12, 732. [Google Scholar] [CrossRef]
- Yao, J.; Zhu, G.; Liang, D.; He, B.; Wang, Y.; Cai, Y.; Zhang, Q. Reference Gene Selection for qPCR Analysis in Schima superba under Abiotic Stress. Genes 2022, 13, 1887. [Google Scholar] [CrossRef]
- Al-Dhubiab, B.E. Pharmaceutical applications and phytochemical profile of Cinnamomum burmannii. Pharmacogn. Rev. 2012, 6, 125–131. [Google Scholar] [CrossRef]
- Muhammad, D.; Lemarcq, V.; Alderweireldt, E.; Vanoverberghe, P.; Dewettinck, K. Antioxidant activity and quality attributes of white chocolate incorporated with Cinnamomum burmannii Blume essential oil. J. Food Sci. Technol. 2019, 57, 1731–1739. [Google Scholar] [CrossRef]
- Muhammad, D.R.A.; Tuenter, E.; Patria, G.D.; Foubert, K.; Pieters, L.; Dewettinck, K. Phytochemical composition and antioxidant activity of Cinnamomum burmannii Blume extracts and their potential application in white chocolate. Food Chem. 2021, 340, 127983. [Google Scholar] [CrossRef]
- Shan, B.; Cai, Y.; Brooks, J.; Corke, H. Antibacterial properties and major bioactive components of cinnamon stick (Cinnamomum burmannii): Activity against foodborne pathogenic bacteria. J. Agric. Food Chem. 2007, 55, 5484–5490. [Google Scholar] [CrossRef]
- Chen, L.; Su, J.; Li, L.; Li, B.; Li, W. A new source of natural D-borneol and its characteristic. J. Med. Plant Res. 2010, 5, 7. [Google Scholar]
- Ji, X.D.; Pu, Q.L.; Garraffo, H.M.; Pannell, L.K. Essential Oils of the Leaf, Bark and Branch of Cinnamomum buramannii Blume. J. Essent. Oil Res. 1991, 3, 373–375. [Google Scholar] [CrossRef]
- Liu, Z.; Li, H.; Cui, G.; Wei, M.; Ni, H. Efficient extraction of essential oil from Cinnamomum burmannii leaves using enzymolysis pretreatment and followed by microwave-assisted method. LWT- Food Sci. Technol. 2021, 147, 111497. [Google Scholar] [CrossRef]
- Wang, R. Extraction of essential oils from five cinnamon leaves and identification of their volatile compound compositions. Innov. Food Sci. Emerg. Technol. 2009, 10, 289–292. [Google Scholar] [CrossRef]
- Hou, C.; Zhang, Q.; Xie, P.; Lian, H.; Wang, Y.; Liang, D.; Cai, Y.; He, B. Full-length transcriptome sequencing reveals the molecular mechanism of monoterpene and sesquiterpene biosynthesis in Cinnamomum burmannii. Front. Genet. 2022, 13, 1087495. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Ma, R.; Su, P.; Jin, B.; Guo, J.; Tang, J.; Chen, T.; Zeng, W.; Lai, C.; Ling, F. Elucidation of the essential oil biosynthetic pathways in Cinnamomum burmannii through identification of six terpene synthases. Plant Sci. Int. J. Exp. Plant Biol. 2022, 317, 111203. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; An, W.; Liu, S.; Huang, Y.; Zheng, X. Mining of candidate genes involved in the biosynthesis of dextrorotatory borneol in Cinnamomum burmannii by transcriptomic analysis on three chemotypes. PeerJ 2020, 8, e9311. [Google Scholar] [CrossRef]
- Derveaux, S.; Vandesompele, J.; Hellemans, J. How to do successful gene expression analysis using real-time PCR. Methods A Companion Methods Enzymol. 2010, 50, 227–230. [Google Scholar] [CrossRef]
- Huggett, J.; Dheda, K.; Bustin, S.; Zumla, A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005, 6, 279–284. [Google Scholar] [CrossRef]
- Pan, X.; Li, Y.; Pan, G.; Yang, A. Bioinformatics study of 1-deoxy-D-xylulose-5-phosphate synthase (DXS) genes in Solanaceae. Mol. Biol. Rep. 2019, 46, 5175–5184. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.; Wan, W.; Yin, D.; Deng, X.; Ma, Z.; Gao, T.; Cao, X. Genome-wide analysis of WRKY transcription factor genes in Toona sinensis: An insight into evolutionary characteristics and terpene synthesis. Front. Plant Sci. 2022, 13, 1063850. [Google Scholar] [CrossRef] [PubMed]
- Muhlemann, J.K.; Klempien, A.; Dudareva, N. Floral volatiles: From biosynthesis to function. Plant Cell Environ. 2014, 37, 1936–1949. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Yang, J.; Qin, B.; Li, Y.; Sun, Y.; Su, S.; Xian, M. Biosynthesis of isoprene in Escherichia coli via methylerythritol phosphate (MEP) pathway. Appl. Microbiol. Biotechnol. 2011, 90, 1915. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.H.; Wang, J.W.; Wang, S.; Chen, W.X.Y. Characterization of GaWRKY1, a Cotton Transcription Factor That Regulates the Sesquiterpene Synthase Gene (+)-δ-Cadinene Synthase. Plant Physiol. 2004, 135, 507–515. [Google Scholar] [CrossRef]
- Ma, D.; Pu, G.; Lei, C.; Ma, L.; Wang, H.; Guo, Y.; Chen, J.; Du, Z.; Wang, H.; Li, G. Isolation and Characterization of AaWRKY1, an Artemisia annua Transcription Factor that Regulates the Amorpha-4,11-diene Synthase Gene, a Key Gene of Artemisinin Biosynthesis. Plant Cell Physiol. 2009, 50, 2146–2161. [Google Scholar] [CrossRef]
- Ye, M.; Gao, R.; Chen, S.; Wei, M.; Wang, J.; Zhang, B.; Wu, S.; Xu, Y.; Wu, P.; Chen, X.; et al. Downregulation of MEG3 and upregulation of EZH2 cooperatively promote neuroblastoma progression. J. Cell. Mol. Med. 2022, 26, 2377–2391. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.C.; Nadeau, K.; Abbasi, M.; Lachance, C.; Nguyen, M.; Fenrich, J. The Ultimate qPCR Experiment: Producing Publication Quality, Reproducible Data the First Time. Trends Biotechnol. 2019, 37, 761–774. [Google Scholar] [CrossRef]
- Kozera, B.; Rapacz, M. Reference genes in real-time PCR. J. Appl. Genet. 2013, 54, 391–406. [Google Scholar] [CrossRef]
- Liu, H.; Lu, Y.; Wang, X.; Wang, X.; Li, R.; Lu, C.; Lan, X.; Chen, Y. Selection and Validation of Reference Genes for RT-qPCR Analysis in Tibetan Medicinal Plant Saussurea Laniceps Callus under Abiotic Stresses and Hormone Treatments. Genes 2022, 13, 904. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Hu, S.; Cao, Y.; Chen, R.; Wang, Z.; Cao, X. Selection and evaluation of reference genes for qRT-PCR of Scutellaria baicalensis Georgi under different experimental conditions. Mol. Biol. Rep. 2021, 48, 1115–1126. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Mao, W.; Duan, Z.; Que, Q.; Li, P. Selection and validation of reference genes for measuring gene expression in Toona ciliata under different experimental conditions by quantitative real-time PCR analysis. BMC Plant Biol. 2020, 20, 450. [Google Scholar] [CrossRef] [PubMed]
- Silver, N.; Best, S.; Jiang, J.; Thein, S. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef]
- Vandesompele, J.; Preter, K.D.; Pattyn, F.; Poppe, B.; Roy, N.V.; Paepe, A.D.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Rntoft, T.F. Normalization of Real-Time Quantitative Reverse Transcription-PCR Data: A Model-Based Variance Estimation Approach to Identify Genes Suited for Normalization, Applied to Bladder and Colon Cancer Data Sets. Am. Assoc. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Ward, D.S.; Jutta, D.W.; Roswitha, W.; Valérie, S.; Hubert, S.; Daniel, N.; Martin, B.; Ralph, B.; Sabine, K.; Linos, V. Reference Gene Validation for RT-qPCR, a Note on Different Available Software Packages. PLoS ONE 2015, 10, e0122515. [Google Scholar]
- Chen, M.; Wang, B.; Li, Y.; Zeng, M.; Wen, Q. Reference Gene Selection for qRT-PCR Analyses of Luffa (Luffa cylindrica) Plants Under Abiotic Stress Conditions. Sci. Rep. 2021, 11, 3161. [Google Scholar] [CrossRef] [PubMed]
- Yi, S.; Lin, Q.; Zhang, X.; Wang, J.; Miao, Y.; Tan, N. Selection and Validation of Appropriate Reference Genes for Quantitative RT-PCR Analysis in Rubia yunnanensis Diels Based on Transcriptome Data. Biomed. Res. Int. 2020, 2020, 5824841. [Google Scholar] [CrossRef]
- Nicot, N.; Hausman, J.-F.; Hoffmann, L.; Evers, D. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J. Exp. Bot. 2005, 56, 2907–2914. [Google Scholar] [CrossRef] [PubMed]
- Chao, J.; Yang, S.; Chen, Y.; Tian, W. Evaluation of Reference Genes for Quantitative Real-Time PCR Analysis of the Gene Expression in Laticifers on the Basis of Latex Flow in Rubber Tree (Hevea brasiliensis Muell. Arg.). Front. Plant Sci. 2016, 7, 1149. [Google Scholar] [CrossRef]
- Li, G.; Zhou, Y.; Zhao, Y.; Liu, Y.; Ma, H. Internal Reference Gene Selection for Quantitative Real-Time RT-PCR Normalization in Potato Tissues. Phyton 2020, 89, 329–344. [Google Scholar] [CrossRef]
- Yin, J.; Sun, L.; Zhang, Q.; Cao, C. Screening and evaluation of the stability of expression of reference genes in Lymantria dispar (Lepidoptera: Erebidae) using qRT-PCR. Gene 2020, 749, 144712. [Google Scholar] [CrossRef]
- Hunter, T.; Garrels, J.I. Characterization of the mRNAs for alpha-, beta- and gamma-actin. Cell 1977, 12, 767–781. [Google Scholar] [CrossRef]
- Wang, B.; Du, H.; Yao, Z.; Ren, C.; Ma, L.; Wang, J.; Zhang, H.; Ma, H. Validation of reference genes for accurate normalization of gene expression with quantitative real-time PCR in Haloxylon ammodendron under different abiotic stresses. Physiol. Mol. Biol. Plants 2018, 24, 455. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Han, F.; Yang, L.; Wang, J.; Jin, F.; Luo, A.; Zhao, F. Identification of Reference Genes for RT-qPCR Analysis in Gleditsia microphylla under Abiotic Stress and Hormone Treatment. Genes 2022, 13, 1227. [Google Scholar] [CrossRef]
- Yi, S.; Lu, H.; Tian, C.; Xu, T.; Song, C.; Wang, W.; Wei, P.; Gu, F.; Liu, D.; Cai, Y.; et al. Selection of Suitable Reference Genes for Gene Expression Normalization Studies in Dendrobium huoshanense. Genes 2022, 13, 1486. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Mao, Y.; Huang, S.; Ni, J.; Lu, W.; Hou, J.; Wang, Y.; Zhao, W.; Li, M.; Wang, Q.; et al. Selection of Suitable Reference Genes for Quantitative Real-time PCR in Sapium sebiferum. Front. Plant Sci. 2017, 8, 637. [Google Scholar] [CrossRef]
- Li, M.; Li, X.; Wang, C.; Li, Q.; Zhu, S.; Zhang, Y.; Li, X.; Yang, F.; Zhu, X. Selection and Validation of Reference Genes for qRT-PCR Analysis of Rhopalosiphum padi (Hemiptera: Aphididae). Front. Physiol. 2021, 12, 663338. [Google Scholar] [CrossRef] [PubMed]
- Yulia, P.; Arno, G.; Shinji, M.; Watanabe, T.M. Validation of Common Housekeeping Genes as Reference for qPCR Gene Expression Analysis during iPS Reprogramming Process. Sci. Rep. 2018, 8, 8716. [Google Scholar]
- Radoni, A.; Thulke, S.; Mackay, I.M.; Landt, O.; Siegert, W.; Nitsche, A. Guideline to reference gene selection for quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2004, 313, 856–862. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef] [PubMed]
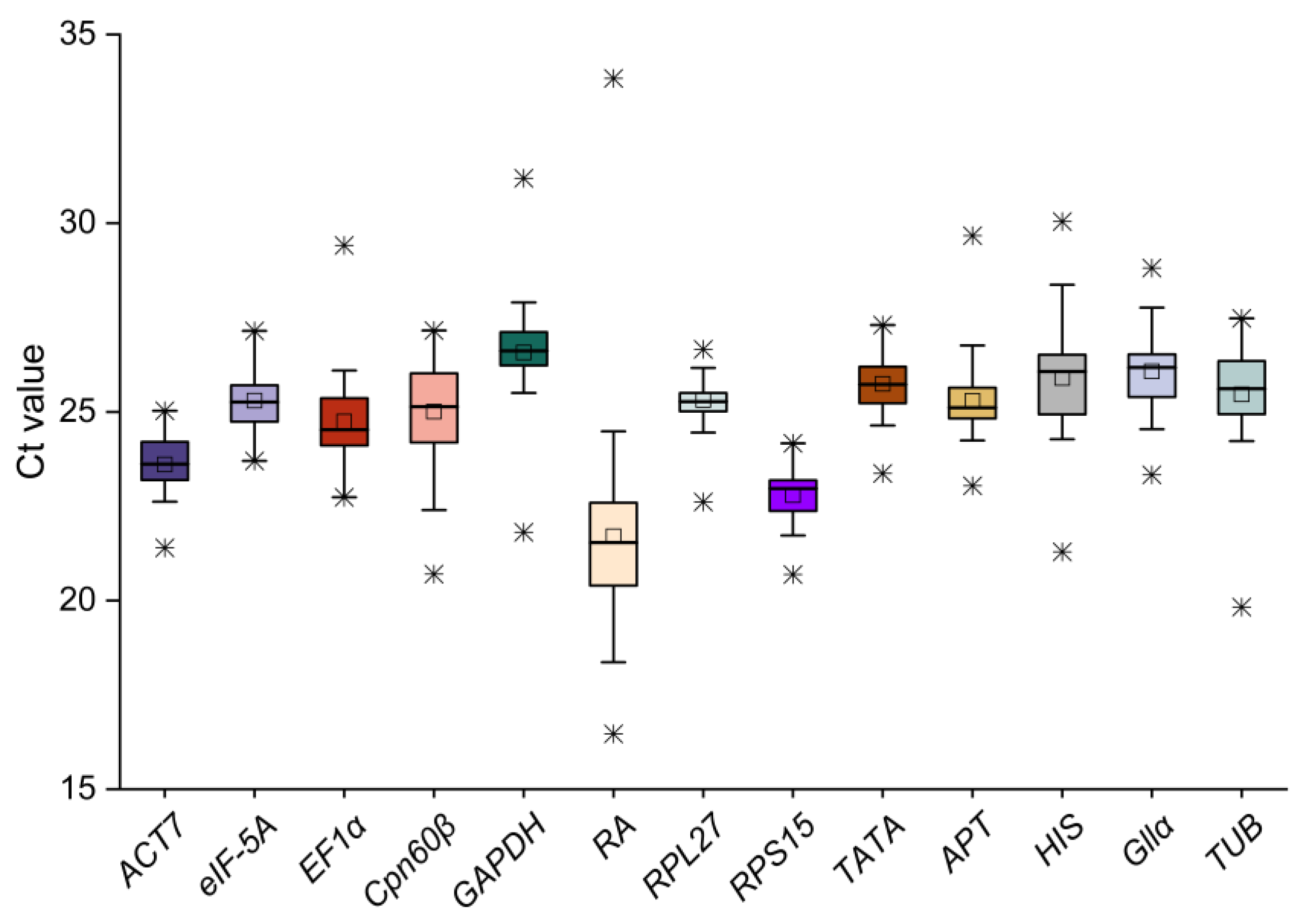
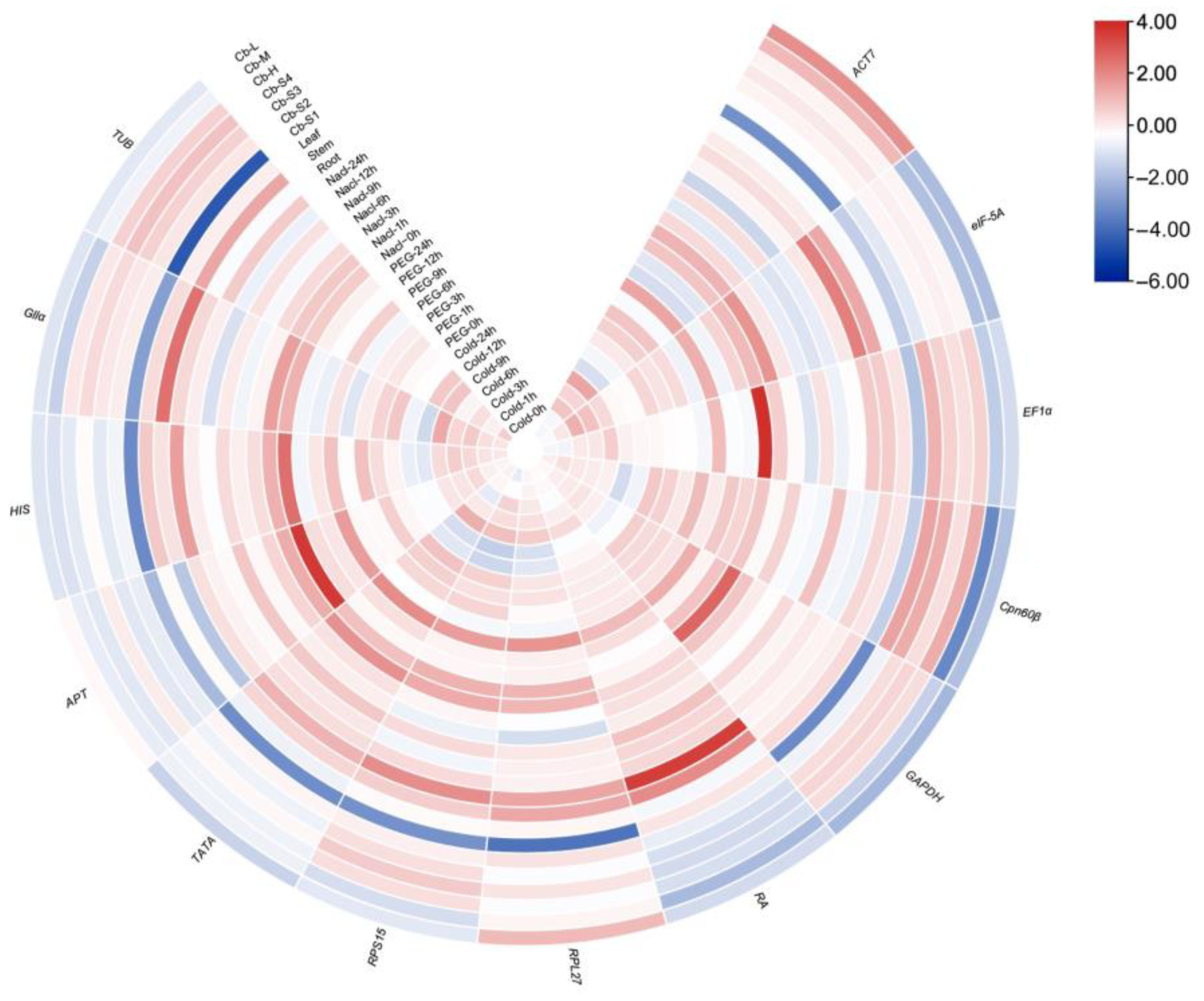

| Gene-ID | Gene Abbreviation | Tentative Annotation | Primer Sequence of Forward | Primer Sequence of Reward | Amplicon Length (bp) | Tm (°C) | E | R2 |
|---|---|---|---|---|---|---|---|---|
| Cbur01G028330 | ACT7 | actin7 | CAACCCAAAAGCCAACAGG | TCACCCGAGTCCAGAACAATAC | 141 | 58.7/59.1 | 98.76% | 0.9968 |
| Cbur02G019900 | Cpn60β | chaperonin 60 subunit beta 2 | CAACAAGGATGGGCTGGCTA | TTGGCCACAGTCACTCCATC | 156 | 60/60 | 98.05% | 0.9979 |
| Cbur01G001170 | EF1α | elongation factor 1-alpha | GGTACAAGGGCCCAACTCTC | CTGGAGAGCTTCATGGTGCA | 236 | 60/60 | 89.99% | 0.9983 |
| Cbur05G032970 | eIF-5A | eukaryotic translation initiation factor 5A | CCAAGTGTCACTTTGTGGCG | AGTGGGGAGCCTCAGATCAT | 191 | 60/60 | 86.05% | 0.9993 |
| Cbur10G024220 | GAPDH | glyceraldehyde-3-phosphate dehydrogenase | AAGGGTGGTGCCAAGAAAGT | GTTGCAGTGATGGAGTGGACAG | 215 | 58.6/60.2 | 92.81% | 0.9917 |
| Cbur06G016220 | GIIα | glucan 1,3-alpha-glucosidase | CCTTATCGCCTTTTCAACCTT | AGCGTATCAATCCGCCCTC | 221 | 58.3/59.9 | 90.63% | 0.9983 |
| Cbur08G011150 | HIS | histone superfamily protein H3 | GGAGGGAAGGCTCCTAGGAA | CAACTGTTCCAGGGCGGTAT | 106 | 60/60 | 96.01% | 0.9985 |
| Cbur10G000690 | RA | rubisco activase | ACAGACCGACAAGGACAAATGG | CGGAGACCCGTGCTCAAGTAT | 168 | 61.3/61.6 | 79.95% | 0.9926 |
| Cbur10G003920 | RPL27 | ribosomal protein L27 | GCCGTCATCGTACGATCCTT | TGCCGTCTTCTTTGCAGAGT | 123 | 60.0/59.9 | 98.39% | 0.9969 |
| Cbur07G013210 | RPS15 | ribosomal protein S15 | GCAGCCGAAGAGGAGAACA | GGCTTCCGCTTCAAACCAC | 144 | 58.4//60.4 | 92.04% | 0.9972 |
| Cbur04G009020 | TATA | TATA-box-binding protein | CCGTAATGCAGAGTATAACCCC | TTTGACATCACAAGAGCCCAC | 146 | 60.1/59.5 | 82.13% | 0.9989 |
| Cbur08G006150 | TUB | tubulin β chain | TGGGAATAACTGGGCTAAGGG | AAGCATCATCCGATCAGGGTA | 205 | 60.9/59.5 | 95.11% | 0.9964 |
| Cbur02G028660 | APT | adenine phosphoribosy ltransferase 1 | TGCTTGATCCCGAGGCATTT | ACTTCGAACCAAGGGCCAAA | 141 | 60.1/60 | 89.03% | 0.9993 |
| Total | Cold-treated | Nacl-treated | PEG-treated | Tissues | Leaves at Different Developmental Stages | Different Borneol Clones | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | mSD | Gene | mSD | Gene | mSD | Gene | mSD | Gene | mSD | Gene | mSD | Gene | mSD |
| ACT7 | 1.21 | ACT7 | 0.51 | ACT7 | 0.87 | ACT7 | 0.62 | ACT7 | 1.30 | ACT7 | 0.94 | ACT7 | 1.56 |
| APT | 1.29 | APT | 0.49 | APT | 1.14 | APT | 0.64 | APT | 1.93 | APT | 1.14 | APT | 1.22 |
| Cpn60β | 1.61 | Cpn60β | 0.63 | Cpn60β | 1.37 | Cpn60β | 1.09 | Cpn60β | 1.81 | Cpn60β | 1.43 | Cpn60β | 2.72 |
| EF1α | 1.22 | EF1α | 0.63 | EF1α | 1.41 | EF1α | 0.49 | EF1α | 1.29 | EF1α | 1.10 | EF1α | 1.10 |
| eIF-5A | 1.10 | eIF-5A | 0.58 | eIF-5A | 0.95 | eIF-5A | 0.55 | eIF-5A | 1.52 | eIF-5A | 1.32 | eIF-5A | 0.94 |
| GAPDH | 1.37 | GAPDH | 0.49 | GAPDH | 1.24 | GAPDH | 0.68 | GAPDH | 1.67 | GAPDH | 1.67 | GAPDH | 1.69 |
| Gllα | 1.13 | Gllα | 0.62 | Gllα | 0.90 | Gllα | 0.75 | Gllα | 1.91 | Gllα | 0.94 | Gllα | 0.93 |
| HIS | 1.31 | HIS | 0.66 | HIS | 1.14 | HIS | 0.81 | HIS | 1.73 | HIS | 1.14 | HIS | 1.01 |
| RA | 3.01 | RA | 0.71 | RA | 1.61 | RA | 0.98 | RA | 6.61 | RA | 3.17 | RA | 1.26 |
| RPL27 | 1.05 | RPL27 | 0.43 | RP L27 | 0.85 | RP L27 | 0.48 | RP L27 | 1.32 | RPL27 | 0.93 | RPL27 | 1.30 |
| RPS15 | 0.98 | RPS15 | 0.46 | RPS15 | 0.83 | RPS15 | 0.47 | RPS15 | 1.36 | RPS15 | 0.93 | RPS15 | 0.90 |
| TATA | 1.04 | TATA | 0.57 | TATA | 0.92 | TATA | 0.49 | TATA | 1.31 | TATA | 0.95 | TATA | 1.11 |
| TUB | 1.34 | TUB | 0.89 | TUB | 0.97 | TUB | 0.51 | TUB | 2.03 | TUB | 2.16 | TUB | 1.05 |
| Total | Cold-treated | Nacl-treated | PEG-treated | Tissues | Leaves at Different Developmental Stages | Different Borneol Clones | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | SV | Gene | SV | Gene | SV | Gene | SV | Gene | SV | Gene | SV | Gene | SV |
| ACT7 | 0.811 | ACT7 | 0.285 | ACT7 | 0.438 | ACT7 | 0.401 | ACT7 | 0.058 | ACT7 | 0.063 | ACT7 | 1.482 |
| APT | 0.877 | APT | 0.247 | APT | 0.934 | APT | 0.448 | APT | 1.380 | APT | 0.547 | APT | 1.007 |
| Cpn60β | 1.314 | Cpn60β | 0.506 | Cpn60β | 1.114 | Cpn60β | 1.005 | Cpn60β | 1.445 | Cpn60β | 1.034 | Cpn60β | 2.660 |
| EF1α | 0.745 | EF1α | 0.476 | EF1α | 1.311 | EF1α | 0.102 | EF1α | 0.058 | EF1α | 0.439 | EF1α | 0.499 |
| eIF-5A | 0.436 | eIF-5A | 0.427 | eIF-5A | 0.559 | eIF-5A | 0.308 | eIF-5A | 0.208 | eIF-5A | 0.834 | eIF-5A | 0.128 |
| GAPDH | 0.995 | GAPDH | 0.255 | GAPDH | 1.082 | GAPDH | 0.521 | GAPDH | 1.205 | GAPDH | 1.448 | GAPDH | 1.440 |
| Gllα | 0.564 | Gllα | 0.473 | Gllα | 0.429 | Gllα | 0.603 | Gllα | 1.390 | Gllα | 0.131 | Gllα | 0.138 |
| HIS | 0.834 | HIS | 0.545 | HIS | 0.899 | HIS | 0.673 | HIS | 0.948 | HIS | 0.617 | HIS | 0.542 |
| RA | 2.918 | RA | 0.593 | RA | 1.499 | RA | 0.887 | RA | 6.568 | RA | 3.132 | RA | 0.760 |
| RPL27 | 0.467 | RPL27 | 0.115 | RPL27 | 0.328 | RPL27 | 0.114 | RPL27 | 0.126 | RPL27 | 0.131 | RPL27 | 1.134 |
| RPS15 | 0.210 | RPS15 | 0.171 | RPS15 | 0.340 | RPS15 | 0.082 | RPS15 | 0.099 | RPS15 | 0.063 | RPS15 | 0.153 |
| TATA | 0.344 | TATA | 0.378 | TATA | 0.570 | TATA | 0.107 | TATA | 0.099 | TATA | 0.119 | TATA | 0.700 |
| TUB | 0.969 | TUB | 0.803 | TUB | 0.601 | TUB | 0.194 | TUB | 1.644 | TUB | 2.086 | TUB | 0.426 |
| Total | Cold-treated | Nacl-treated | PEG-treated | Tissues | Leaves at Different Developmental Stages | Different Borneol Clones | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | SD [±CP] | Gene | SD [±CP] | Gene | SD [±CP] | Gene | SD [±CP] | Gene | SD [±CP] | Gene | SD [±CP] | Gene | SD [±CP] |
| ACT7 | 0.56 | ACT7 | 0.58 | ACT7 | 0.52 | ACT7 | 0.58 | ACT7 | 0.19 | ACT7 | 0.79 | ACT7 | 0.53 |
| APT | 0.73 | APT | 0.27 | APT | 1.13 | APT | 0.54 | APT | 0.85 | APT | 0.60 | APT | 0.32 |
| Cpn60β | 1.00 | Cpn60β | 0.56 | Cpn60β | 0.83 | Cpn60β | 0.38 | Cpn60β | 0.61 | Cpn60β | 1.43 | Cpn60β | 2.37 |
| EF1α | 0.78 | EF1α | 0.36 | EF1α | 1.28 | EF1α | 0.38 | EF1α | 0.26 | EF1α | 1.03 | EF1α | 0.96 |
| eIF-5A | 0.64 | eIF-5A | 0.42 | eIF-5A | 0.66 | eIF-5A | 0.40 | eIF-5A | 0.83 | eIF-5A | 0.46 | eIF-5A | 0.63 |
| GAPDH | 0.96 | GAPDH | 0.37 | GAPDH | 1.21 | GAPDH | 0.57 | GAPDH | 0.32 | GAPDH | 1.80 | GAPDH | 1.45 |
| Gllα | 0.76 | Gllα | 0.67 | Gllα | 0.75 | Gllα | 0.59 | Gllα | 1.04 | Gllα | 1.09 | Gllα | 0.62 |
| HIS | 1.09 | HIS | 0.74 | HIS | 1.21 | HIS | 0.71 | HIS | 0.79 | HIS | 1.38 | HIS | 0.10 |
| RA | 1.87 | RA | 0.53 | RA | 0.68 | RA | 0.80 | RA | 4.74 | RA | 1.20 | RA | 0.89 |
| RPL27 | 0.47 | RPL27 | 0.35 | RPL27 | 0.44 | RPL27 | 0.35 | RPL27 | 0.53 | RPL27 | 1.01 | RPL27 | 0.43 |
| RPS15 | 0.53 | RPS15 | 0.55 | RPS15 | 0.45 | RPS15 | 0.36 | RPS15 | 0.56 | RPS15 | 0.87 | RPS15 | 0.41 |
| TATA | 0.57 | TATA | 0.48 | TATA | 0.38 | TATA | 0.43 | TATA | 0.52 | TATA | 0.79 | TATA | 0.26 |
| TUB | 0.83 | TUB | 0.55 | TUB | 0.65 | TUB | 0.37 | TUB | 1.05 | TUB | 2.38 | TUB | 0.80 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, L.; Cai, Y.; Yao, J.; Zhang, Q.; He, B.; Lin, S. Reference Genes Selection and Validation for Cinnamomum burmanni by Real-Time Quantitative Polymerase Chain Reaction. Int. J. Mol. Sci. 2024, 25, 3500. https://doi.org/10.3390/ijms25063500
Shi L, Cai Y, Yao J, Zhang Q, He B, Lin S. Reference Genes Selection and Validation for Cinnamomum burmanni by Real-Time Quantitative Polymerase Chain Reaction. International Journal of Molecular Sciences. 2024; 25(6):3500. https://doi.org/10.3390/ijms25063500
Chicago/Turabian StyleShi, Lingling, Yanling Cai, Jun Yao, Qian Zhang, Boxiang He, and Shanzhi Lin. 2024. "Reference Genes Selection and Validation for Cinnamomum burmanni by Real-Time Quantitative Polymerase Chain Reaction" International Journal of Molecular Sciences 25, no. 6: 3500. https://doi.org/10.3390/ijms25063500
APA StyleShi, L., Cai, Y., Yao, J., Zhang, Q., He, B., & Lin, S. (2024). Reference Genes Selection and Validation for Cinnamomum burmanni by Real-Time Quantitative Polymerase Chain Reaction. International Journal of Molecular Sciences, 25(6), 3500. https://doi.org/10.3390/ijms25063500





