Analysis of the Conformational Landscape of the N-Domains of the AAA ATPase p97: Disentangling the Continuous Conformational Variability in Partially Symmetrical Complexes
Abstract
1. Introduction
2. Results
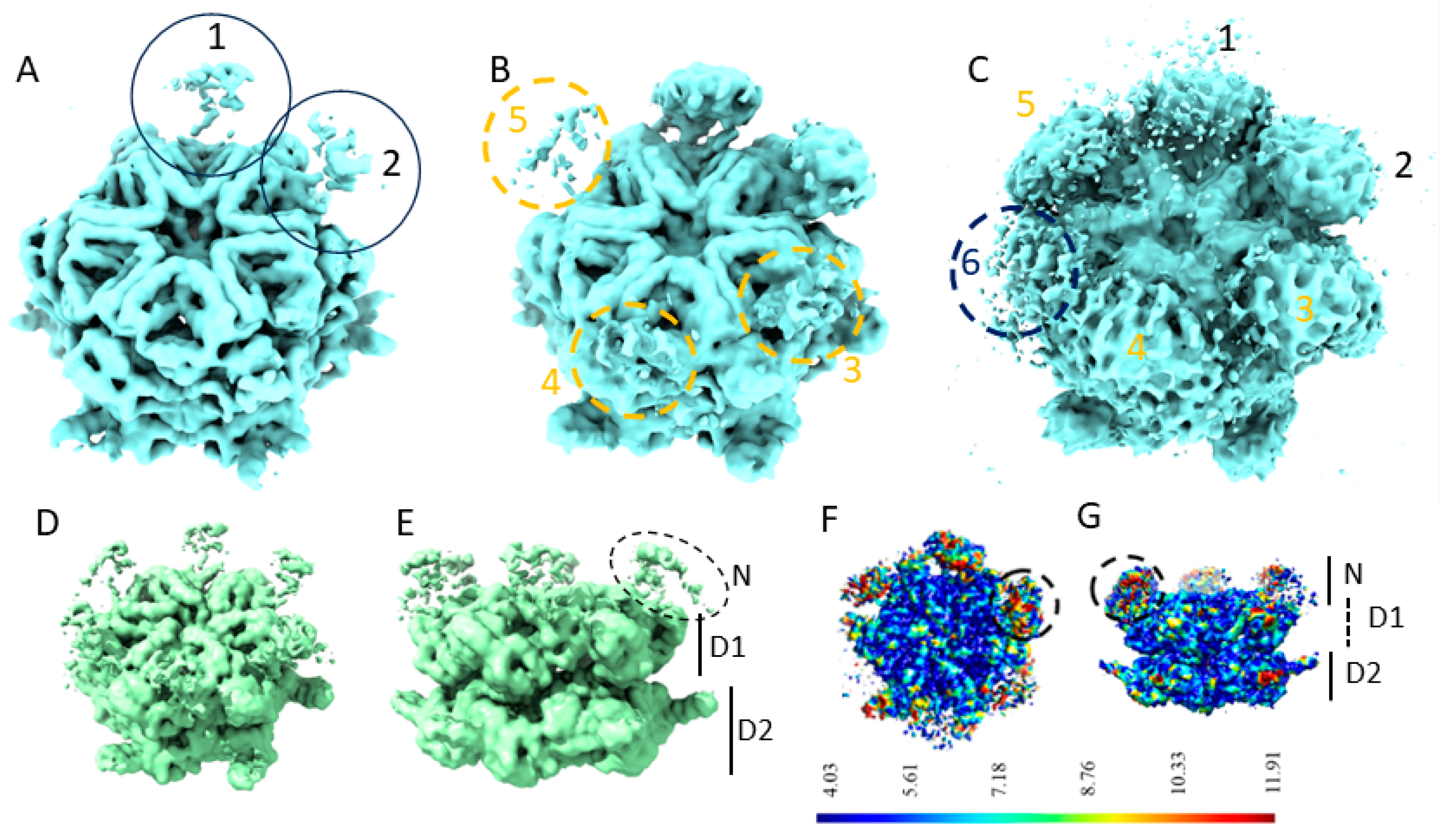
2.1. The N-Domains of p97-R155P-ATPγS Are Partially Visible Using the Methods of Global Reconstruction and Discrete Classification
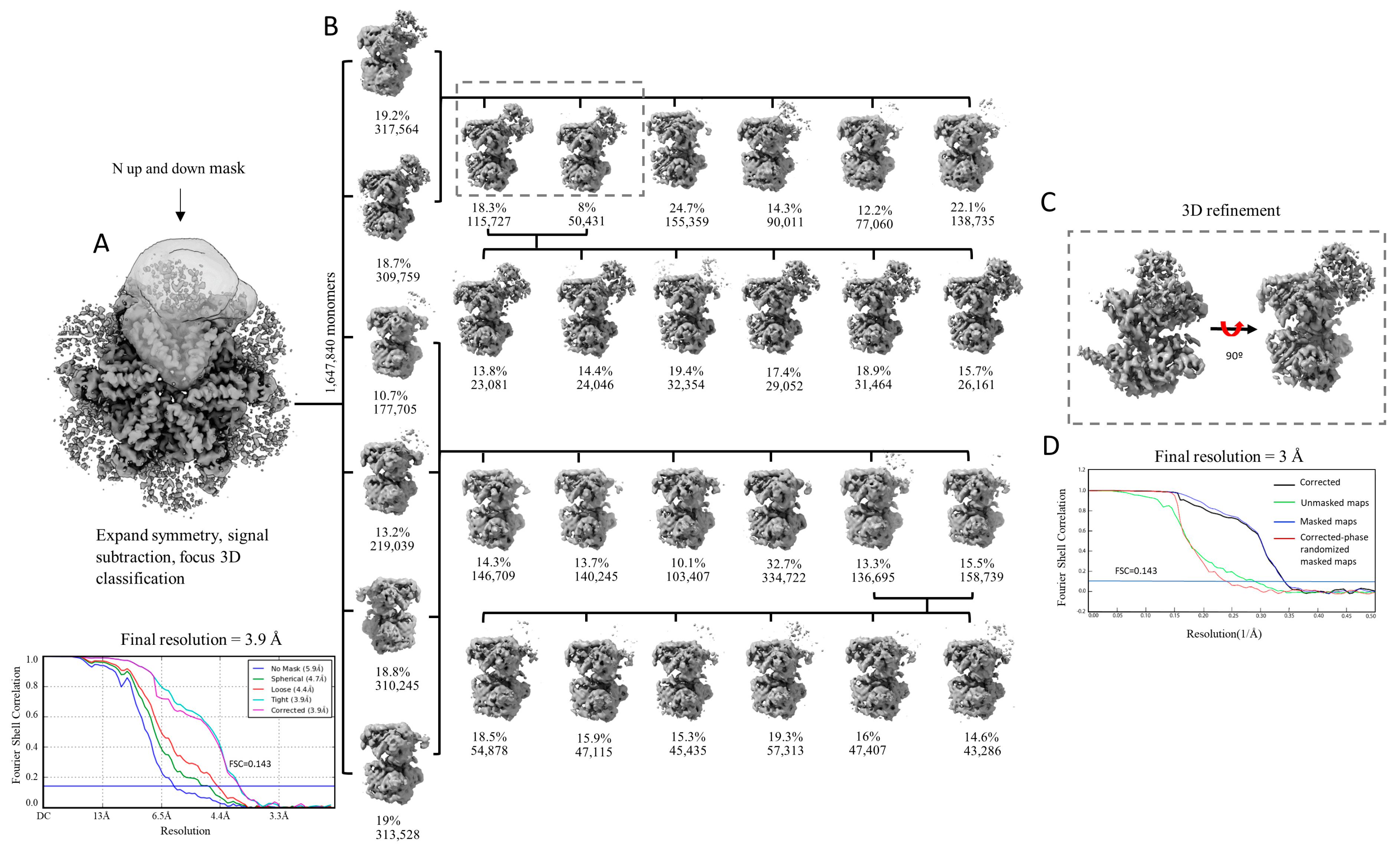
2.2. Focused Classification Failed to Disentangle and Resolve the Different Positions of the N-Domains
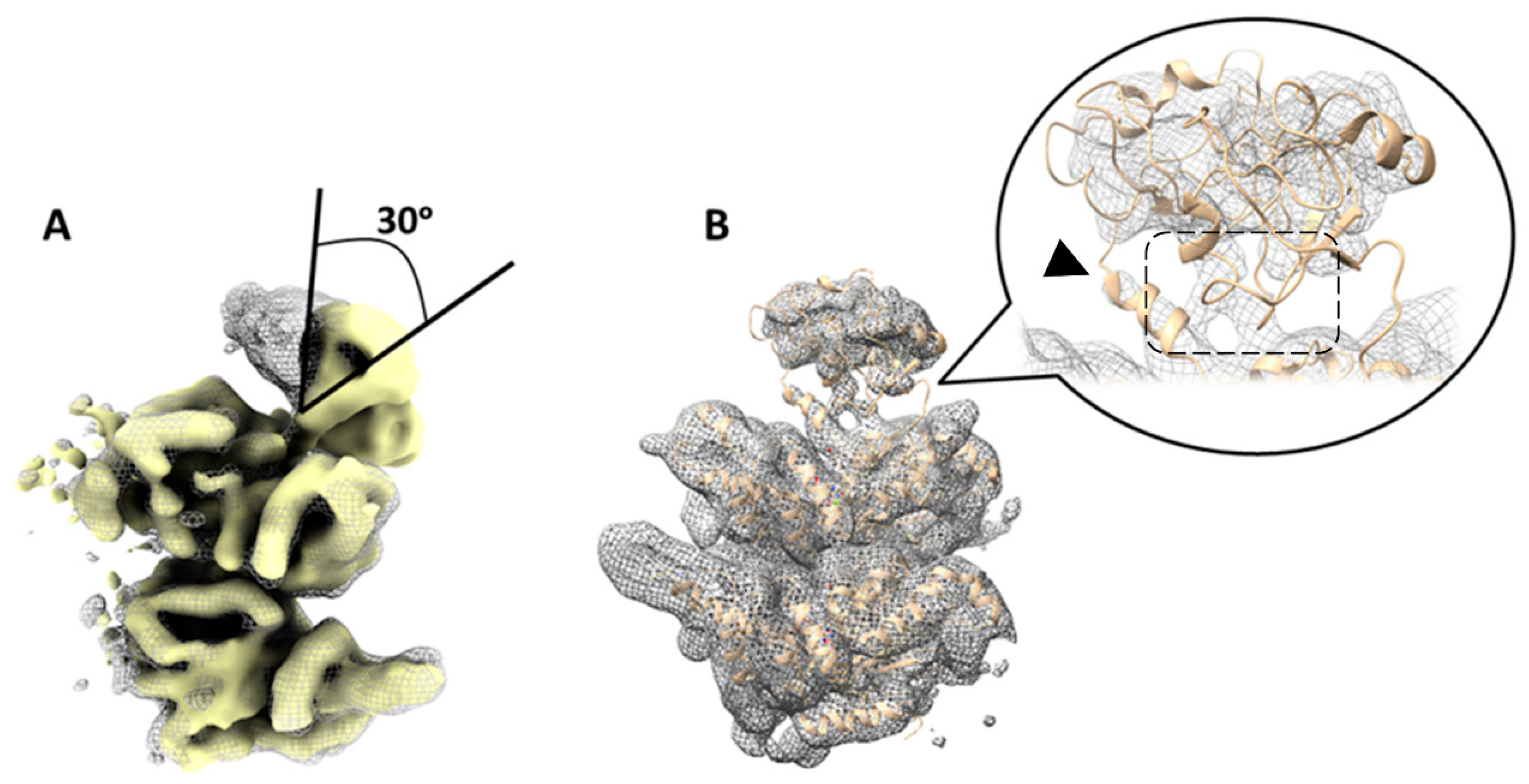
2.3. Focused Classification on Low-Pass Filtered Images Detected Movement of the N-Domain by Up to 30 Degrees
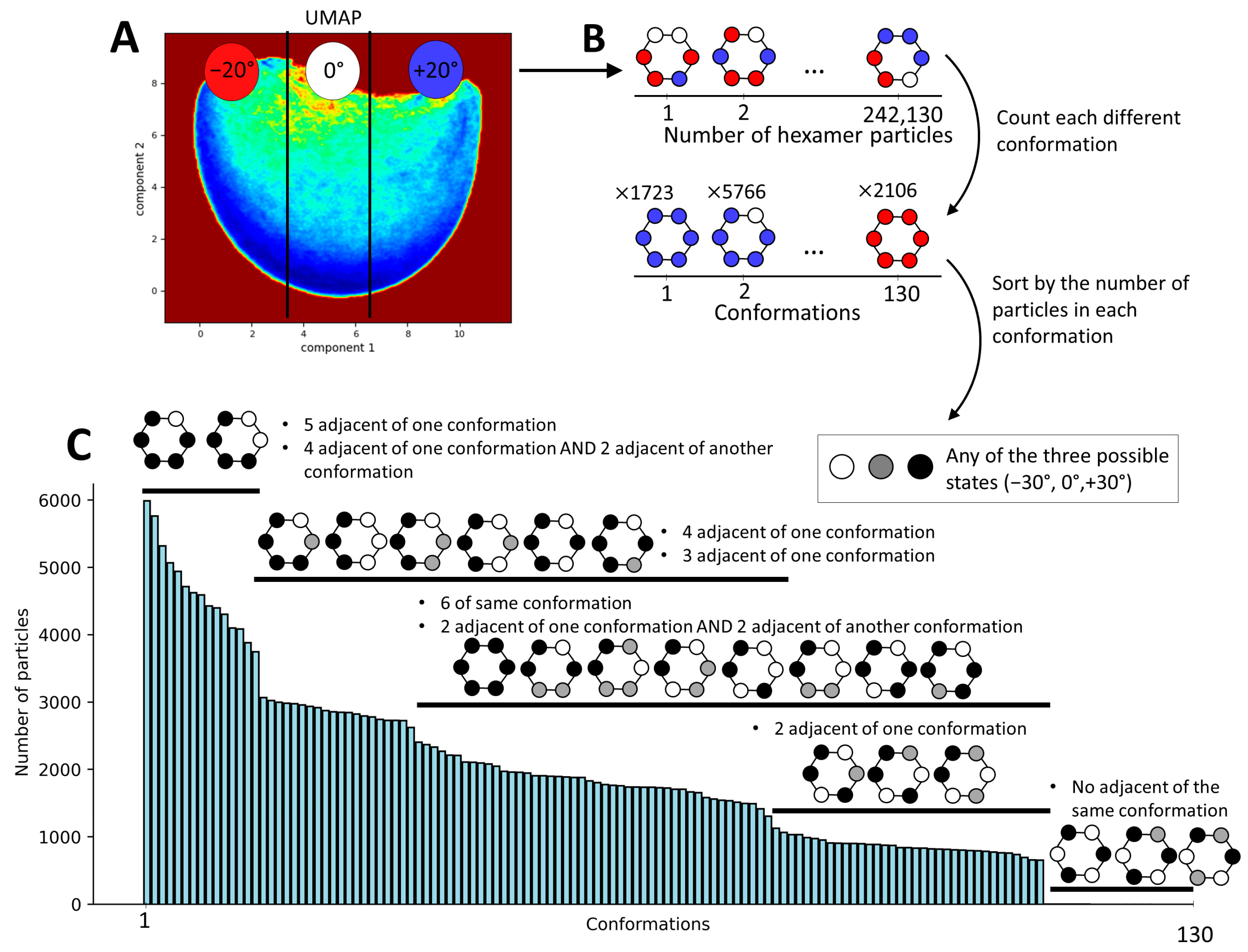
2.4. Continuous Conformational Variability Analysis Using MDSPACE at the Hexameric Level
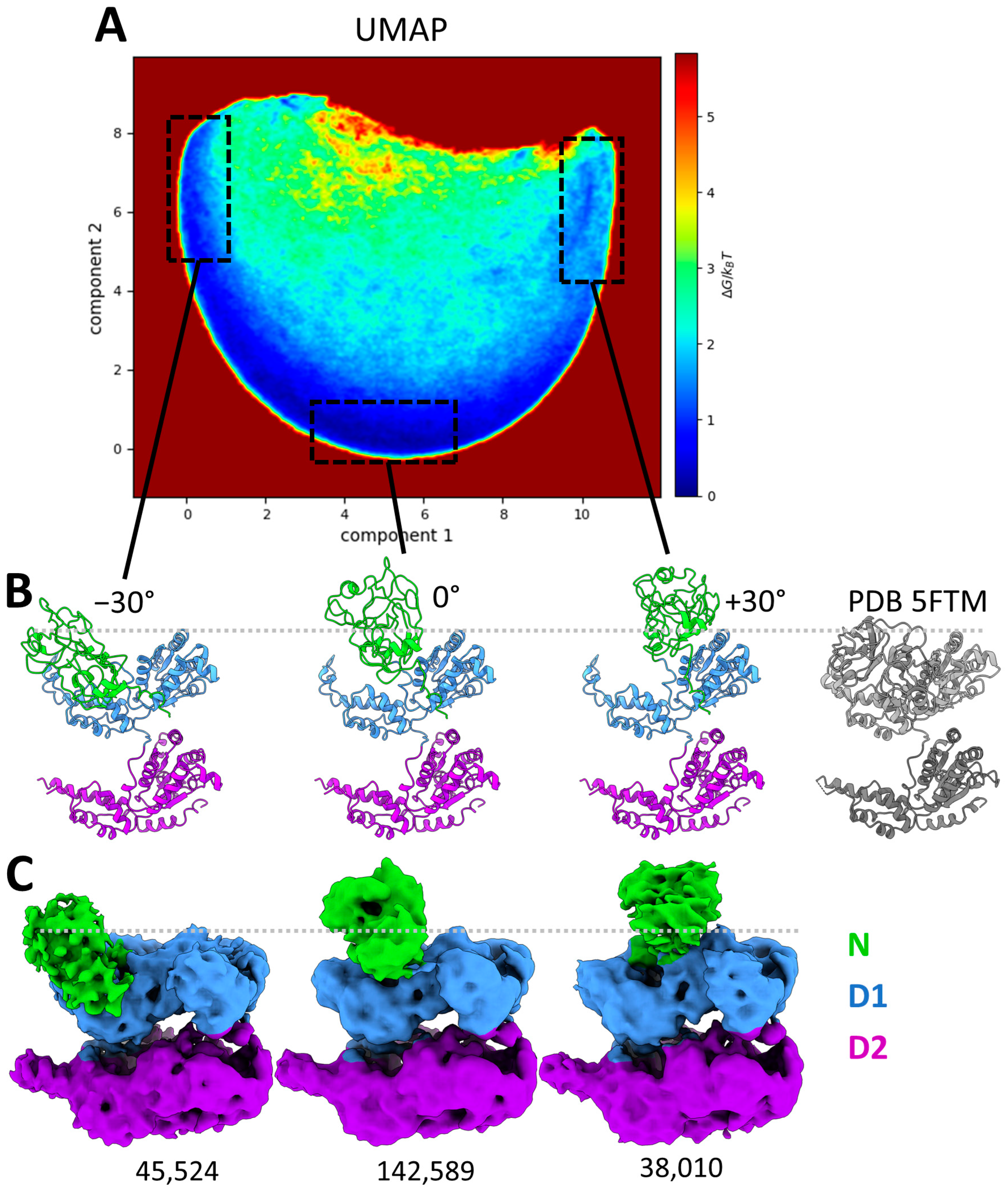
2.5. Analysis of the Continuous Conformational Variability at the Monomeric Level Using MDSPACE
2.6. Retrieving Hexameric Conformations from the Analysis at the Monomeric Level
3. Discussion
4. Materials and Methods
4.1. Sample Preparation
4.2. 3D Reconstruction
4.3. Focused 3D Classification at the Monomeric Level
4.4. Analysis of Conformational Heterogeneity Using MDSPACE
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kuhlbrandt, W. Biochemistry. The resolution revolution. Science 2014, 343, 1443–1444. [Google Scholar] [CrossRef]
- Yip, K.M.; Fischer, N.; Paknia, E.; Chari, A.; Stark, H. Atomic-resolution protein structure determination by cryo-EM. Nature 2020, 587, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Nakane, T.; Kotecha, A.; Sente, A.; McMullan, G.; Masiulis, S.; Brown, P.; Grigoras, I.T.; Malinauskaite, L.; Malinauskas, T.; Miehling, J.; et al. Single-particle cryo-EM at atomic resolution. Nature 2020, 587, 152–156. [Google Scholar] [CrossRef] [PubMed]
- Amunts, A.; Brown, A.; Bai, X.C.; Llacer, J.L.; Hussain, T.; Emsley, P.; Long, F.; Murshudov, G.; Scheres, S.H.; Ramakrishnan, V. Structure of the yeast mitochondrial large ribosomal subunit. Science 2014, 343, 1485–1489. [Google Scholar] [CrossRef] [PubMed]
- Liao, M.; Cao, E.; Julius, D.; Cheng, Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature 2013, 504, 107–112. [Google Scholar] [CrossRef] [PubMed]
- Allegretti, M.; Mills, D.J.; McMullan, G.; Kuhlbrandt, W.; Vonck, J. Atomic model of the F420-reducing [NiFe] hydrogenase by electron cryo-microscopy using a direct electron detector. eLife 2014, 3, e01963. [Google Scholar] [CrossRef]
- Zhou, A.; Rohou, A.; Schep, D.G.; Bason, J.V.; Montgomery, M.G.; Walker, J.E.; Grigorieff, N.; Rubinstein, J.L. Structure and conformational states of the bovine mitochondrial ATP synthase by cryo-EM. eLife 2015, 4, e10180. [Google Scholar] [CrossRef]
- de Oliveira, T.M.; van Beek, L.; Shilliday, F.; Debreczeni, J.E.; Phillips, C. Cryo-EM: The Resolution Revolution and Drug Discovery. SLAS Discov. 2021, 26, 17–31. [Google Scholar] [CrossRef]
- Lees, J.A.; Dias, J.M.; Han, S. Applications of Cryo-EM in small molecule and biologics drug design. Biochem. Soc. Trans. 2021, 49, 2627–2638. [Google Scholar] [CrossRef]
- Wigge, C.; Stefanovic, A.; Radjainia, M. The rapidly evolving role of cryo-EM in drug design. Drug Discov. Today Technol. 2020, 38, 91–102. [Google Scholar] [CrossRef]
- Chari, A.; Stark, H. Prospects and Limitations of High-Resolution Single-Particle Cryo-Electron Microscopy. Annu. Rev. Biophys. 2023, 52, 391–411. [Google Scholar] [CrossRef] [PubMed]
- Scheres, S.H. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012, 180, 519–530. [Google Scholar] [CrossRef] [PubMed]
- Punjani, A.; Rubinstein, J.L.; Fleet, D.J.; Brubaker, M.A. cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 2017, 14, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Rouiller, I.; DeLaBarre, B.; May, A.P.; Weis, W.I.; Brunger, A.T.; Milligan, R.A.; Wilson-Kubalek, E.M. Conformational changes of the multifunction p97 AAA ATPase during its ATPase cycle. Nat. Struct. Biol. 2002, 9, 950–957. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Bartesaghi, A.; Merk, A.; Rao, P.; Bulfer, S.L.; Yan, Y.; Green, N.; Mroczkowski, B.; Neitz, R.J.; Wipf, P.; et al. 2.3 A resolution cryo-EM structure of human p97 and mechanism of allosteric inhibition. Science 2016, 351, 871–875. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Ripstein, Z.A.; Rubinstein, J.L.; Kay, L.E. Probing Cooperativity of N-Terminal Domain Orientations in the p97 Molecular Machine: Synergy between NMR Spectroscopy and Cryo-EM. Angew. Chem. Int. Ed. Engl. 2020, 59, 22423–22426. [Google Scholar] [CrossRef]
- Xu, Y.; Han, H.; Cooney, I.; Guo, Y.; Moran, N.G.; Zuniga, N.R.; Price, J.C.; Hill, C.P.; Shen, P.S. Active conformation of the p97-p47 unfoldase complex. Nat. Commun. 2022, 13, 2640. [Google Scholar] [CrossRef]
- van den Boom, J.; Marini, G.; Meyer, H.; Saibil, H.R. Structural basis of ubiquitin-independent PP1 complex disassembly by p97. EMBO J. 2023, 42, e113110. [Google Scholar] [CrossRef]
- Caffrey, B.; Zhu, X.; Berezuk, A.; Tuttle, K.; Chittori, S.; Subramaniam, S. AAA+ ATPase p97/VCP mutants and inhibitor binding disrupt inter-domain coupling and subsequent allosteric activation. J. Biol. Chem. 2021, 297, 101187. [Google Scholar] [CrossRef] [PubMed]
- Valimehr, S.; Sethi, A.; Shukla, M.; Bhattacharyya, S.; Kazemi, M.; Rouiller, I. Molecular Mechanisms Driving and Regulating the AAA+ ATPase VCP/p97, an Important Therapeutic Target for Treating Cancer, Neurological and Infectious Diseases. Biomolecules 2023, 13, 737. [Google Scholar] [CrossRef]
- Meyer, H.; van den Boom, J. Targeting of client proteins to the VCP/p97/Cdc48 unfolding machine. Front. Mol. Biosci. 2023, 10, 1142989. [Google Scholar] [CrossRef] [PubMed]
- Hanzelmann, P.; Galgenmuller, C.; Schindelin, H. Structure and Function of the AAA+ ATPase p97, a Key Player in Protein Homeostasis. Sub-Cell. Biochem. 2019, 93, 221–272. [Google Scholar] [CrossRef]
- Braxton, J.R.; Southworth, D.R. Structural insights of the p97/VCP AAA+ ATPase: How adapter interactions coordinate diverse cellular functionality. J. Biol. Chem. 2023, 299, 105182. [Google Scholar] [CrossRef] [PubMed]
- Buchberger, A.; Schindelin, H.; Hanzelmann, P. Control of p97 function by cofactor binding. FEBS Lett. 2015, 589, 2578–2589. [Google Scholar] [CrossRef] [PubMed]
- Watts, G.D.; Wymer, J.; Kovach, M.J.; Mehta, S.G.; Mumm, S.; Darvish, D.; Pestronk, A.; Whyte, M.P.; Kimonis, V.E. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat. Genet. 2004, 36, 377–381. [Google Scholar] [CrossRef] [PubMed]
- Darwich, N.F.; Phan, J.M.; Kim, B.; Suh, E.; Papatriantafyllou, J.D.; Changolkar, L.; Nguyen, A.T.; O’Rourke, C.M.; He, Z.; Porta, S.; et al. Autosomal dominant VCP hypomorph mutation impairs disaggregation of PHF-tau. Science 2020, 370, eaay8826. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.O.; Mandrioli, J.; Benatar, M.; Abramzon, Y.; Van Deerlin, V.M.; Trojanowski, J.Q.; Gibbs, J.R.; Brunetti, M.; Gronka, S.; Wuu, J.; et al. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron 2010, 68, 857–864. [Google Scholar] [CrossRef]
- Boock, V.; Roy, B.; Pfeffer, G.; Kimonis, V. Therapeutic developments for valosin-containing protein mediated multisystem proteinopathy. Curr. Opin. Neurol. 2023, 36, 432–440. [Google Scholar] [CrossRef]
- Kobakhidze, G.; Sethi, A.; Valimehr, S.; Ralph, S.A.; Rouiller, I. The AAA+ ATPase p97 as a novel parasite and tuberculosis drug target. Trends Parasitol. 2022, 38, 572–590. [Google Scholar] [CrossRef]
- Huryn, D.M.; Kornfilt, D.J.P.; Wipf, P. p97: An Emerging Target for Cancer, Neurodegenerative Diseases, and Viral Infections. J. Med. Chem. 2020, 63, 1892–1907. [Google Scholar] [CrossRef]
- Del Rio Oliva, M.; Basler, M. Valosin-containing protein (VCP/p97) inhibition reduces viral clearance and induces toxicity associated with muscular damage. Cell Death Dis. 2022, 13, 1015. [Google Scholar] [CrossRef] [PubMed]
- Das, P.; Dudley, J.P. How Viruses Use the VCP/p97 ATPase Molecular Machine. Viruses 2021, 13, 1881. [Google Scholar] [CrossRef]
- Kilgas, S.; Ramadan, K. Inhibitors of the ATPase p97/VCP: From basic research to clinical applications. Cell Chem. Biol. 2023, 30, 3–21. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Wang, F.; Lin, X.; Li, Q.; Lu, Y.; Zhang, J.; Shen, X.; Tan, J.; Qin, Z.; Chen, J.; et al. Nuclear VCP drives colorectal cancer progression by promoting fatty acid oxidation. Proc. Natl. Acad. Sci. USA 2023, 120, e2221653120. [Google Scholar] [CrossRef]
- Anderson, D.J.; Le Moigne, R.; Djakovic, S.; Kumar, B.; Rice, J.; Wong, S.; Wang, J.; Yao, B.; Valle, E.; Kiss von Soly, S.; et al. Targeting the AAA ATPase p97 as an Approach to Treat Cancer through Disruption of Protein Homeostasis. Cancer Cell 2015, 28, 653–665. [Google Scholar] [CrossRef] [PubMed]
- Vekaria, P.H.; Home, T.; Weir, S.; Schoenen, F.J.; Rao, R. Targeting p97 to Disrupt Protein Homeostasis in Cancer. Front. Oncol. 2016, 6, 181. [Google Scholar] [CrossRef]
- DeLaBarre, B.; Brunger, A.T. Complete structure of p97/valosin-containing protein reveals communication between nucleotide domains. Nat. Struct. Biol. 2003, 10, 856–863. [Google Scholar] [CrossRef]
- DeLaBarre, B.; Brunger, A.T. Nucleotide dependent motion and mechanism of action of p97/VCP. J. Mol. Biol. 2005, 347, 437–452. [Google Scholar] [CrossRef]
- Hanzelmann, P.; Schindelin, H. Structural Basis of ATP Hydrolysis and Intersubunit Signaling in the AAA+ ATPase p97. Structure 2016, 24, 127–139. [Google Scholar] [CrossRef]
- Le, L.T.; Kang, W.; Kim, J.Y.; Le, O.T.; Lee, S.Y.; Yang, J.K. Structural Details of Ufd1 Binding to p97 and Their Functional Implications in ER-Associated Degradation. PLoS ONE 2016, 11, e0163394. [Google Scholar] [CrossRef]
- Schuetz, A.K.; Kay, L.E. A Dynamic molecular basis for malfunction in disease mutants of p97/VCP. eLife 2016, 5, e20143. [Google Scholar] [CrossRef]
- Briggs, L.C.; Baldwin, G.S.; Miyata, N.; Kondo, H.; Zhang, X.; Freemont, P.S. Analysis of nucleotide binding to P97 reveals the properties of a tandem AAA hexameric ATPase. J. Biol. Chem. 2008, 283, 13745–13752. [Google Scholar] [CrossRef]
- Tang, W.K.; Li, D.; Li, C.C.; Esser, L.; Dai, R.; Guo, L.; Xia, D. A novel ATP-dependent conformation in p97 N-D1 fragment revealed by crystal structures of disease-related mutants. EMBO J. 2010, 29, 2217–2229. [Google Scholar] [CrossRef]
- Bulfer, S.L.; Chou, T.F.; Arkin, M.R. p97 Disease Mutations Modulate Nucleotide-Induced Conformation to Alter Protein-Protein Interactions. ACS Chem. Biol. 2016, 11, 2112–2116. [Google Scholar] [CrossRef] [PubMed]
- Ritz, D.; Vuk, M.; Kirchner, P.; Bug, M.; Schutz, S.; Hayer, A.; Bremer, S.; Lusk, C.; Baloh, R.H.; Lee, H.; et al. Endolysosomal sorting of ubiquitylated caveolin-1 is regulated by VCP and UBXD1 and impaired by VCP disease mutations. Nat. Cell Biol. 2011, 13, 1116–1123. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Saiz, V.; Buchberger, A. Imbalances in p97 co-factor interactions in human proteinopathy. EMBO Rep. 2010, 11, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Mountassif, D.; Fabre, L.; Zaid, Y.; Halawani, D.; Rouiller, I. Cryo-EM of the pathogenic VCP variant R155P reveals long-range conformational changes in the D2 ATPase ring. Biochem. Biophys. Res. Commun. 2015, 468, 636–641. [Google Scholar] [CrossRef] [PubMed]
- Blythe, E.E.; Gates, S.N.; Deshaies, R.J.; Martin, A. Multisystem Proteinopathy Mutations in VCP/p97 Increase NPLOC4.UFD1L Binding and Substrate Processing. Structure 2019, 27, 1820–1829.e4. [Google Scholar] [CrossRef] [PubMed]
- Blythe, E.E.; Olson, K.C.; Chau, V.; Deshaies, R.J. Ubiquitin- and ATP-dependent unfoldase activity of P97/VCP*NPLOC4*UFD1L is enhanced by a mutation that causes multisystem proteinopathy. Proc. Natl. Acad. Sci. USA 2017, 114, E4380–E4388. [Google Scholar] [CrossRef] [PubMed]
- Schutz, A.K.; Rennella, E.; Kay, L.E. Exploiting conformational plasticity in the AAA+ protein VCP/p97 to modify function. Proc. Natl. Acad. Sci. USA 2017, 114, E6822–E6829. [Google Scholar] [CrossRef]
- Cooney, I.; Han, H.; Stewart, M.G.; Carson, R.H.; Hansen, D.T.; Iwasa, J.H.; Price, J.C.; Hill, C.P.; Shen, P.S. Structure of the Cdc48 segregase in the act of unfolding an authentic substrate. Science 2019, 365, 502–505. [Google Scholar] [CrossRef] [PubMed]
- Twomey, E.C.; Ji, Z.; Wales, T.E.; Bodnar, N.O.; Ficarro, S.B.; Marto, J.A.; Engen, J.R.; Rapoport, T.A. Substrate processing by the Cdc48 ATPase complex is initiated by ubiquitin unfolding. Science 2019, 365, eaax1033. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Wu, D.; Chen, Q.; Gao, N. Structural dynamics of AAA + ATPase Drg1 and mechanism of benzo-diazaborine inhibition. Nat. Commun. 2022, 13, 6765. [Google Scholar] [CrossRef] [PubMed]
- Pan, M.; Yu, Y.; Ai, H.; Zheng, Q.; Xie, Y.; Liu, L.; Zhao, M. Mechanistic insight into substrate processing and allosteric inhibition of human p97. Nat. Struct. Mol. Biol. 2021, 28, 614–625. [Google Scholar] [CrossRef]
- Klaholz, B.P. Structure Sorting of Multiple Macromolecular States in Heterogeneous Cryo-EM Samples by 3D Multivariate Statistical Analysis. Open J. Stat. 2015, 5, 820–826. [Google Scholar] [CrossRef]
- Klaholz, B.P.; Myasnikov, A.G.; Van Heel, M. Visualization of release factor 3 on the ribosome during termination of protein synthesis. Nature 2004, 427, 862–865. [Google Scholar] [CrossRef] [PubMed]
- Penczek, P.A.; Frank, J.; Spahn, C.M. A method of focused classification, based on the bootstrap 3D variance analysis, and its application to EF-G-dependent translocation. J. Struct. Biol. 2006, 154, 184–194. [Google Scholar] [CrossRef]
- Loerke, J.; Giesebrecht, J.; Spahn, C.M. Multiparticle cryo-EM of ribosomes. Methods Enzymol. 2010, 483, 161–177. [Google Scholar] [CrossRef]
- Simonetti, A.; Marzi, S.; Myasnikov, A.G.; Fabbretti, A.; Yusupov, M.; Gualerzi, C.O.; Klaholz, B.P. Structure of the 30S translation initiation complex. Nature 2008, 455, 416–420. [Google Scholar] [CrossRef]
- Scheres, S.H. Processing of Structurally Heterogeneous Cryo-EM Data in RELION. Methods Enzymol. 2016, 579, 125–157. [Google Scholar] [CrossRef]
- Dempster, A.P.; Laird, N.M.; Rubin, D.B. Maximum Likelihood from Incomplete Data Via Em Algorithm. J. Roy. Stat. Soc. B Met. 1977, 39, 1–38. [Google Scholar] [CrossRef]
- Scheres, S.H. Classification of structural heterogeneity by maximum-likelihood methods. Methods Enzymol. 2010, 482, 295–320. [Google Scholar] [CrossRef] [PubMed]
- Jonic, S. Computational methods for analyzing conformational variability of macromolecular complexes from cryo-electron microscopy images. Curr. Opin. Struct. Biol. 2017, 43, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Frank, J.; Ourmazd, A. Continuous changes in structure mapped by manifold embedding of single-particle data in cryo-EM. Methods 2016, 100, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Dashti, A.; Schwander, P.; Langlois, R.; Fung, R.; Li, W.; Hosseinizadeh, A.; Liao, H.Y.; Pallesen, J.; Sharma, G.; Stupina, V.A.; et al. Trajectories of the ribosome as a Brownian nanomachine. Proc. Natl. Acad. Sci. USA 2014, 111, 17492–17497. [Google Scholar] [CrossRef] [PubMed]
- Jin, Q.; Sorzano, C.O.; de la Rosa-Trevin, J.M.; Bilbao-Castro, J.R.; Nunez-Ramirez, R.; Llorca, O.; Tama, F.; Jonic, S. Iterative elastic 3D-to-2D alignment method using normal modes for studying structural dynamics of large macromolecular complexes. Structure 2014, 22, 496–506. [Google Scholar] [CrossRef] [PubMed]
- Zhong, E.D.; Bepler, T.; Berger, B.; Davis, J.H. CryoDRGN: Reconstruction of heterogeneous cryo-EM structures using neural networks. Nat. Methods 2021, 18, 176–185. [Google Scholar] [CrossRef]
- Punjani, A.; Fleet, D.J. 3DFlex: Determining structure and motion of flexible proteins from cryo-EM. Nat. Methods 2023, 20, 860–870. [Google Scholar] [CrossRef]
- Hamitouche, I.; Jonic, S. DeepHEMNMA: ResNet-based hybrid analysis of continuous conformational heterogeneity in cryo-EM single particle images. Front. Mol. Biosci. 2022, 9, 965645. [Google Scholar] [CrossRef]
- Vuillemot, R.; Mirzaei, A.; Harastani, M.; Hamitouche, I.; Frechin, L.; Klaholz, B.P.; Miyashita, O.; Tama, F.; Rouiller, I.; Jonic, S. MDSPACE: Extracting Continuous Conformational Landscapes from Cryo-EM Single Particle Datasets Using 3D-to-2D Flexible Fitting based on Molecular Dynamics Simulation. J. Mol. Biol. 2023, 9, 167951. [Google Scholar] [CrossRef]
- Punjani, A.; Fleet, D.J. 3D variability analysis: Resolving continuous flexibility and discrete heterogeneity from single particle cryo-EM. J. Struct. Biol. 2021, 213, 107702. [Google Scholar] [CrossRef]
- Kinman, L.F.; Powell, B.M.; Zhong, E.D.; Berger, B.; Davis, J.H. Uncovering structural ensembles from single-particle cryo-EM data using cryoDRGN. Nat. Protoc. 2023, 18, 319–339. [Google Scholar] [CrossRef] [PubMed]
- Harastani, M.; Sorzano, C.O.S.; Jonić, S. Hybrid Electron Microscopy Normal Mode Analysis with Scipion. Protein Sci. 2020, 29, 223–236. [Google Scholar] [CrossRef] [PubMed]
- Tama, F.; Gadea, F.X.; Marques, O.; Sanejouand, Y.H. Building-block approach for determining low-frequency normal modes of macromolecules. Proteins 2000, 41, 107702. [Google Scholar] [CrossRef]
- Suhre, K.; Sanejouand, Y.H. ElNemo: A normal mode web server for protein movement analysis and the generation of templates for molecular replacement. Nucleic Acids Res. 2004, 32, W610–W614. [Google Scholar] [CrossRef]
- Vuillemot, R.; Miyashita, O.; Tama, F.; Rouiller, I.; Jonic, S. NMMD: Efficient Cryo-EM Flexible Fitting Based on Simultaneous Normal Mode and Molecular Dynamics atomic displacements. J. Mol. Biol. 2022, 434, 167483. [Google Scholar] [CrossRef]
- Orzechowski, M.; Tama, F. Flexible fitting of high-resolution x-ray structures into cryoelectron microscopy maps using biased molecular dynamics simulations. Biophys. J. 2008, 95, 5692–5705. [Google Scholar] [CrossRef] [PubMed]
- Miyashita, O.; Kobayashi, C.; Mori, T.; Sugita, Y.; Tama, F. Flexible fitting to cryo-EM density map using ensemble molecular dynamics simulations. J. Comput. Chem. 2017, 38, 1447–1461. [Google Scholar] [CrossRef]
- de la Rosa-Trevin, J.M.; Quintana, A.; Del Cano, L.; Zaldivar, A.; Foche, I.; Gutierrez, J.; Gomez-Blanco, J.; Burguet-Castell, J.; Cuenca-Alba, J.; Abrishami, V.; et al. Scipion: A software framework toward integration, reproducibility and validation in 3D electron microscopy. J. Struct. Biol. 2016, 195, 93–99. [Google Scholar] [CrossRef]
- Gao, H.; Li, F.; Ji, Z.; Shi, Z.; Li, Y.; Yu, H. Cryo-EM structures of human p97 double hexamer capture potentiated ATPase-competent state. Cell Discov. 2022, 8, 19. [Google Scholar] [CrossRef]
- Bai, X.C.; Rajendra, E.; Yang, G.; Shi, Y.; Scheres, S.H. Sampling the conformational space of the catalytic subunit of human gamma-secretase. eLife 2015, 4, e11182. [Google Scholar] [CrossRef]
- de la Rosa-Trevin, J.M.; Oton, J.; Marabini, R.; Zaldivar, A.; Vargas, J.; Carazo, J.M.; Sorzano, C.O. Xmipp 3.0: An improved software suite for image processing in electron microscopy. J. Struct. Biol. 2013, 184, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Baker, M.L.; Ju, T.; Chiu, W. Identification of secondary structure elements in intermediate-resolution density maps. Structure 2007, 15, 7–19. [Google Scholar] [CrossRef] [PubMed]
- von Loeffelholz, O.; Natchiar, S.K.; Djabeur, N.; Myasnikov, A.G.; Kratzat, H.; Menetret, J.F.; Hazemann, I.; Klaholz, B.P. Focused classification and refinement in high-resolution cryo-EM structural analysis of ribosome complexes. Curr. Opin. Struct. Biol. 2017, 46, 140–148. [Google Scholar] [CrossRef] [PubMed]
- Braxton, J.R.; Altobelli, C.R.; Tucker, M.R.; Tse, E.; Thwin, A.C.; Arkin, M.R.; Southworth, D.R. The p97/VCP adaptor UBXD1 drives AAA+ remodeling and ring opening through multi-domain tethered interactions. Nat. Struct. Mol. Biol. 2023, 30, 2009–2019. [Google Scholar] [CrossRef] [PubMed]
- Conicella, A.E.; Huang, R.; Ripstein, Z.A.; Nguyen, A.; Wang, E.; Lohr, T.; Schuck, P.; Vendruscolo, M.; Rubinstein, J.L.; Kay, L.E. An intrinsically disordered motif regulates the interaction between the p47 adaptor and the p97 AAA+ ATPase. Proc. Natl. Acad. Sci. USA 2020, 117, 26226–26236. [Google Scholar] [CrossRef] [PubMed]
- Blueggel, M.; Kroening, A.; Kracht, M.; van den Boom, J.; Dabisch, M.; Goehring, A.; Kaschani, F.; Kaiser, M.; Bayer, P.; Meyer, H.; et al. The UBX domain in UBXD1 organizes ubiquitin binding at the C-terminus of the VCP/p97 AAA-ATPase. Nat. Commun. 2023, 14, 3258. [Google Scholar] [CrossRef] [PubMed]
- Rao, B.; Wang, Q.; Yao, D.; Xia, Y.; Li, W.; Xie, Y.; Li, S.; Cao, M.; Shen, Y.; Qin, A.; et al. The cryo-EM structure of the human ERAD retrotranslocation complex. Sci. Adv. 2023, 9, eadi5656. [Google Scholar] [CrossRef] [PubMed]
- Halawani, D.; LeBlanc, A.C.; Rouiller, I.; Michnick, S.W.; Servant, M.J.; Latterich, M. Hereditary inclusion body myopathy-linked p97/VCP mutations in the NH2 domain and the D1 ring modulate p97/VCP ATPase activity and D2 ring conformation. Mol. Cell. Biol. 2009, 29, 4484–4494. [Google Scholar] [CrossRef]
- Goddard, T.D.; Huang, C.C.; Meng, E.C.; Pettersen, E.F.; Couch, G.S.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. A Publ. Protein Soc. 2018, 27, 14–25. [Google Scholar] [CrossRef]
- Strelak, D.; Jimenez-Moreno, A.; Vilas, J.L.; Ramirez-Aportela, E.; Sanchez-Garcia, R.; Maluenda, D.; Vargas, J.; Herreros, D.; Fernandez-Gimenez, E.; de Isidro-Gomez, F.P.; et al. Advances in Xmipp for Cryo-Electron Microscopy: From Xmipp to Scipion. Molecules 2021, 26, 6224. [Google Scholar] [CrossRef] [PubMed]
- Sorzano, C.O.; Marabini, R.; Velazquez-Muriel, J.; Bilbao-Castro, J.R.; Scheres, S.H.; Carazo, J.M.; Pascual-Montano, A. XMIPP: A new generation of an open-source image processing package for electron microscopy. J. Struct. Biol. 2004, 148, 194–204. [Google Scholar] [CrossRef]
- Harastani, M.; Vuillemot, R.; Hamitouche, I.; Moghadam, N.B.; Jonic, S. ContinuousFlex: Software package for analyzing continuous conformational variability of macromolecules in cryo electron microscopy and tomography data. J. Struct. Biol. 2022, 214, 107906. [Google Scholar] [CrossRef] [PubMed]
- McInnes, L.; Healy, J.; Melville, J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. ArXiv 2018, arXiv:1802.03426. [Google Scholar]
- Tipping, M.E.; Bishop, C.M. Probabilistic principal component analysis. J. R. Stat. Soc. B 1999, 61, 611–622. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valimehr, S.; Vuillemot, R.; Kazemi, M.; Jonic, S.; Rouiller, I. Analysis of the Conformational Landscape of the N-Domains of the AAA ATPase p97: Disentangling the Continuous Conformational Variability in Partially Symmetrical Complexes. Int. J. Mol. Sci. 2024, 25, 3371. https://doi.org/10.3390/ijms25063371
Valimehr S, Vuillemot R, Kazemi M, Jonic S, Rouiller I. Analysis of the Conformational Landscape of the N-Domains of the AAA ATPase p97: Disentangling the Continuous Conformational Variability in Partially Symmetrical Complexes. International Journal of Molecular Sciences. 2024; 25(6):3371. https://doi.org/10.3390/ijms25063371
Chicago/Turabian StyleValimehr, Sepideh, Rémi Vuillemot, Mohsen Kazemi, Slavica Jonic, and Isabelle Rouiller. 2024. "Analysis of the Conformational Landscape of the N-Domains of the AAA ATPase p97: Disentangling the Continuous Conformational Variability in Partially Symmetrical Complexes" International Journal of Molecular Sciences 25, no. 6: 3371. https://doi.org/10.3390/ijms25063371
APA StyleValimehr, S., Vuillemot, R., Kazemi, M., Jonic, S., & Rouiller, I. (2024). Analysis of the Conformational Landscape of the N-Domains of the AAA ATPase p97: Disentangling the Continuous Conformational Variability in Partially Symmetrical Complexes. International Journal of Molecular Sciences, 25(6), 3371. https://doi.org/10.3390/ijms25063371







