The TAM Subfamily of Receptor Tyrosine Kinases: The Early Years
Abstract
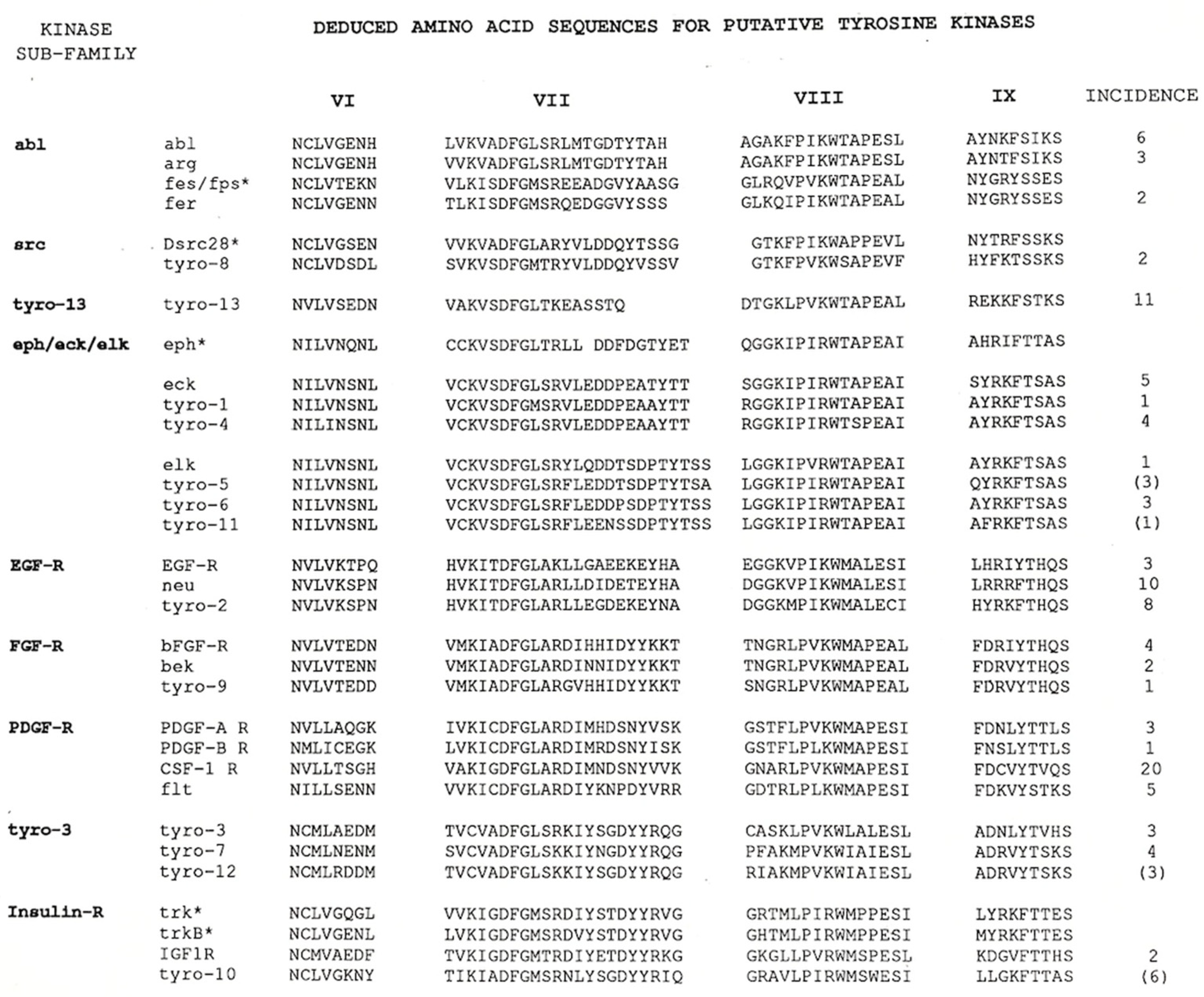
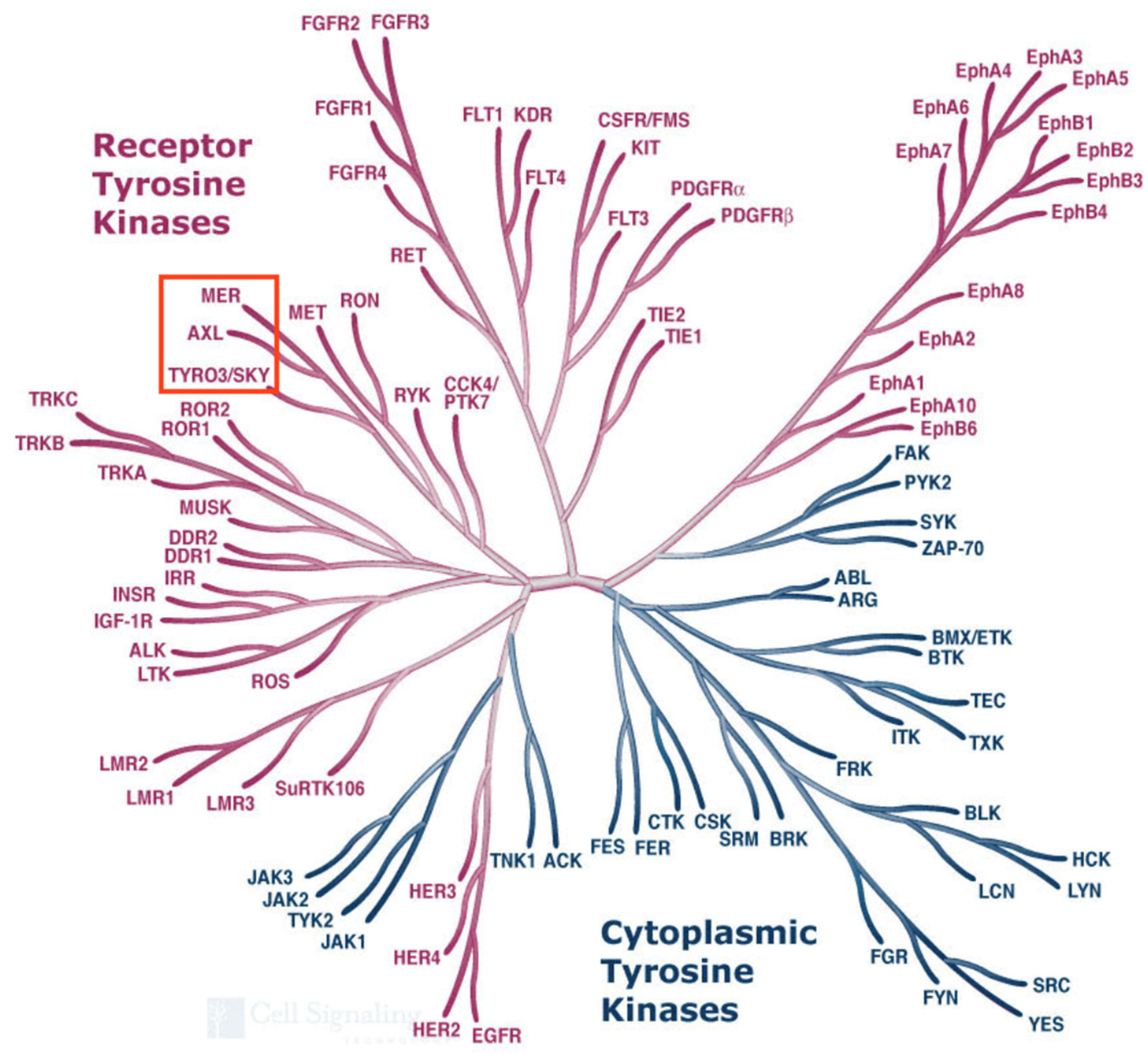
1. The Cloning of the “Tyros”
2. The Cloning of Axl
3. The Cloning of Mer (MERTK)
4. The Cloning of Tyro3
5. The Identification of the TAM Subfamily Ligands
6. The Lemke Lab Has Played a Major Role in Elucidating the Biological Relevance of the TAMs
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CML | chronic myelogenous leukemia |
| FN | Fibronectin |
| Gas6 | growth arrest-specific gene 6 |
| Ig | Immunoglobulin |
| P | Postnatal |
| PCR | polymerase chain reaction |
| ProS | protein S |
| RTK | receptor protein tyrosine kinase |
| TAM | Tyro3 Axl and Mer |
References
- Shafit-Zagardo, B.; Gruber, R.C.; DuBois, J.C. The role of TAM family receptors and ligands in the nervous system: From development to pathobiology. Pharmacol. Ther. 2018, 188, 97–117. [Google Scholar] [CrossRef]
- Lemke, G. Biology of the TAM receptors. Cold Spring Harb. Perspect. Biol. 2013, 5, a009076. [Google Scholar] [CrossRef]
- DeRyckere, D.; Huelse, J.M.; Earp, H.S.; Graham, D.K. TAM family kinases as therapeutic targets at the interface of cancer and immunity. Nat. Rev. Clin. Oncol. 2023, 20, 755–779. [Google Scholar] [CrossRef]
- Burstyn-Cohen, T. TAM receptor signaling in development. Int. J. Dev. Biol. 2017, 61, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Rothlin, C.V.; Carrera-Silva, E.A.; Bosurgi, L.; Ghosh, S. TAM receptor signaling in immune homeostasis. Annu. Rev. Immunol. 2015, 33, 355–391. [Google Scholar] [CrossRef]
- Basler, K.; Hafen, E. Ubiquitous expression of sevenless: Position-dependent specification of cell fate. Science 1989, 243, 931–934. [Google Scholar] [CrossRef] [PubMed]
- Lindberg, R.A.; Hunter, T. cDNA cloning and characterization of eck, an epithelial cell receptor protein-tyrosine kinase in the eph/elk family of protein kinases. Mol. Cell. Biol. 1990, 10, 6316–6324. [Google Scholar]
- Wilks, A.F. Two putative protein-tyrosine kinases identified by application of the polymerase chain reaction. Proc. Natl. Acad. Sci. USA 1989, 86, 1603–1607. [Google Scholar] [CrossRef]
- Hedrick, S.M.; Cohen, D.I.; Nielsen, E.A.; Davis, M.M. Isolation of cDNA clones encoding T cell-specific membrane-associated proteins. Nature 1984, 308, 149–153. [Google Scholar] [CrossRef]
- Lai, C.; Lemke, G. An extended family of protein-tyrosine kinase genes differentially expressed in the vertebrate nervous system. Neuron 1991, 6, 691–704. [Google Scholar] [CrossRef] [PubMed]
- Klein, R.; Jing, S.Q.; Nanduri, V.; O’Rourke, E.; Barbacid, M. The trk proto-oncogene encodes a receptor for nerve growth factor. Cell 1991, 65, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava, A.; Radziejewski, C.; Campbell, E.; Kovac, L.; McGlynn, M.; Ryan, T.E.; Davis, S.; Goldfarb, M.P.; Glass, D.J.; Lemke, G.; et al. An orphan receptor tyrosine kinase family whose members serve as nonintegrin collagen receptors. Mol. Cell 1997, 1, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Vogel, W.; Gish, G.D.; Alves, F.; Pawson, T. The discoidin domain receptor tyrosine kinases are activated by collagen. Mol. Cell 1997, 1, 13–23. [Google Scholar] [CrossRef]
- Manning, G.; Whyte, D.B.; Martinez, R.; Hunter, T.; Sudarsanam, S. The protein kinase complement of the human genome. Science 2002, 298, 1912–1934. [Google Scholar] [CrossRef]
- Gassmann, M.; Casagranda, F.; Orioli, D.; Simon, H.; Lai, C.; Klein, R.; Lemke, G. Aberrant neural and cardiac development in mice lacking the ErbB4 neuregulin receptor. Nature 1995, 378, 390–394. [Google Scholar] [CrossRef]
- Reber, M.; Hindges, R.; Lemke, G. Eph receptors and ephrin ligands in axon guidance. Adv. Exp. Med. Biol. 2007, 621, 32–49. [Google Scholar]
- Partanen, J.; Makela, T.P.; Eerola, E.; Korhonen, J.; Hirvonen, H.; Claesson-Welsh, L.; Alitalo, K. FGFR-4, a novel acidic fibroblast growth factor receptor with a distinct expression pattern. EMBO J. 1991, 10, 1347–1354. [Google Scholar] [CrossRef]
- Partanen, J.; Makela, T.P.; Alitalo, R.; Lehvaslaiho, H.; Alitalo, K. Putative tyrosine kinases expressed in K-562 human leukemia cells. Proc. Natl. Acad. Sci. USA 1990, 87, 8913–8917. [Google Scholar] [CrossRef]
- Fujimoto, J.; Yamamoto, T. brt, a mouse gene encoding a novel receptor-type protein-tyrosine kinase, is preferentially expressed in the brain. Oncogene 1994, 9, 693–698. [Google Scholar]
- Crosier, K.E.; Hall, L.R.; Lewis, P.M.; Morris, C.M.; Wood, C.R.; Morris, J.C.; Crosier, P.S. Isolation and characterization of the human DTK receptor tyrosine kinase. Growth Factors 1994, 11, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Crosier, P.S.; Lewis, P.M.; Hall, L.R.; Vitas, M.R.; Morris, C.M.; Beier, D.R.; Wood, C.R.; Crosier, K.E. Isolation of a receptor tyrosine kinase (DTK) from embryonic stem cells: Structure, genetic mapping and analysis of expression. Growth Factors 1994, 11, 125–136. [Google Scholar] [CrossRef] [PubMed]
- Biscardi, J.S.; Denhez, F.; Buehler, G.F.; Chesnutt, D.A.; Baragona, S.C.; O’Bryan, J.P.; Der, C.J.; Fiordalisi, J.J.; Fults, D.W.; Maness, P.F. Rek, a gene expressed in retina and brain, encodes a receptor tyrosine kinase of the Axl/Tyro3 family. J. Biol. Chem. 1996, 271, 29049–29059. [Google Scholar] [CrossRef][Green Version]
- Mark, M.R.; Scadden, D.T.; Wang, Z.; Gu, Q.; Goddard, A.; Godowski, P.J. rse, a novel receptor-type tyrosine kinase with homology to Axl/Ufo, is expressed at high levels in the brain. J. Biol. Chem. 1994, 269, 10720–10728. [Google Scholar] [CrossRef]
- Ohashi, K.; Mizuno, K.; Kuma, K.; Miyata, T.; Nakamura, T. Cloning of the cDNA for a novel receptor tyrosine kinase, Sky, predominantly expressed in brain. Oncogene 1994, 9, 699–705. [Google Scholar]
- Dai, W.; Pan, H.; Hassanain, H.; Gupta, S.L.; Murphy, M.J., Jr. Molecular cloning of a novel receptor tyrosine kinase, tif, highly expressed in human ovary and testis. Oncogene 1994, 9, 975–979. [Google Scholar] [PubMed]
- Rescigno, J.; Mansukhani, A.; Basilico, C. A putative receptor tyrosine kinase with unique structural topology. Oncogene 1991, 6, 1909–1913. [Google Scholar] [PubMed]
- O’Bryan, J.P.; Frye, R.A.; Cogswell, P.C.; Neubauer, A.; Kitch, B.; Prokop, C.; Espinosa, R., 3rd; Le Beau, M.M.; Earp, H.S.; Liu, E.T. axl, a transforming gene isolated from primary human myeloid leukemia cells, encodes a novel receptor tyrosine kinase. Mol. Cell. Biol. 1991, 11, 5016–5031. [Google Scholar]
- Janssen, J.W.; Schulz, A.S.; Steenvoorden, A.C.; Schmidberger, M.; Strehl, S.; Ambros, P.F.; Bartram, C.R. A novel putative tyrosine kinase receptor with oncogenic potential. Oncogene 1991, 6, 2113–2120. [Google Scholar]
- Jia, R.; Hanafusa, H. The proto-oncogene of v-eyk (v-ryk) is a novel receptor-type protein tyrosine kinase with extracellular Ig/GN-III domains. J. Biol. Chem. 1994, 269, 1839–1844. [Google Scholar] [CrossRef]
- Graham, D.K.; Bowman, G.W.; Dawson, T.L.; Stanford, W.L.; Earp, H.S.; Snodgrass, H.R. Cloning and developmental expression analysis of the murine c-mer tyrosine kinase. Oncogene 1995, 10, 2349–2359. [Google Scholar]
- Graham, D.K.; Dawson, T.L.; Mullaney, D.L.; Snodgrass, H.R.; Earp, H.S. Cloning and mRNA expression analysis of a novel human protooncogene, c-mer. Cell Growth Differ. 1994, 5, 647–657. [Google Scholar]
- Ling, L.; Kung, H.J. Mitogenic signals and transforming potential of Nyk, a newly identified neural cell adhesion molecule-related receptor tyrosine kinase. Mol. Cell. Biol. 1995, 15, 6582–6592. [Google Scholar] [CrossRef]
- Liu, E.; Hjelle, B.; Bishop, J.M. Transforming genes in chronic myelogenous leukemia. Proc. Natl. Acad. Sci. USA 1988, 85, 1952–1956. [Google Scholar] [CrossRef]
- Jia, R.; Mayer, B.J.; Hanafusa, T.; Hanafusa, H. A novel oncogene, v-ryk, encoding a truncated receptor tyrosine kinase is transduced into the RPL30 virus without loss of viral sequences. J. Virol. 1992, 66, 5975–5987. [Google Scholar] [CrossRef]
- Hovens, C.M.; Stacker, S.A.; Andres, A.C.; Harpur, A.G.; Ziemiecki, A.; Wilks, A.F. RYK, a receptor tyrosine kinase-related molecule with unusual kinase domain motifs. Proc. Natl. Acad. Sci. USA 1992, 89, 11818–11822. [Google Scholar] [CrossRef]
- Edelman, G.M.; Crossin, K.L. Cell adhesion molecules: Implications for a molecular histology. Annu. Rev. Biochem. 1991, 60, 155–190. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.; Gore, M.; Lemke, G. Structure, expression, and activity of Tyro 3, a neural adhesion-related receptor tyrosine kinase. Oncogene 1994, 9, 2567–2578. [Google Scholar] [PubMed]
- Caberoy, N.B.; Zhou, Y.; Li, W. Tubby and tubby-like protein 1 are new MerTK ligands for phagocytosis. EMBO J. 2010, 29, 3898–3910. [Google Scholar] [CrossRef] [PubMed]
- Caberoy, N.B.; Alvarado, G.; Bigcas, J.L.; Li, W. Galectin-3 is a new MerTK-specific eat-me signal. J. Cell Physiol. 2012, 227, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Manfioletti, G.; Brancolini, C.; Avanzi, G.; Schneider, C. The protein encoded by a growth arrest-specific gene (gas6) is a new member of the vitamin K-dependent proteins related to protein S, a negative coregulator in the blood coagulation cascade. Mol. Cell. Biol. 1993, 13, 4976–4985. [Google Scholar] [PubMed]
- Dahlback, B. Protein S and C4b-binding protein: Components involved in the regulation of the protein C anticoagulant system. Thromb. Haemost. 1991, 66, 49–61. [Google Scholar] [CrossRef]
- Heeb, M.J. Role of the PROS1 gene in thrombosis: Lessons and controversies. Expert. Rev. Hematol. 2008, 1, 9–12. [Google Scholar] [CrossRef]
- Hafizi, S.; Dahlback, B. Gas6 and protein S. FEBS J. 2006, 273, 5231–5244. [Google Scholar] [CrossRef] [PubMed]
- Nakano, T.; Ishimoto, Y.; Kishino, J.; Umeda, M.; Inoue, K.; Nagata, K.; Ohashi, K.; Mizuno, K.; Arita, H. Cell adhesion to phosphatidylserine mediated by a product of growth arrest-specific gene 6. J. Biol. Chem. 1997, 272, 29411–29414. [Google Scholar] [CrossRef] [PubMed]
- Lew, E.D.; Oh, J.; Burrola, P.G.; Lax, I.; Zagorska, A.; Traves, P.G.; Schlessinger, J.; Lemke, G. Differential TAM receptor-ligand-phospholipid interactions delimit differential TAM bioactivities. eLife 2014, 3, e03385. [Google Scholar] [CrossRef] [PubMed]
- Lemke, G. Phosphatidylserine Is the Signal for TAM Receptors and Their Ligands. Trends Biochem. Sci. 2017, 42, 738–748. [Google Scholar] [CrossRef] [PubMed]
- Varnum, B.C.; Young, C.; Elliott, G.; Garcia, A.; Bartley, T.D.; Fridell, Y.W.; Hunt, R.W.; Trail, G.; Clogston, C.; Toso, R.J.; et al. Axl receptor tyrosine kinase stimulated by the vitamin K-dependent protein encoded by growth-arrest-specific gene 6. Nature 1995, 373, 623–626. [Google Scholar] [CrossRef] [PubMed]
- Stitt, T.N.; Conn, G.; Gore, M.; Lai, C.; Bruno, J.; Radziejewski, C.; Mattsson, K.; Fisher, J.; Gies, D.R.; Jones, P.F.; et al. The anticoagulation factor protein S and its relative, Gas6, are ligands for the Tyro 3/Axl family of receptor tyrosine kinases. Cell 1995, 80, 661–670. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, K.; Nagata, K.; Toshima, J.; Nakano, T.; Arita, H.; Tsuda, H.; Suzuki, K.; Mizuno, K. Stimulation of sky receptor tyrosine kinase by the product of growth arrest-specific gene 6. J. Biol. Chem. 1995, 270, 22681–22684. [Google Scholar] [CrossRef]
- Godowski, P.J.; Mark, M.R.; Chen, J.; Sadick, M.D.; Raab, H.; Hammonds, R.G. Reevaluation of the roles of protein S and Gas6 as ligands for the receptor tyrosine kinase Rse/Tyro 3. Cell 1995, 82, 355–358. [Google Scholar] [CrossRef]
- Mark, M.R.; Chen, J.; Hammonds, R.G.; Sadick, M.; Godowsk, P.J. Characterization of Gas6, a member of the superfamily of G domain-containing proteins, as a ligand for Rse and Axl. J. Biol. Chem. 1996, 271, 9785–9789. [Google Scholar] [CrossRef]
- Nagata, K.; Ohashi, K.; Nakano, T.; Arita, H.; Zong, C.; Hanafusa, H.; Mizuno, K. Identification of the product of growth arrest-specific gene 6 as a common ligand for Axl, Sky, and Mer receptor tyrosine kinases. J. Biol. Chem. 1996, 271, 30022–30027. [Google Scholar] [CrossRef]
- Nyberg, P.; He, X.; Hardig, Y.; Dahlback, B.; Garcia de Frutos, P. Stimulation of Sky tyrosine phosphorylation by bovine protein S--domains involved in the receptor-ligand interaction. Eur. J. Biochem. FEBS 1997, 246, 147–154. [Google Scholar] [CrossRef]
- Burstyn-Cohen, T.; Heeb, M.J.; Lemke, G. Lack of protein S in mice causes embryonic lethal coagulopathy and vascular dysgenesis. J. Clin. Investig. 2009, 119, 2942–2953. [Google Scholar] [CrossRef]
- Burstyn-Cohen, T.; Lew, E.D.; Traves, P.G.; Burrola, P.G.; Hash, J.C.; Lemke, G. Genetic dissection of TAM receptor-ligand interaction in retinal pigment epithelial cell phagocytosis. Neuron 2012, 76, 1123–1132. [Google Scholar] [CrossRef] [PubMed]
- Dransfield, I.; Zagorska, A.; Lew, E.D.; Michail, K.; Lemke, G. Mer receptor tyrosine kinase mediates both tethering and phagocytosis of apoptotic cells. Cell Death Dis. 2015, 6, e1646. [Google Scholar] [CrossRef] [PubMed]
- Fourgeaud, L.; Traves, P.G.; Tufail, Y.; Leal-Bailey, H.; Lew, E.D.; Burrola, P.G.; Callaway, P.; Zagorska, A.; Rothlin, C.V.; Nimmerjahn, A.; et al. TAM receptors regulate multiple features of microglial physiology. Nature 2016, 532, 240–244. [Google Scholar] [CrossRef] [PubMed]
- Happonen, K.E.; Burrola, P.G.; Lemke, G. Regulation of brain endothelial cell physiology by the TAM receptor tyrosine kinase Mer. Commun. Biol. 2023, 6, 916. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Happonen, K.E.; Burrola, P.G.; O’Connor, C.; Hah, N.; Huang, L.; Nimmerjahn, A.; Lemke, G. Microglia use TAM receptors to detect and engulf amyloid beta plaques. Nat. Immunol. 2021, 22, 586–594. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Lemke, G. Early death in a mouse model of Alzheimer’s disease exacerbated by microglial loss of TAM receptor signaling. Proc. Natl. Acad. Sci. USA 2022, 119, e2204306119. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Gore, M.; Zhang, Q.; Camenisch, T.; Boast, S.; Casagranda, F.; Lai, C.; Skinner, M.K.; Klein, R.; Matsushima, G.K.; et al. Tyro-3 family receptors are essential regulators of mammalian spermatogenesis. Nature 1999, 398, 723–728. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Lemke, G. Homeostatic regulation of the immune system by receptor tyrosine kinases of the Tyro 3 family. Science 2001, 293, 306–311. [Google Scholar] [CrossRef] [PubMed]
- Miao, S.; Fourgeaud, L.; Burrola, P.G.; Stern, S.; Zhang, Y.; Happonen, K.E.; Novak, S.W.; Gage, F.H.; Lemke, G. Tyro3 promotes the maturation of glutamatergic synapses. Front. Neurosci. 2024, 18, 1327423. [Google Scholar] [CrossRef]
- Prasad, D.; Rothlin, C.V.; Burrola, P.; Burstyn-Cohen, T.; Lu, Q.; Garcia de Frutos, P.; Lemke, G. TAM receptor function in the retinal pigment epithelium. Mol. Cell. Neurosci. 2006, 33, 96–108. [Google Scholar] [CrossRef] [PubMed]
- Roth, C.; Rothlin, C.; Riou, S.; Raulet, D.H.; Lemke, G. Stromal-cell regulation of natural killer cell differentiation. J. Mol. Med. 2007, 85, 1047–1056. [Google Scholar] [CrossRef]
- Rothlin, C.V.; Ghosh, S.; Zuniga, E.I.; Oldstone, M.B.; Lemke, G. TAM receptors are pleiotropic inhibitors of the innate immune response. Cell 2007, 131, 1124–1136. [Google Scholar] [CrossRef]
- Zagorska, A.; Traves, P.G.; Lew, E.D.; Dransfield, I.; Lemke, G. Diversification of TAM receptor tyrosine kinase function. Nat. Immunol. 2014, 15, 920–928. [Google Scholar] [CrossRef] [PubMed]
- Rothlin, C.V.; Lemke, G. TAM receptor signaling and autoimmune disease. Curr. Opin. Immunol. 2010, 22, 740–746. [Google Scholar] [CrossRef]
- Lemke, G.; Huang, Y. The dense-core plaques of Alzheimer’s disease are granulomas. J. Exp. Med. 2022, 219, e20212477. [Google Scholar] [CrossRef]
| Subfamily | Human | Rodent | Avian |
|---|---|---|---|
| Tyro3 | RSE, SKY, DTK, TIF | Tyro3, Dtk, Brt | REK |
| Axl | AXL, UFO | Tyro7, Ark | |
| Mer | MERTK, NYK | Tyro12, Mertk | EYK |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prieto, A.L.; Lai, C. The TAM Subfamily of Receptor Tyrosine Kinases: The Early Years. Int. J. Mol. Sci. 2024, 25, 3369. https://doi.org/10.3390/ijms25063369
Prieto AL, Lai C. The TAM Subfamily of Receptor Tyrosine Kinases: The Early Years. International Journal of Molecular Sciences. 2024; 25(6):3369. https://doi.org/10.3390/ijms25063369
Chicago/Turabian StylePrieto, Anne L., and Cary Lai. 2024. "The TAM Subfamily of Receptor Tyrosine Kinases: The Early Years" International Journal of Molecular Sciences 25, no. 6: 3369. https://doi.org/10.3390/ijms25063369
APA StylePrieto, A. L., & Lai, C. (2024). The TAM Subfamily of Receptor Tyrosine Kinases: The Early Years. International Journal of Molecular Sciences, 25(6), 3369. https://doi.org/10.3390/ijms25063369





