Abstract
Strigolactones (SLs) were recently defined as a novel class of plant hormones that act as key regulators of diverse developmental processes and environmental responses. Much research has focused on SL biosynthesis and signaling in roots and shoots, but little is known about whether SLs are produced in early developing seeds and about their roles in ovule development after fertilization. This study revealed that the fertilized ovules and early developing pericarp in Xanthoceras sorbifolium produced minute amounts of two strigolactones: 5-deoxystrigol and strigol. Their content decreased in the plants with the addition of exogenous phosphate (Pi) compared to those without the Pi treatment. The exogenous application of an SL analog (GR24) and a specific inhibitor of SL biosynthesis (TIS108) affected early seed development and fruit set. In the Xanthoceras genome, we identified 69 potential homologs of genes involved in SL biological synthesis and signaling. Using RNA-seq to characterize the expression of these genes in the fertilized ovules, 37 genes were found to express differently in the fertilized ovules that were aborting compared to the normally developing ovules. A transcriptome analysis also revealed that in normally developing ovules after fertilization, 12 potential invertase genes were actively expressed. Hexoses (glucose and fructose) accumulated at high concentrations in normally developing ovules during syncytial endosperm development. In contrast, a low ratio of hexose and sucrose levels was detected in aborting ovules with a high strigolactone content. XsD14 virus-induced gene silencing (VIGS) increased the hexose content in fertilized ovules and induced the proliferation of endosperm free nuclei, thereby promoting early seed development and fruit set. We propose that the crosstalk between sugar and strigolactone signals may be an important part of a system that accurately regulates the abortion of ovules after fertilization. This study is useful for understanding the mechanisms underlying ovule abortion, which will serve as a guide for genetic or chemical approaches to promote seed yield in Xanthoceras.
1. Introduction
Strigolactones (SLs) are a group of carotenoid-derived compounds that were recently defined as a novel class of plant hormones [1]. These signaling molecules were found to regulate diverse developmental processes [2,3,4,5,6] and environmental responses [7,8], such as inhibiting the outgrowth of axillary buds, regulating root development, accelerating leaf senescence, and mediating plants’ adaptation to nutrient deficiency. SLs were initially discovered because of their ability to stimulate the seed germination of root-parasitic plants such as Striga lutea. As a germination stimulant for S. lutea, strigol was the first SL that was isolated from the root exudate of cotton [9]. To date, more than 20 SLs have been extracted and identified in a variety of plant species [10]. Strigolactones are produced at high levels in the roots of various plant species under phosphate (Pi)-deficient conditions, stimulating changes in shoot and root architecture that enable them to adapt to environmental stress and improve their ability to acquire Pi [11,12].
SL-related mutants have been identified in several plant species, such as in Arabidopsis more axillary growth (max) mutants [13,14,15,16], lateral branching oxidoreductase (lbo) mutants [16], suppressor of more axillary growth2-like 6,7,8 (smaxl 6,7,8) mutants [17], pea (Pisum sativum) ramosus (rms) mutants [18,19,20], petunia (Petunia hybrida) decreased apical dominance (dad) mutants [21,22], and rice (Oryza sativa) dwarf (d) or high-tillering dwarf (htd) mutants [23]. Based on studies with mutants, eight genes required for SL biosynthesis and signaling have been identified to date [24]. Among them, five have been shown to be involved in SL biosynthesis, including D27, MAX3/D17/HTD1/RMS5/DAD3, MAX4/D10/RMS1/DAD1, MAX1, and LBO, which encode β-carotene isomerase, carotenoid cleavage dioxygenase 7 (CCD7), CCD8, cytochrome P450 monooxygenase, 2-oxoglutarate, and Fe(II)-dependent dioxygenase (LBO), respectively. D27, CCD7, CCD8, and MAX1 have been identified as canonical SL biosynthetic enzymes [25]. The two-oxoglutarate-dependent dioxygenase LBO is considered to be a late-acting, noncanonical SL biosynthetic enzyme in Arabidopsis [17].
Three genes encoding an α/β fold hydrolase, an F-box leucine-rich protein, and a repressor protein known as D53 mediate the perception and signaling pathway of SLs. D14 in rice [26], AtD14 in Arabidopsis [27], RMS3 in pea [28], and DAD2 in petunia [29] encode an α/β-fold hydrolase that cleaves SLs and covalently binds to one of the cleavage products. This binding induces a conformational change in the receptor protein structure, leading to the activation of D14 [26,30]. The activated D14 interacts with the leucine-rich-repeat F-box protein encoded by D3/MAX2/PhMAX2/RMS4 (in rice/Arabidopsis/petunia/pea, respectively) to form an Skp-Cullin-F-box (SCF) complex, SCF-D3-D14 [31]. This complex further activates the 26S proteasome and degrades transcription repressors encoded by D53 in rice (SMXL6/7/8 in Arabidopsis) in the presence of SL to trigger various SL-regulated plant responses [23].
Sugars not only provide a source of cellular energy and metabolic precursors for numerous biosynthetic pathways, but also function as signaling molecules to regulate gene expression in response to developmental and environmental cues [32,33]. A number of plant metabolic, physiological, and developmental processes are regulated in response to changing levels of soluble sugars [34]. Recent studies have provided evidence of interactions between sugar and the strigolactone response [35]. Increased sugar availability alleviates the inhibitory effect of strigolactone on shoot branching in pea, rose, chrysanthemum, and rice [36,37,38]. In many species studied, the rate of unloading and utilization of sucrose has been found to be correlated with the activity of invertases in sink organs [39]. Invertase activity is regulated by internal and environmental factors including plant hormones, carbohydrates, and mineral nutrition [40].
Xanthoceras sorbifolium, a tree species of the Sapindaceae family, is an emerging valuable oilseed crop in Northern China [41]. The plant has attracted considerable interest due to the high oil content (from 30% to 36% on a dry matter basis) in its seeds and its favorable oil qualities. The oil is considered an excellent source of unsaturated fatty acids because of its high oleic acid (ca. 28% of the total fatty acids) and linoleic acid (ca. 46%) contents [42]. Xanthoceras oil also contains high levels of vitamin E and nervonic acid, which have various health benefits. Despite its high-quality nutritional oil, its large-scale commercial cultivation has been hampered by low seed productivity. More than 95% of young fruits cannot reach maturity due to the abortion of ovules after fertilization within them. Although cytological observations of the development of fertilized ovules have been detailed in X. sorbifolium [41,43], much less attention has been paid to the regulatory mechanisms of ovule abortion in the plant. Strigolactones have been suggested to be important regulators of signaling and responses related to nutritional deficiencies [7]. This enticed us to examine whether SLs are produced in fertilized ovules and to assess whether SL signaling is correlated with Pi deficiency, invertase activity, and sugar availability in ovules after fertilization. Using multiple approaches, we attempted to determine whether SLs are involved in the abortion of fertilized ovules in X. sorbifolium. A better understanding of the mechanisms underlying ovule abortion will serve as a guide for genetic or chemical approaches to promote seed yield in Xanthoceras.
2. Results
2.1. The Presence of Strigolactones in Ovules after Fertilization and the Effect of Exogenous Pi Application on Their Content
Most SLs have been identified and quantified in root exudates and extracts, whereas studies for SL identification and quantification in other tissues are rare. Here, we determined the presence and level of endogenous SLs in early developing seeds and pericarps using LC-MS/MS and compared the differences between plants with or without an exogenous Pi application treatment. The results indicated that the fertilized ovules and early developing pericarps produce minute amounts of two strigolactones: 5-deoxystrigol and strigol (Figure 1). The contents of 5-deoxystrigol and strigol decreased by 54.39% and 61.03%, respectively, in the fertilized ovules of the plants with an exogenous Pi supply compared with those in untreated Pi-deficient plants (Figure 2). The levels of 5-deoxystrigol and strigol in the young pericarps seemed to change in a similar manner to the fertilized ovules within them (Figure 2). Upon Pi application, the content of strigol was reduced more obviously than that of 5-deoxystrigol in the young pericarps. Pi starvation-induced (PSI) genes are commonly used as markers for the response of plants to low-Pi conditions. PHOSPHATE2 (PHO2), encoding a ubiquitin-conjugating E2 enzyme, is considered a PSI gene in Xanthoceras [43]. To confirm that the plants without the Pi application were under low-Pi conditions, we examined the expression of PHO2 in fertilized ovules. The results showed that transcript levels of PHO2 were increased in the plants without the Pi application in comparison to the plants with the Pi application (Figure 3). These investigations suggest that Pi availability is associated with the production of SLs in early developing seeds and young fruits.
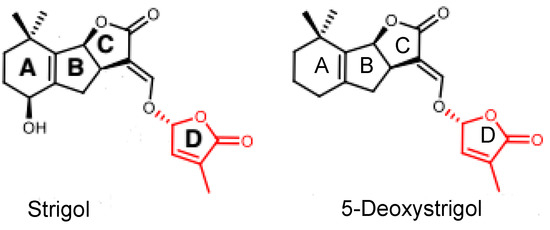
Figure 1.
Chemical structures of two natural strigolactones (SLs). Both strigol and 5-deoxystrigol contain a methylbutenolide moiety (D-ring) connected to a variable tricyclic lactone (ABC-ring), which are termed canonical SLs.
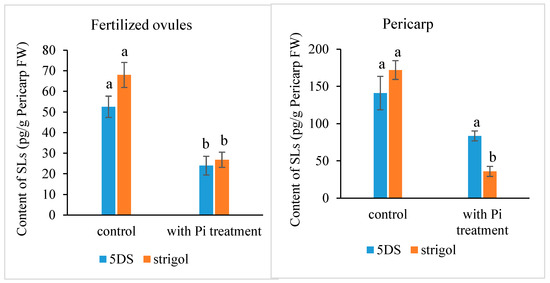
Figure 2.
Strigolactone content in the fertilized ovule and the pericarp in the Xanthoceras plants under phosphate (Pi)-deficient conditions (control) and with Pi treatment. The values are the means of three biological replicates with standard error bars. Different letters represent significant differences at p < 0.05 (ANOVA, Tukey’s HSD test). 5DS: 5-deoxystrigol.
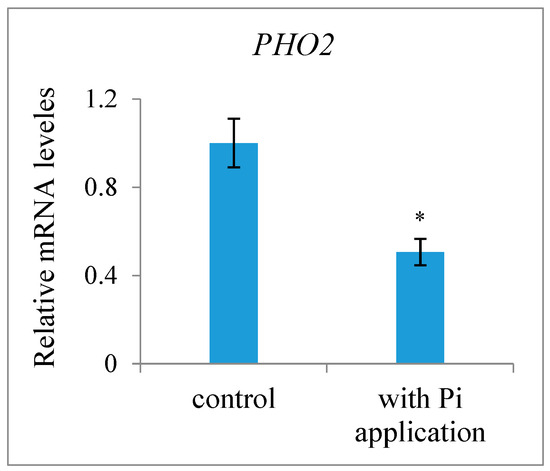
Figure 3.
Effect of exogenous phosphate (Pi) application on expression level of the Pi starvation-induced gene PHO2 in fertilized ovules of the Xanthoceras plants without Pi application and with Pi application, which were normalized to the expression level of actin-2 as a reference. The values are the means of five biological replicates with standard error bars. Asterisks indicate significant difference at p < 0.05 (ANOVA, Tukey’s HSD test).
To elucidate the effect of SL levels on the development of fertilized ovules, we performed comparative cytological studies on the ovules after fertilization. In the fertilized ovules showing a decreased SL content, the embryo sac volume was more quickly enlarged relative to that of the fertilized ovules with high SL levels (Figure 4A,B). The extent of normal nucellar degeneration coincided with the progression of embryo sac growth in fertilized ovules with low SL levels. In contrast, cells in the nucellus tissue were less likely to collapse, embryo sac growth was retarded or completely ceased, and free endosperm nuclei were lower in number and much further apart from each other in the ovules with a high SL content compared with the ovules with reduced SL levels (Figure 4A,B). The resting zygote collapsed in the fertilized ovules with high SL levels, whereas it remained alive in the fertilized ovules with decreased SL contents. These morphological observations show that the abortion process occurs in fertilized ovules with high SL levels, suggesting roles for SLs in the regulation of ovule development after fertilization.
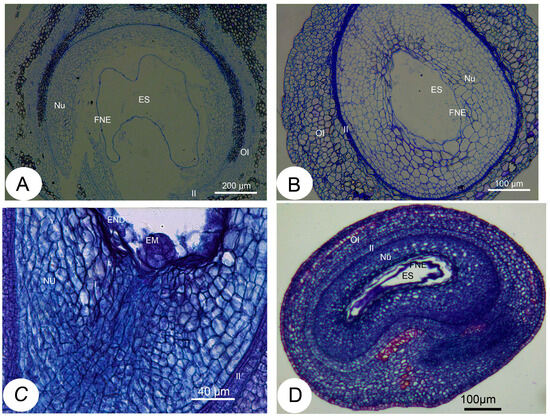
Figure 4.
Histological sections of the ovules after fertilization in Xanthoceras sorbifolium. (A): A portion of a transverse resin section of the normal ovules with low SL levels, showing a rapidly expanding embryo sac and a normally developing syncytial endosperm; (B): A portion of a transverse resin section of the abnormal ovules with high SL contents, showing that development of the embryo sac was arrested and syncytial endosperm degenerated. (C): A many-celled proembryo was formed at the micropylar end of embryo sac in the normally developing ovule 18 days after pollination (DAP). (D). The longitudinal section of the XsD14-silenced ovule 7 DAP, showing delayed degeneration. EM: embryo, END: endosperm, ES: embryo sac, FNE: free nuclear endosperm, II: inner integument, NU: nucellus, OI: outer integument.
2.2. Effect of Exogenous Application of GR24 and TIS108 on Fruit Set and Early Seed Development
Only a terminal racemose inflorescence in a branch can bear functionally female flowers in X. sorbifolium, an andromonoecious species. Although a single terminal inflorescence may produce as many as 30 functionally female flowers, only a very small percentage of them (usually less than 5%) may set fruits due to various limiting factors, including nutrient deficiency [43]. To determine whether strigolactones affect the fruit set and development of fertilized ovules in X. sorbifolium, we treated two groups of terminal inflorescences in each genotype with GR24, a synthetic strigolactone analog, and TIS108, a specific inhibitor of SL biosynthesis. The exogenous application of GR24 decreased the number of mature fruits per inflorescence, but there were some differences among the genotypes (Figure 5). Treatment with GR24 accelerated the abortion process of aberrantly developing fruits and the fertilized ovules within them. The inflorescences treated with TIS108 produced more mature fruits than those in the mock treatment. Treatment with TIS108 delayed the abortion process of the fertilized ovules in the aberrantly developing fruits. We examined the expression levels of the genes encoding Xanthoceras homologs of ACC oxidase (1-aminocyclopropane-1-carboxylic acid oxidase, ACO) ACO2 and ACO3, key enzymes in ethylene biosynthesis [44], in the fertilized ovules of the inflorescences with the GR24 treatment. The results indicated that the GR24 treatment increased the transcript levels of these two ACO genes (Figure 6), suggesting the promotion of ethylene biosynthesis. Additionally, an RT-PCR analysis showed that the ovules treated with GR24 had higher expression levels of genes encoding vacuolar processing enzymes (VPEs), which are executors of vacuole-triggered programmed cell death (PCD), including VPE2 and VPE4 [45]. This suggests that ethylene-mediated PCD may be the cause of the cell degeneration in the GR24-treated ovules. All of these findings point to the possibility that SLs influence ovule abortion following fertilization in Xanthoceras.
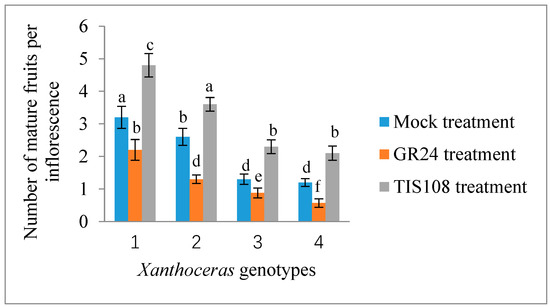
Figure 5.
Response of fruit set to treatment with exogenous GR24, a synthetic strigolactone analog, and TIS108, a specific inhibitor of SL biosynthesis, in inflorescences of Xanthoceras sorbifolium plants of different genotypes. The same concentration of acetone was used as mock treatment. The values are the means of three biological replicates with standard error bars. Different letters represent significant differences at p < 0.05 (ANOVA, Tukey’s HSD test).
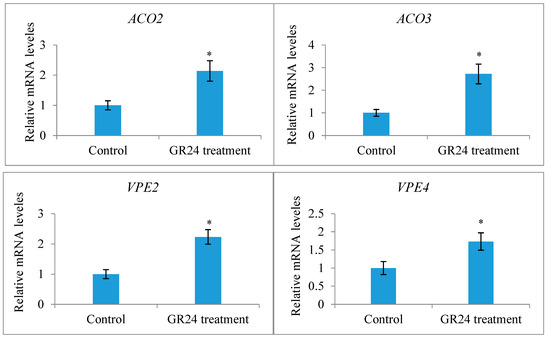
Figure 6.
Expression analysis of the genes encoding Xanthoceras homologs of ACC oxidase (1-aminocyclopropane-1-carboxylic acid oxidase, ACC oxidase, ACO) ACO2 and ACO3 and encoding vacuolar processing enzyme (VPE) VPE2 and VPE4 in the fertilized ovules of the inflorescences with the GR24 or control treatment. The values are the means of three biological replicates with standard error bars. Asterisks indicate significant difference at p < 0.05 (ANOVA, Tukey’s HSD test).
2.3. Genome-Wide Identification of SL Biosynthesis Genes and Their Expression in Normally and Abnormally Developing Ovules after Fertilization
To determine whether the morphological changes in the fertilized ovules mentioned above are associated with the extensive reprogramming of gene expression involved in SL biosynthesis and signaling pathways, next-generation RNA sequencing was performed on the mRNAs obtained from normally (with low levels of SLs) and abnormally (with high levels of SLs) developing ovules 10 DAP, which were collected from the plants with the exogenous Pi supply and the ones without the Pi treatment (under Pi deficiency conditions), respectively [43]. Protein sequences of CCD7 (AT2G44990) and CCD8 (AT4G32810) in Arabidopsis were used to search against the Xanthoceras genome database (http://gigadb.org/dataset/100606, accessed on 14 May 2019) and the transcriptomic database (this study) using the BLAST tool. Subsequently, each subject sequence was blasted against the NCBI (https://blast.ncbi.nlm.nih.gov/, accessed on 23 January 2021) database to confirm that it contained the conserved domain REP65 or PLN02258 [46]. As a result, we identified a total of 19 candidate CCD family genes, including sixteen members of the CCD subfamily and three members of the 9-cis epoxycarotenoid dioxygenase (NCED) subfamily (Table S1). A phylogenetic tree was constructed using the protein sequences of CCDs and NCEDs from Xanthoceras, Arabidopsis, rice, and Forsythia suspensa. The results showed that the CCD family proteins were divided into two clades: CCD and NCED (Figure S1).
The transcriptomic analysis indicated that 13 of the 19 CCD genes were expressed (FPKM > 0.1) in the fertilized ovules (Table S1). Of these, nine genes showed differential gene expression between the normal and abnormal ovules (log2 fold change ≥ 1.5 or ≤−1.5; p ≤ 0.05), and six genes (including XsCCD8) were upregulated in the abnormal ovules. Among the identified CCD family genes, all three NCED genes showed significantly increased gene expression in the abnormally developing ovules compared with the normally developing ovules. One of these NCED genes, termed XsNCED3-2 (EVM0019606), showed an extremely high abundance of transcripts in both abnormal (FPKM = 645.27) and normal ovules (FPKM = 136.97) (Table S1).
Using the Arabidopsis D27, MAX1, and LBO protein sequences as queries, we identified 4 D27, 19 MAX1, and 16 LBO homologs in the Xanthoceras genome, respectively (Table S1). The transcriptomic analysis revealed that 4 D27, 16 MAX1, and 13 LBO Xanthoceras homolog genes were expressed in the fertilized ovules (FPKM > 0.1). Among them, 2 D27, 6 MAX1, and 12 LBO genes were differentially expressed between the normally developing ovules and aborting ovules (Table S1).
To validate the quantification of the transcript levels of Xanthoceras genes obtained from the RNA-seq approach, the relative gene expression of six selected SL biosynthesis-related candidate genes, including D27, CCD, NCED, MAX1, and LBO, was measured by quantitative RT-PCR (Figure 7). As shown in the presented data, there was agreement between the results of the qPCR and RNA-seq tests for all six candidate genes examined.
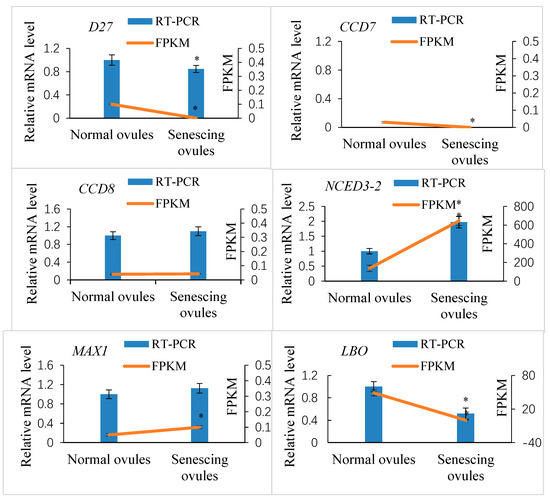
Figure 7.
Quantitative real-time PCR (qRT-PCR) verification of expression of the selected SL biosynthesis-related candidate genes in Xanthoceras fertilized ovules and comparisons between the qRT-PCR and RNA-Seq data analysis. The CCD7 gene was not expressed at detectable levels in the qRT-PCR analysis. The values are the means of three biological replicates with standard error bars. Asterisks indicate significant difference at p < 0.05 (ANOVA, Tukey’s HSD test).
2.4. Genome-Wide Identification of Genes Related to SL Perception and Signaling and Their Expression in Fertilized Ovules
The perception and signaling pathways of SLs in rice and Arabidopsis involve three highly conserved components: D14/AtD14, D3/MAX2, and D53/SMXL6/7/8 [47]. The D14 gene, first characterized in rice [26], encodes a protein of the α/β hydrolase superfamily with a strictly conserved Ser-His-Asp catalytic triad necessary for hydrolase activity. We used the protein sequence of rice D14 (Os03g0203200) as a query to search for D14 homologs in the Xanthoceras genome database. As a result, five Xanthoceras D14 homologs (EVM0010622, EVM0016131, EVM0023764, EVM0021522, and EVM0018761) were identified (Table S2). The sequences of these proteins share the putative hydrolase catalytic triad of Ser-Asp-His residues (Figure S2). A phylogenetic analysis of D14 proteins from rice, Arabidopsis, petunia, and Xanthoceras indicated that the Xanthoceras protein EVM0010622.1 was in the same clade as D14, AtD14, and DAD2 (Figure 8). EVM0010622.1 was the most similar to AtD14, sharing 81.4% of its protein sequence identity; therefore, we designated EVM0010622.1 as XsD14.
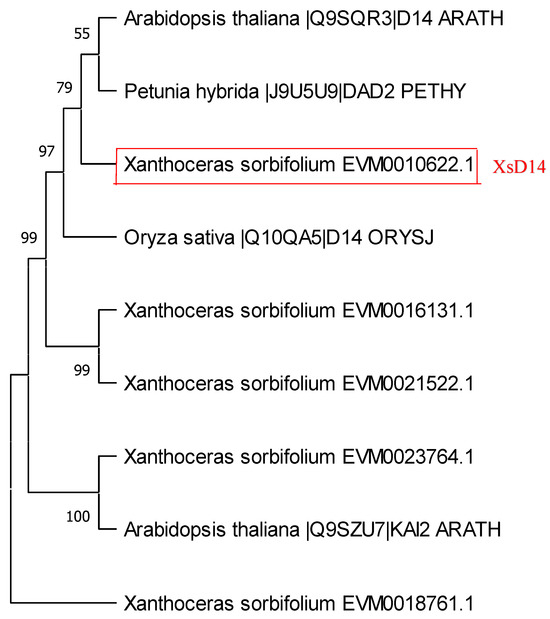
Figure 8.
Phylogenetic analysis of DWARF14 (D14) homologs. Maximum likelihood and Bayesian inference phylogenies were produced from an alignment of D14 hydrolases from Arabidopsis, rice, petunia, and Xanthoceras. A bootstrap analysis was performed using 1000 replications.
A transcriptomic analysis indicated that four putative Xanthoceras D14 homologous genes were expressed in the fertilized ovules, of which EVM0016131 and XsD14 showed very high levels of transcription in the aborting ovules (FPKM = 919.2 and FPKM = 225.42, respectively). Among these four genes, three were significantly differentially expressed between the aborting and normally developing ovules (Table S2). The EVM0016131 and XsD14 genes were upregulated in the aborting ovules, whereas EVM0021522 was upregulated in the normally developing ovules. To validate the expression profile of the Xanthoceras D14 homologous genes, we selected two differentially expressed genes, XsD14 and EVM0016131, for our qRT-PCR analysis. As shown in Figure 9, these two genes showed similar expression patterns to those observed in the RNA-seq data.
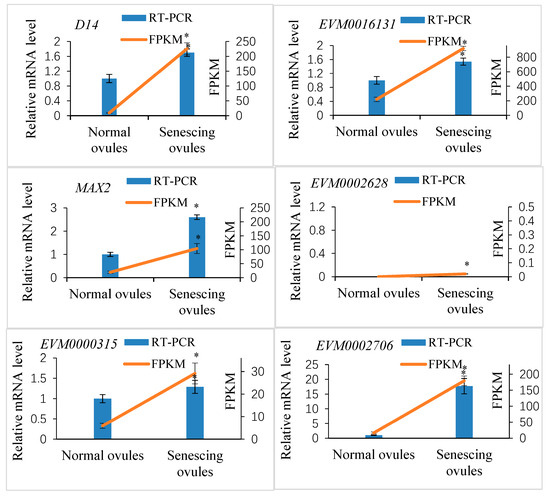
Figure 9.
Quantitative real-time PCR (qRT-PCR) verification of expression of the selected SL perception- and signaling-related candidate genes in Xanthoceras fertilized ovules and comparisons between the qRT-PCR and RNA-seq data. The values are the means of three biological replicates with standard error bars. Asterisks indicate significant difference at p < 0.05 (ANOVA, Tukey’s HSD test).
Four putative Xanthoceras MAX2 homologs (EVM0023678, EVM0002628, EVM0020331, and EVM0007925) were identified with BLASTP searches against the genomic database of Xanthoceras using the Arabidopsis MAX2 (AT2G42620) protein sequence as a query (Table S2). The EVM0023678 protein was most similar to AtMAX2; therefore, we designated EVM0023678 as XsMAX2, which was 63.7%, 60.7%, and 45.5% identical at the amino acid level to the MAX2 proteins from P. hybrida, A. thaliana, and O. sativa subsp. japonica, respectively. The transcriptome analysis showed that the EVM0002628 and EVM0020331 genes were not expressed in the fertilized ovules, whereas the XsMAX2 gene showed very high transcript levels and was significantly upregulated in the aborting ovules compared with the normally developing ovules (Table S2). The results were confirmed by a qRT-PCR analysis of the expression of two selected MAX2 homologs (Figure 9).
Six putative Xanthoceras SMXL homologs were identified by BLASTP against the Xanthoceras genome database and the transcriptomic database (this study) using the AtSMXL6 (AT1G07200) protein sequence from Arabidopsis. A phylogenetic analysis of D53 in rice and SMXL proteins in Arabidopsis, pea, and Xanthoceras indicated that the Xanthoceras protein EVM0012767 was in the same clade as AtSMXL8; therefore, we designated EVM0012767 as XsSMXL8 (Figure S3). This study did not determine the relationship between the other four Xanthoceras SMXL homologs and Arabidopsis AtSMXL6 and AtSMXL7. The protein sequence analysis revealed that the XsSMXL8 protein contains an RGKT motif, which is highly conserved in rice D53, Arabidopsis AtSMXL6/7/8, and pea PsSMXL8 and considered to be essential for SL-mediated D53/SMXL protein degradation [48]. The transcriptomic analysis indicated that all six Xanthoceras SMXL homolog genes were expressed in the early developing seeds. Among them, four showed significantly differential expression between the degenerating and normally developing ovules after fertilization, and all were upregulated in the aborting ovules compared with the normal ovules. The qRT-PCR analysis of two selected DEGs validated the differences in transcript abundance from the RNA-seq analysis (Figure 9).
2.5. Genome-Wide Identification of Xanthoceras Invertase Genes and Crosstalk between Sugar and Strigolactone in the Development of Fertilized Ovules
Based on their pH optima, invertases are classified into acid invertase (A-Inv) and alkaline/neutral invertase (A/N-Inv) [49]. According to their subcellular localization, acid invertases are further subdivided into cell-wall-bound invertase (CWIN) and vacuolar invertase (VIN). Alkaline/neutral invertases have different subcellular localizations, including cytosol, mitochondria, chloroplast, and nuclei. Based on the sequences of six cell wall invertase (AtcwInv1–6), two vacuolar invertase (AtvaInv1–2), and eleven neutral/alkaline invertase (At-A/N-InvA-K) proteins from Arabidopsis and a BLASTP search, we identified six cell wall, two vacuolar, and eight alkaline/neutral invertase homologs in the X. sorbifolium genome (Table S3). The alignment analysis of the deduced protein sequences of the Xanthoceras CWINs and VINs indicated that they contain a β-fructofuranosidase motif (NDPNG/A), an RDP motif, and a cysteine catalytic domain (WECP/VD), as well as four putative enzyme active site residues [49] (Figure S4). A valine residue in the ‘WEC-P/V-D’ box was found for two vacuolar invertases, whereas six cell wall invertases were characterized by the presence of a proline residue (Figure S4). Twelve conserved domains, which have been identified in rice A/N-Invs [49], could be detected in all eight Xanthoceras alkaline/neutral invertases (Figure S5). Consistently, a phylogenetic analysis showed that the invertase proteins from Xanthoceras, Arabidopsis, Litchi (Sapindaceae), and Dimocarpus (Sapindaceae) were clustered into two subgroups: acid invertases and alkaline/neutral invertases (Figure S6). The acid invertase clade contained cell-wall- and vacuole-targeted subgroups.
Our transcriptome data analysis revealed that 12 putative invertase genes were actively expressed in normally developing ovules at 10 DAP (FPKM ≥ 10; Table S3). Of these, four genes showed significantly differential expression between the abnormally and normally developing ovules (log2-fold change ≥1.5 or ≤−1.5; p ≤ 0.05). The cell wall invertase gene XsCWIN2 (EVM0004408) was upregulated in the normally developing ovules, whereas two vacuolar invertase genes, XsVIN1 (EVM0014291) and XsVIN2 (EVM0018616), and an alkaline/neutral invertase gene, XsA/N-Inv1 (EVM0006730), were downregulated in the normally developing ovules. The transcript abundance of these four DEGs was validated by a qRT-PCR analysis (Figure 10).
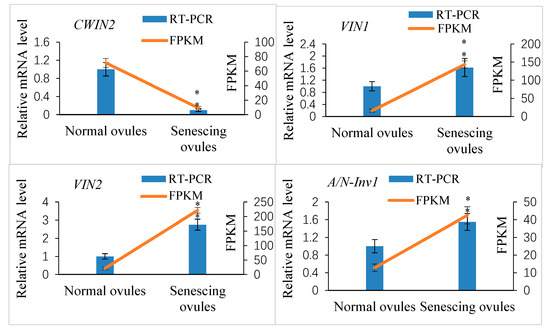
Figure 10.
Quantitative real-time PCR (qRT-PCR) verification of expression of the selected invertase candidate genes in Xanthoceras fertilized ovules and comparisons between the qRT-PCR and RNA-seq data. The values are the means of three biological replicates with standard error bars. Asterisks indicate significant difference at p < 0.05 (ANOVA, Tukey’s HSD test).
Based on the data above, we hypothesized that strigolactone may regulate early seed development by interacting with sugars in Xanthoceras and that the crosstalk between sugar and strigolactone may occur via sucrose metabolic processes. To preliminarily test this idea, we measured the endogenous content of soluble sugars in various developmental stages of the ovules after pollination using LC-MS/MS. The results indicated that the sucrose levels rose steadily in the normally developing ovules before 20 DAP and then increased dramatically during the formation and early development of cotyledons between 20 and 28 DAP (Table 1). The embryo at the middle stage of development between 40 and 48 DAP contained high levels of sucrose. The fructose and glucose contents also increased progressively before 13 DAP, while the free nuclei of the developing endosperm divided rapidly and accumulated on the embryo sac periphery. The ratio of hexose to sucrose reached the highest value at 13 DAP and then decreased following the formation of many-celled proembryos at the micropylar end of the embryo sac (Figure 4C). Oligosaccharide (raffinose) was not detected in the fertilized ovules until 28 DAP, and its content was high in the embryo at 48 DAP. Very high levels of sucrose and relatively low levels of hexose were found in the aborting ovules with a high strigolactone content at 12 DAP, whereas only traces of sucrose could be detected in the aborting ovules at 20 DAP (Table 1).

Table 1.
Soluble sugar content in fertilized ovules and early embryos in Xanthoceras sorbifolium. Values (mg g−1 dry weight) are the means ± SD of four biological replicates.
Further experimental studies revealed that the exogenous treatment of the ovaries at 1 DAP with 100 mM sucrose reduced the transcript levels of the putative strigolactone biosynthesis genes CCD8 and NCED3-2 in the ovules at 7 DAP compared to the control, indicating the likelihood that the strigolactone decreased (Figure 11). This exogenous sucrose application either delayed the abortion process of early aberrantly developing fruits and the fertilized ovules within them or partially restored the development of these fruits and ovules. Additionally, we assessed the expression of the CWIN2 gene and the invertase enzyme activity in ovules 9 DAP treated with exogenous strigolactone (GR24) 1 DAP. The results showed that both the invertase activity and CWIN2 transcript amounts in the fertilized ovules with the GR24 treatment were lower than those in the control ovules (Figure 12), in parallel with the low accumulation of glucose and fructose in the aborting ovules, suggesting a potential inhibitory effect of strigolactone on invertase activity.
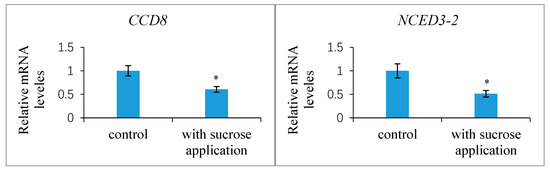
Figure 11.
Expression levels of CCD8 and NCED3-2 genes in the Xanthoceras fertilized ovules with sucrose application and control treatment, respectively, relative to the housekeeping gene actin-2. The values are the means of three biological replicates with standard error bars. Asterisks indicate significant difference at p < 0.05 (ANOVA, Tukey’s HSD test).
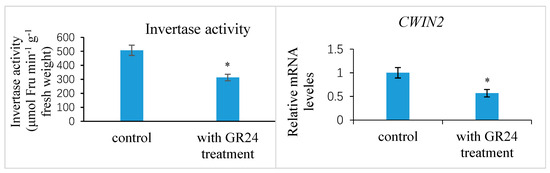
Figure 12.
Invertase activity and relative expression levels of CWIN2 gene in the Xanthoceras fertilized ovules with a synthetic strigolactone analog GR24 treatment and control treatment, respectively. The values are the means of three biological replicates with standard error bars. Asterisks indicate significant difference at p < 0.05 (ANOVA, Tukey’s HSD test).
2.6. Silencing of the XsD14 Gene Promoted Early Seed Development
As mentioned previously, the XsD14 gene showed very high transcript abundance in the fertilized ovules and was upregulated in the aborting ovules, so we further characterized the role of this gene during early seed development. TRV-based VIGS has been successfully applied to induce the silencing of endogenous genes in young fruits and fertilized ovules in X. sorbifolium [43]. The present study also used this approach to silence the expression of the XsD14 gene in young fruits and fertilized ovules by injecting an Agrobacterium tumefaciens suspension containing the pTRV2-XsD14 vector into terminal inflorescences 3 d before anthesis. Our phenotypic observations showed that pTRV2-XsD14 infiltration led to higher fruit retention in the inflorescences than the pTRV2-empty vector control treatment, but there were differences between genotypes in terms of the silencing effects (Figure 13). The qRT-PCR analyses indicated that XsD14 mRNA levels were significantly reduced in the fertilized ovules within the pTRV2-XsD14-infiltrated fruits, compared with those in the control (Figure 14). The development of the fertilized ovules was arrested 7 DAP in untreated aberrantly developing fruits, whereas the silencing of the XsD14 gene resulted in a delay in the arrest and abortion process of the fertilized ovules. The results of a cytological investigation and sugar quantification showed that the XsD14-silenced ovules had a larger hexose content (Figure 15) and more durable proliferation of endosperm free nuclei (Figure 4D) than the control ovules.
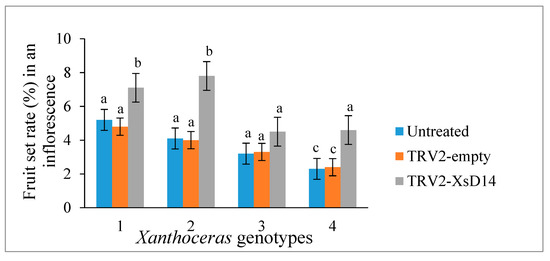
Figure 13.
Effect of VIGS-mediated XsD14 gene silencing on fruit set in terminal inflorescences of 4 Xanthoceras sorbifolium genotypes. The values are the means of five biological replicates with standard error bars. Different letters represent significant differences at p < 0.05 (ANOVA, Tukey’s HSD test).
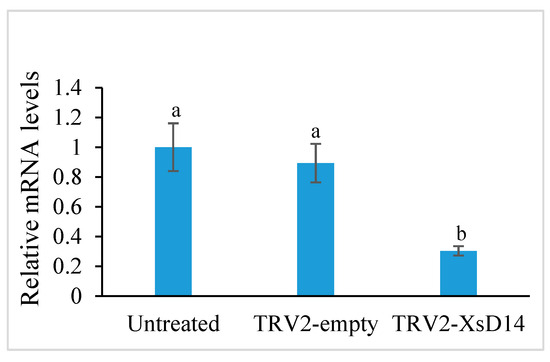
Figure 14.
Expression levels of XsD14 gene in the fertilized ovules with VIGS-mediated XsD14 gene silencing treatment and control treatment in relation to the expression levels of actin-2. The values are the means of three biological replicates with standard error bars. Different letters represent significant differences at p < 0.05 (ANOVA, Tukey’s HSD test).
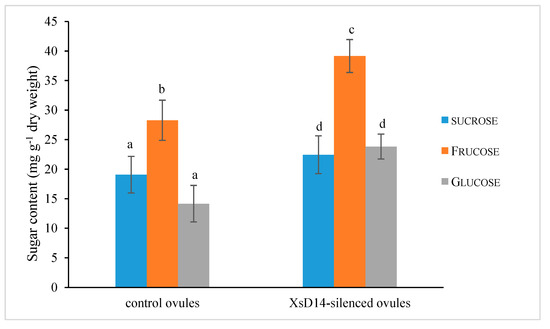
Figure 15.
Soluble sugar content in the fertilized ovules with VIGS-mediated XsD14 gene silencing treatment and control treatment. The values are the means of three biological replicates with standard error bars. Different letters represent significant differences at p < 0.05 (ANOVA, Tukey’s HSD test).
3. Discussion
Most studies on SL biosynthesis and signaling have focused on roots and shoots, but little is known about whether SLs are produced in early developing seeds and about their roles in ovule development after fertilization [1,47]. This study revealed that SLs were also generated in early developing seeds and that the aborting ovules after fertilization contained a high accumulation of SLs during the early stages of endosperm development in X. sorbifolium plants under low-Pi conditions. Enhanced SL biosynthesis was associated with higher expression levels of PHO2, a PSI gene, in the plants exposed to Pi starvation than in the plants with the Pi application. The inorganic form of phosphorus (phosphate, Pi) that is available to plants is a severely limiting factor for the fruit and seed production of Xanthoceras due to its low availability in most soils [43]. The requirement of Xanthoceras plants for phosphorus nutrients may be quite high during early seed development because of the active proliferation and growth of the syncytial endosperm and maternal tissues, including the nucellus and integuments, followed by the rapid expansion of the large embryo sac after fertilization [43,50]. We assume that the fertilized ovules in X. sorbifolium are the most sensitive to Pi deficiency. Plants have evolved elaborate mechanisms for adapting to low-Pi conditions and for maintaining cellular Pi homeostasis [5,12,51]. Many lines of evidence support the involvement of hormone-dependent signaling pathways, including SL pathways, in Pi starvation responses that lead to increased Pi uptake and reallocation within the plant [12,52]. Based on the present and previous studies, we propose that the development of fertilized ovules during Pi deficiency might be influenced by SL signaling and could be utilized during the process of Pi starvation responses as a way to fine-tune the adaptations of Xanthoceras plants to environmental stress conditions.
Here, we showed that there was an obvious difference in the progression of abortion between ovules treated with GR24 and those with the control treatment. The abortion process of the fertilized ovules in the aberrantly developing fruits was accelerated by the treatment with GR24; in contrast, the ovule abortion process was delayed by the treatment with TIS108, a triazole derivative that inhibits SL biosynthesis. The accelerated abortion process of the ovules with the GR24 treatment was associated with the degeneration of free endosperm nuclei and rapid reduction of the embryo sac. Our previous study identified a number of ethylene biosynthesis and signaling genes whose transcript abundances were significantly elevated in aborting ovules after fertilization [43]. The present study indicated that the GR24 treatment significantly induced the expression of Xanthoceras homologs of ACO genes, key components involved in ethylene biosynthesis, in fertilized ovules. This was consistent with findings of Lee and Yoon (2020) [53], whose treatment of etiolated Arabidopsis seedlings with GR24 increased the transcript levels of ACO genes, thereby enhancing ethylene biosynthesis. Our studies imply that SLs possibly act as abortion signals of fertilized ovules in X. sorbifolium and that potential crosstalk between SLs and ethylene may be involved in the control of ovule abortion after fertilization.
Our transcriptome analysis identified a total of 62 putative Xanthoceras homologs of the proposed SL biosynthesis genes in the Xanthoceras genome, including the D27, CCD, MAX1, and LBO genes. CCD7/MAX3 and CCD8/MAX4 have been shown to be expressed mainly in roots and lower stems in various examined species [15,46,54,55]. We found that several CCD family genes, including CCD8 and three NCED subfamily genes, were also expressed in the early developing seeds, of which some genes showed increased transcript levels in the aborting ovules after fertilization. The most similar Xanthoceras homolog to AtMAX1 was only weakly expressed in the fertilized ovules, but several other MAX1 homologues showed abundant transcripts and differential expression between the normal and aborting ovules. The expression patterns observed for multiple SL biosynthesis genes seem to support their activities in determining the amount of SLs (including 5-deoxystrigol and strigol) accumulated in early developing seeds, in which their upregulated expression might increase the SL levels in the aborting ovules after fertilization. These transcripts likely contribute to the key traits required to enhance the abortion process of fertilized ovules in the plant.
Substantial evidence has established that the D14 α/β-fold hydrolase functions as an SL receptor and is required for the perception of the SL signal in petunia, rice, Arabidopsis, pea, and tomato [47]. The D14 gene was transcribed at high levels in rosette and cauline leaves and at lower levels in axillary buds, inflorescences, stems, and roots in Arabidopsis plants [56]. In Brassica napus, CRISPR/Cas9-mediated knockout lines of the genes encoding BnD14 showed the characteristic feedback upregulation of CCD7 and CCD8 transcripts [57]. The lack of BnD14 function resulted in a prolific branching phenotype and an increase in total flower and total pod weight per plant. Our study found that the D14 gene was also expressed in fertilized ovules in which Xanthoceras homologs of MAX2 and SLs coincide. This is consistent with the proposed interaction between D14 and MAX2 in the presence of SLs [29], suggesting that the SL response takes place in the fertilized ovules where these factors coincide. We used a VIGS approach to characterize XsD14 functions during reproductive processes in X. sorbifolium. The results demonstrated that XsD14 gene silencing could alleviate fruit abortion and delay the abortion process of fertilized ovules, suggesting an important role of this gene in regulating the early development of fruits and seeds.
Xanthoceras sorbifolium follows a nuclear-type endosperm development, in which the endosperm initially develops as a syncytium, and cellularization is triggered after the formation of globular embryos [41]. The fertilized egg cell does not start to divide until 11 d after pollination, unlike the central cell, which quickly undergoes many mitotic cycles after fertilization to form the syncytial endosperm. Zygote division is a crucial developmental transition, which, in the case of failure, leads to ovule abortion [58]. It is likely that the syncytial endosperm produces structural and signaling substances such as sugars, which could activate the resting zygote and act as a nutritive component supporting early embryo growth. Fruit growth and ovule development after fertilization show great plasticity in X. sorbifolium, as they are regulated by pollen sources and nutrient availability [50]. Under low-nutrient conditions, up to 70% of the young fruits are aborted within 4 to 11 d following pollination. On average, the endosperm develops for 3 to 8 days before ovule abortion and the zygote degenerates during this period. The early collapse of the zygote can be used for identifying abortive ovules. By applying a sucrose treatment, the growth of young fruits and fertilized ovules within them is partially restored from an early abnormality. When nutrients, such as sugar and Pi, are limiting, increased SLs possibly inhibit fruit development to avoid the nutrient-demanding growth of the aborting fruits and fertilized ovules within them, thereby reducing sink tissue. We assume that the SL-mediated promotion of fruit and ovule abortion in X. sorbifolium might allow for a reallocation of resources to healthy fruits or other parts of the plant under nutrient-deficient conditions.
We found that hexoses (glucose and fructose) accumulated at high concentrations in fertilized ovules during the syncytial endosperm development, whereas sucrose increased during the mid- and late-development stages. High ratios of hexose and sucrose during early seed development might promote the proliferation of endosperm free nuclei and syncytial endosperm development in X. sorbifolium. When the ratio of hexose to sucrose levels decreased significantly, the growth of the fertilized ovules might be arrested and their abortion process would be initiated. The development of the fertilized ovules might be positively correlated with the hexose content but negatively correlated with SL levels. SL accumulation caused changes in the ratio of hexose to sucrose during ovule degeneration, suggesting the possibility that SLs may control the development of ovules after fertilization through their effects on the activity of invertases. The invertase-mediated release of hexoses is likely critical for appropriate carbon partitioning and normal seed development in X. sorbifolium.
4. Materials and Methods
4.1. Plant Materials
Seven-year-old Xanthoceras sorbifolium trees that were used in this study were grown at the experimental farm of the Institute of Botany, Chinese Academy of Sciences (Xiangshan, Beijing, China). Functional female flowers were pollinated on the first day of anthesis. Ovules were isolated from the ovaries at various developmental stages after pollination. Isolated ovules were flash frozen in liquid nitrogen and stored at −80 °C until further use. For cytological studies, some collected ovules were immediately fixed with FAA (formaldehyde acetic acid) solution containing 50% (v/v) ethanol, 5% (v/v) acetic acid, and 3.7% (w/v) formaldehyde for at least 24 h and then stored at 4 °C.
4.2. Histological Analysis
Fixed ovules were dehydrated, infiltrated, and embedded in either Paraplast Plus or Spurr resin. Paraplast Plus sections were cut to 6–10 μm using a steel knife and stained with safranin and fast green. Resin sections were cut to be 1–1.5 μm thick with a diamond knife and stained with 0.5% toluidine blue O in 0.1% sodium carbonate (pH 11.1) for general observations. Stained sections were observed with a Carl Zeiss microscope (Carl Zeiss Microscopy GmbH, Jena, Germany).
4.3. Strigolactone Analysis
Frozen ovules were homogenized to a fine powder under liquid nitrogen, and SL extraction was performed using 1 mL acetone containing d1-epi-5DS (100 pg) at −20 °C overnight. After the samples were filtered, and the filtrates were dried under nitrogen gas and then dissolved in 10% acetone. The extracts were loaded onto Oasis HLB (Waters Corporation, Micromass, TA, USA) and Sep-Pak Silica (Waters, Micromass, TA, USA) cartridges for purification and then washed with water, eluted with acetone, and dried under nitrogen gas. The SL-containing fraction was eluted with acetone and then evaporated until dry. The sample was re-dissolved in 50% acetonitrile (acetonitrile:water = 50:50, v:v) and filtered through a filter for liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis. SLs were analyzed using a high-pressure liquid chromatography (HPLC) Q-Trap-MS/MS in MRM (multiple reaction monitoring) mode.
4.4. Soluble Sugar Analysis
Ovules at various developmental stages after pollination were sampled for soluble sugar analysis. The samples were lyophilized with a freeze drier (SYHX, Beijing, China) and subsequently ground to a powder in liquid nitrogen. Sugars were extracted with an 80% aqueous methanol (v/v) solution and centrifuged for 15 min at 20 °C. The supernatant was collected and evaporated in a centrifugal vacuum evaporator (CV100-DNA, Aijimu, Beijing, China). Chloroform and distilled water were added to the samples and then centrifuged. The supernatant was dried by a vacuum concentrator at room temperature. The dried samples were oximated with methoxyamination reagent at 37 °C for 2 h. Then, the silylation reagent, MSTFA, was added to each oximated sample. The mixture was incubated at 60 °C for 30 min. Finally, the derivatized samples were transferred to glass vials for GC-MS analyses using an Agilent 7890A GC system equipped with an Agilent 7693 autosampler and Agilent 5975C-inert MSD with Triple-Axis Detector (Agilent, Atlanta, GA, USA). Three replicates were used for each sample at the same stages.
4.5. Invertase Enzyme Assay
Ovules were ground to powder in liquid nitrogen, followed by the addition of 150 mM Tris-HCl (pH 8.0) extraction buffer containing 2 mM EDTA, 10 mM MgCl2, 0.2% (v/v) 2-mercaptoethanol, 0.1 mM phenylmethyl sulfonyl fluoride, 1 mM benzamidine, and 10 mM ascorbic acid. Homogenates were centrifuged at 15,000× g rpm for 20 min at 4 °C. The sediments were homogenized with 150 mM Tris-HCl (at pH of 8.0) extraction buffer (the same as above) and subsequently centrifuged at 15,000× g rpm for 20 min at 4 °C. The supernatants were desalted twice using Sephadex G-25 (Pharmacia PD-10) and kept on ice until use. The mixture was incubated at 37 °C for 120 min. The desalted extracts and glucose were used as background control and standard, respectively. The reaction was stopped by adding the stop solution containing 1 M sodium potassium tartrate, 1% 3,5-dinitrosalicylic acid, and 0.5 M KOH. The liberated reducing sugars were quantified by measuring the absorbance at 540 nm.
4.6. Assays of GR24 and TIS108 Treatment
Concentrations for GR24 at 2.5 μM, an SL analog, and TIS108 at 1.5 μM, an SL biosynthetic inhibitor, were empirically determined. Both compounds were initially dissolved in acetone and volume was then adjusted with double-distilled sterile water. Young fruits at 1 DAP were dipped in the GR24 or TIS108 solution every other day for 6 days of the induction process. Treatments with the same concentration of acetone (diluted with water) were used as the experimental control. Each experiment was performed using three biological replicates.
4.7. Assays of Sucrose Treatment
Concentration for sucrose at 100 mM was empirically determined. Ovaries at 1 DAP were dipped in the sucrose solutions every other day over 6 days of the induction process. Effects of sucrose supply on fruit growth and ovule development were compared with mannitol, an osmotic control. Each experiment was performed using three biological replicates.
4.8. Phosphorus (P) Applications
Phosphorus application experiments were conducted with seven-year-old trees of 4 Xanthoceras genotypes (5 trees per genotype). Either phosphorus was not applied (control) or phosphorus fertilization dose of 150 g P2O5 per tree was used. Triple superphosphate (46% P2O5) was used as the source of phosphorus. Applications were realized in the form of a circle (40 cm radius) in front of roots. Plants received P applications 10 days before anthesis.
4.9. RNA Extraction, Library Construction, and Illumina Sequencing
Based on histological observations of ovules after fertilization, normal and abnormal ovules were determined and each was sampled for RNA-seq analysis. The total RNA from the samples was extracted using a Plant Total RNA Isolation Kit (Huayueyang, Beijing, China). A total of 6 RNA preparations (3 biological replicates for each sample) were used to increase sequencing coverage. The quantity and quality of extracted RNA were evaluated using a Qubit fluorometer (Invitrogen Inc., Carlsbad, CA, USA) and bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA), respectively. Oligo-dT (Qiagen, Valencia, CA, USA) was used to to isolate mRNA. The mRNA was fragmented using fragmentation buffer. cDNA was then synthesized using the mRNA fragments as templates. The cDNA fragments were purified using Qiaquick PCR Purification Kits (Qiagen, Valencia, CA, USA). The purified cDNA fragments were end repaired, added to poly (A), and ligated to Illumina sequencing adapters. Suitable fragments were selected for the PCR amplification and sequenced using BGISEQ-500 at BGI (Shenzhen, China).
4.10. RNA-Seq Data Analysis
Raw RNA-seq reads were filtered using a Perl program to obtain high-quality clean reads by removing low-quality sequences (having more than 20% bases with quality lower than 15 in one sequence), reads with more than 5% N bases (bases unknown), and reads containing Illumina sequencing adapters. The raw RNA-seq data have been deposited in the publicly accessible NCBI Sequence Read Archive (SRA) database under Bioproject PRJNA966773 (https://www.ncbi.nlm.nih.gov/sra/PRJNA966773, accessed on 3 May 2023). The filtered sequence pairs were aligned to the Xanthoceras sorbifolium reference genome sequences, using the alignment tool HISAT2 version 2.1.0. The transcript levels of individual transcripts in each sample were normalized as fragments per kilobase of transcript per million mapped reads (FPKM). Differentially expressed genes (DEGs) were identified using DEseq2.
4.11. Real-Time qRT-PCR Analysis
Real-time qRT-PCR was used to estimate the accuracy of RNA-seq data. The same RNA samples used for RNA-seq were used for qRT-PCR. RNA was used for synthesis of cDNA with the PrimeScript RT Master Mix Perfect Real Time Kit (Takara, San Jose, CA, USA). Real-time qRT-PCR was performed on the StepOnePLUSTM real-time PCR System (Applied Biosystems, Waltham, MA, USA). The actin gene (EVM0010329) was used as an internal reference, and relative expression was calculated using the 2−△△Ct method. Each sample was analyzed using 3 independent biological replicates. All of the primers used in this study are listed in Table S4.
4.12. Virus-Induced Gene Silence (VIGS) Assays
A 370 bp cDNA fragment of XsD14 gene was amplified using the primers presented in Table S4. The purified PCR products were digested with EcoRI and KpnI and ligated to pTRV2, resulting in the plasmid TRV2-XsD14. The plasmids were sequenced to verify correct insertion of the fragment and were then transformed into Agrobacterium tumefaciens strain GV3101. The cultures containing pTRV1 and pTRV2/pTRV2-XsD14 vector were mixed in a 1:1 ratio, and then 1 mL of culture was injected into the base of an inflorescence axial. Five independent biological replicates were performed for each treatment.
4.13. Statistical Analysis
All the data were analyzed statistically using SPSS 25.0 (SPSS Inc., Chicago, IL, USA) software. One-way analysis of variance (ANOVA) test was performed to assess the statistical differences. Data are shown as the means ± standard error of at least three biological replicates, and the mean differences were evaluated by Tukey’s honestly significant difference test at p < 0.05.
5. Conclusions
After its ovules are fertilized, the Xanthoceras plant decides whether or not to proceed with seed development. Signals from the environment, including the Pi nutritional state, and endogenous production, such as metabolite and hormone levels, influence these decisions. The genes and chemicals that participate in this finely tuned network of communication must be identified and characterized. This study identified a total of 69 putative Xanthoceras homologs of genes related to SL biosynthesis and signaling in the Xanthoceras genome; among them, 37 genes were found to be differently expressed in the fertilized ovules that were aborting compared to the normally developing ovules, suggesting their important roles in seed development and environmental responses. These genes could be ideal candidates for further studies and Xanthoceras germplasm genetic engineering in the future. The present study indicated that the production and signaling pathways of SLs are likely correlated with Pi deficiency, invertase activity, and sugar availability in ovules after fertilization. The crosstalk between sugar and strigolactone signals may be an important part of a system that accurately regulates ovule development after fertilization in X. sorbifolium.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms25063276/s1.
Author Contributions
Q.Z. conceived the research. Q.Z. and Q.C. performed the experiments. Q.Z. and L.Z. analyzed the data and wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by National Natural Science Foundation of China (32171834, U1903103, and 30972344) and Beijing Natural Science Foundation (6172028).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data generated and used in this study are included in this article and its additional files. The raw RNA-seq data have been deposited in the publicly accessible NCBI Sequence Read Archive (SRA) database under Bioproject PRJNA966773 (https://www.ncbi.nlm.nih.gov/sra/PRJNA966773, accessed on 3 May 2023).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Brewer, P.B.; Koltai, H.; Beveridge, C.A. Diverse roles of strigolactones in plant development. Mol. Plant 2013, 6, 18–28. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Roldan, V.; Fermas, S.; Brewer, P.B.; Puech-Pagès, V.; Dun, E.A.; Pillot, J.-P.; Letisse, F.; Matusova, R.; Danoun, S.; Portais, J.-C.; et al. Strigolactone inhibition of shoot branching. Nature 2008, 455, 189–194. [Google Scholar] [CrossRef]
- Marzec, M.; Melzer, M. Regulation of root development and architecture by strigolactones under optimal and nutrient deficiency conditions. Int. J. Mol. Sci. 2018, 19, 1887. [Google Scholar] [CrossRef]
- Elhiti, M.; Mira, M.M.; So, K.K.Y.; Stasolla, C.; Hebelstrup, K.H. Synthetic strigolactone GR24 improves Arabidopsis somatic embryogenesis through changes in auxin responses. Plants 2021, 10, 2720. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Furusawa, S.; Nagasaka, S.; Shimomura, K.; Yamaguchi, S.; Umehara, M. Strigolactone signaling regulates rice leaf senescence in response to a phosphate deficiency. Planta 2014, 240, 399–408. [Google Scholar] [CrossRef]
- Xu, X.; Jibran, R.; Wang, Y.T.; Dong, L.M.; Flokova, K.; Esfandiari, A.; McLachlan, A.R.G.; Heiser, A.; Sutherland-Smith, A.J.; Brummell, D.A.; et al. Strigolactones regulate sepal senescence in Arabidopsis. J. Exp. Bot. 2021, 72, 5462–5477. [Google Scholar] [CrossRef]
- Yoneyama, K.; Xie, X.N.; Kim, H.I.; Kisugi, T.; Nomura, T.; Sekimoto, H.; Yokota, T.; Yoneyama, K. How do nitrogen and phosphorus deficiencies affect strigolactone production and exudation? Planta 2012, 235, 1197–1207. [Google Scholar] [CrossRef] [PubMed]
- Van Ha, C.; Leyva-González, M.A.; Osakabe, Y.; Tran, U.T.; Nishiyama, R.; Watanabe, Y.; Tanaka, M.; Seki, M.; Yamaguchi, S.; Van Dong, N.; et al. Positive regulatory role of strigolactone in plant responses to drought and salt stress. Proc. Natl. Acad. Sci. USA 2014, 111, 851–856. [Google Scholar]
- Cook, C.E.; Whichard, L.P.; Turner, B.; Wall, M.E.; Egley, G.H. Germination of witchweed (Striga lutea Lour.): Isolation and properties of a potent stimulant. Science 1966, 154, 1189–1190. [Google Scholar] [CrossRef]
- Wakabayashi, T.; Ishiwa, S.; Shida, K.; Motonami, N.; Suzuki, H.; Takikawa, H.; Mizutani, M.; Sugimoto, Y. Identification and characterization of sorgomol synthase in sorghum strigolactone biosynthesis. Plant Physiol. 2021, 185, 902–913. [Google Scholar] [CrossRef]
- Czarnecki, O.; Yang, J.; Weston, D.J.; Tuskan, G.A.; Chen, J.G. A dual role of strigolactones in phosphate acquisition and utilization in plants. Int. J. Mol. Sci. 2013, 14, 7681–7701. [Google Scholar] [CrossRef]
- Gamir, J.; Torres-Vera, R.; Rial, C.; Berrio, E.; de Souza Campos, P.M.; Varela, R.M.; Macías, F.A.; Pozo, M.J.; Flors, V.; López-Ráez, J.A. Exogenous strigolactones impact metabolic profiles and phosphate starvation signalling in roots. Plant Cell Environ. 2020, 43, 1655–1668. [Google Scholar] [CrossRef]
- Woo, H.R.; Chung, K.M.; Park, J.H.; Oh, S.A.; Ahn, T.; Hong, S.H.; Jang, S.K.; Nam, H.G. ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. Plant Cell 2001, 13, 1779–1790. [Google Scholar] [CrossRef]
- Stirnberg, P.; van De Sande, K.; Leyser, H.M. MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development 2002, 129, 1131–1141. [Google Scholar] [CrossRef]
- Booker, J.; Auldridge, M.; Wills, S.; McCarty, D.; Klee, H.; Leyser, O. MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Curr. Biol. 2004, 14, 1232–1238. [Google Scholar] [CrossRef]
- Yao, R.; Ming, Z.; Yan, L.; Li, S.; Wang, F.; Ma, S.; Yu, C.; Yang, M.; Chen, L.; Chen, L.; et al. DWARF14 is a non-canonical hormone receptor for strigolactone. Nature 2016, 536, 469–473. [Google Scholar] [CrossRef]
- Brewer, P.B.; Yoneyama, K.; Filardo, F.; Meyers, E.; Scaffidi, A.; Frickey, T.; Akiyama, K.; Seto, Y.; Dun, E.A.; Cremer, J.E.; et al. LATERAL BRANCHING OXIDOREDUCTASE acts in the final stages of strigolactone biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2016, 113, 6301–6306. [Google Scholar] [CrossRef]
- Beveridge, C.A.; Ross, J.J.; Murfet, I.C. Branching mutant rms-2 in Pisum sativun. Plant Physiol. 1994, 104, 953–959. [Google Scholar] [CrossRef]
- Beveridge, C.A.; Symons, G.M.; Murfet, I.C.; Ross, J.J.; Rameau, C. The rms1 mutant of pea has elevated indole-3-acetic acid levels and reduced root-sap zeatin riboside content but increased branching controlled by graft-transmissible signal(s). Plant Physiol. 1997, 115, 1251–1258. [Google Scholar] [CrossRef]
- Morris, S.E.; Turnbull, C.G.; Murfet, I.C.; Beveridge, C.A. Mutational analysis of branching in pea. Evidence that Rms1 and Rms5 regulate the same novel signal. Plant Physiol. 2001, 126, 1205–1213. [Google Scholar] [CrossRef]
- Napoli, C. Highly branched phenotype of the petunia dad1-1 mutant is reversed by grafting. Plant Physiol. 1996, 111, 27–37. [Google Scholar] [CrossRef]
- Snowden, K.C.; Simkin, A.J.; Janssen, B.J.; Templeton, K.R.; Loucas, H.M.; Simons, J.L.; Karunairetnam, S.; Gleave, A.P.; Clark, D.G.; Klee, H.J. The decreased apical dominance1/Petunia hybrida carotenoid cleavage dioxygenase8 gene affects branch production and plays a role in leaf senescence, root growth, and flower development. Plant Cell 2005, 17, 746–759. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Lin, Q.B.; Zhu, L.H.; Ren, Y.L.; Zhou, K.N.; Shabek, N.; Wu, F.; Mao, H.; Dong, W.; Gan, L.; et al. D14-SCFD3-dependent degradation of D53 regulates strigolactone signalling. Nature 2013, 504, 406–410. [Google Scholar] [CrossRef]
- Haider, I.; Andreo-Jimenez, B.; Bruno, M.; Bimbo, A.; Floková, K.; Abuauf, H.; Ntui, V.O.; Guo, X.; Charnikhova, T.; Al-Babili, S.; et al. The interaction of strigolactones with abscisic acid during the drought response in rice. J. Exp. Bot. 2018, 69, 2403–2414. [Google Scholar] [CrossRef]
- Flematti, G.R.; Scaffidi, A.; Waters, M.T.; Smith, S.M. Stereospecificity in strigolactone biosynthesis and perception. Planta 2016, 243, 1361–1373. [Google Scholar] [CrossRef]
- Arite, T.; Umehara, M.; Ishikawa, S.; Hanada, A.; Maekawa, M.; Yamaguchi, S.; Kyozuka, J. d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol. 2009, 50, 1416–1424. [Google Scholar] [CrossRef]
- Waters, M.T.; Nelson, D.C.; Scaffidi, A.; Flematti, G.R.; Sun, Y.K.; Dixon, K.W.; Smith, S.M. Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development 2012, 139, 1285–1295. [Google Scholar] [CrossRef]
- de Saint Germain, A.; Clavé, G.; Badet-Denisot, M.-A.; Pillot, J.-P.; Cornu, D.; Le Caer, J.-P.; Burger, M.; Pelissier, F.; Retailleau, P.; Turnbull, C.; et al. An histidine covalent receptor and butenolide complex mediates strigolactone perception. Nature Chem. Biol. 2016, 12, 787–794. [Google Scholar] [CrossRef] [PubMed]
- Hamiaux, C.; Drummond, R.S.; Janssen, B.J.; Ledger, S.E.; Cooney, J.M.; Newcomb, R.D.; Snowden, K.C. DAD2 is an alpha/beta hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr. Biol. 2012, 22, 2032–2036. [Google Scholar] [CrossRef]
- Yao, R.; Wang, L.; Li, Y.; Chen, L.; Li, S.; Du, X.; Wang, B.; Yan, J.; Li, J.; Xie, D. Rice DWARF14 acts as an unconventional hormone receptor for strigolactone. J. Exp. Bot. 2018, 69, 2355–2365. [Google Scholar] [CrossRef] [PubMed]
- Shabek, N.; Ticchiarelli, F.; Mao, H.B.; Hinds, T.R.; Leyser, O.; Zheng, N. Structural plasticity of D3-D14 ubiquitin ligase in strigolactone signalling. Nature 2018, 563, 652–656. [Google Scholar] [CrossRef]
- Jeandet, P.; Formela-Luboińska, M.; Labudda, M.; Morkunas, I. The role of sugars in plant responses to stress and their regulatory function during development. Int. J. Mol. Sci. 2022, 23, 5161. [Google Scholar] [CrossRef]
- Fichtner, F.; Barbier, F.F.; Annunziata, M.G.; Feil, R.; Olas, J.J.; Mueller-Roeber, B.; Stitt, M.; Beveridge, C.A.; Lunn, J.E. Regulation of shoot branching in arabidopsis by trehalose 6-phosphate. New Phytol. 2021, 229, 2135–2151. [Google Scholar] [CrossRef] [PubMed]
- Mishra, B.S.; Singh, M.; Aggrawal, P.; Laxmi, A. Glucose and auxin signaling interaction in controlling Arabidopsis thaliana seedlings root growth and development. PLoS ONE 2009, 4, e4502. [Google Scholar] [CrossRef] [PubMed]
- Barbier, F.F.; Cao, D.; Fichtner, F.; Weiste, C.; Perez-Garcia, M.D.; Caradeuc, M.; Le Gourrierec, J.; Sakr, S.; Beveridge, C.A. HEXOKINASE1 signalling promotes shoot branching and interacts with cytokinin and strigolactone pathways. New Phytol. 2021, 231, 1088–1104. [Google Scholar] [CrossRef] [PubMed]
- Dierck, R.; Dhooghe, E.; Van Huylenbroeck, J.; De Riek, J.; De Keyser, E.; Van Der Straeten, D. Response to strigolactone treatment in chrysanthemum axillary buds is influenced by auxin transport inhibition and sucrose availability. Acta Physiol. Plant 2016, 38, 271. [Google Scholar] [CrossRef]
- Bertheloot, J.; Barbier, F.; Boudon, F.; Perez-Garcia, M.D.; Thomas, P.; Citerne, S.; Dun, E.; Beveridge, C.; Godin, C.; Sakr, S. Sugar availability suppresses the auxin-induced strigolactone pathway to promote bud outgrowth. New Phytol. 2020, 225, 866–879. [Google Scholar] [CrossRef] [PubMed]
- Patil, S.B.; Barbier, F.F.; Zhao, J.; Zafar, S.A.; Uzair, M.; Sun, Y.L.; Fang, J.; Perez-Garcia, M.; Bertheloot, J.; Sakr, S.; et al. Sucrose promotes D53 accumulation and tillering in rice. New Phytol. 2022, 234, 122–136. [Google Scholar] [CrossRef]
- Soltani, A.; Weraduwage, S.M.; Sharkey, T.S.; Lowry, D.B. Elevated temperatures cause loss of seed set in common bean (Phaseolus vulgaris L.) potentially through the disruption of source-sink relationships. BMC Genom. 2019, 20, 312. [Google Scholar] [CrossRef]
- Trouverie, J.; Chateau-Joubert, S.; Thévenot, C.; Jacquemot, M.P.; Prioul, J.L. Regulation of vacuolar invertase by abscisic acid or glucose in leaves and roots from maize plantlets. Planta 2004, 219, 894–905. [Google Scholar] [CrossRef]
- Zhou, Q.Y.; Liu, G.S. The embryology of Xanthoceras and its phylogenetic implications. Plant Syst. Evol. 2012, 298, 457–468. [Google Scholar] [CrossRef]
- Gu, L.B.; Zhang, G.J.; Du, L.; Du, J.; Qi, K.; Zhu, X.L.; Zhang, X.Y.; Jiang, Z.H. Comparative study on the extraction of Xanthoceras sorbifolia Bunge (yellow horn) seed oil using subcritical n-butane, supercritical CO2, and the Soxhlet method. LWT-Food Sci. Technol. 2019, 111, 548–554. [Google Scholar] [CrossRef]
- Zhou, Q.Y.; Cai, Q. Role of ethylene in the regulatory mechanism underlying the abortion of ovules after fertilization in Xanthoceras sorbifolium. Plant Mol. Biol. 2021, 106, 67. [Google Scholar] [CrossRef]
- Linkies, A.; Müller, K.; Morris, K.; Turečková, V.; Wenk, M.; Cadman, C.S.; Corbineau, F.; Strnad, M.; Lynn, J.R.; Finch-Savage, W.E.; et al. Ethylene interacts with abscisic acid to regulate endosperm rupture during germination: A comparative approach using Lepidium sativum and Arabidopsis thaliana. Plant Cell 2009, 21, 3803–3822. [Google Scholar] [CrossRef] [PubMed]
- Radchuk, V.; Weier, D.; Radchuk, R.; Weschke, W.; Weber, H. Development of maternal seed tissue in barley is mediated by regulated cell expansion and cell disintegration and coordinated with endosperm growth. J. Exp. Bot. 2011, 62, 1217–1227. [Google Scholar] [CrossRef]
- Zhao, X.L.; Yang, Y.L.; Xia, H.X.; Li, Y. Genome-wide analysis of the carotenoid cleavage dioxygenases gene family in Forsythia suspensa: Expression profile and cold and drought stress responses. Front. Plant Sci. 2022, 13, 998911. [Google Scholar] [CrossRef] [PubMed]
- Mashiguchi, K.; Seto, Y.; Yamaguchi, S. Strigolactone biosynthesis, transport and perception. Plant J. 2021, 105, 335–350. [Google Scholar] [CrossRef] [PubMed]
- Kerr, S.C.; Patil, S.B.; de Saint Germain, A.; Pillot, J.P.; Saffar, J.; Ligerot, Y.; Aubert, G.; Citerne, S.; Bellec, Y.; Dun, E.A.; et al. Integration of the SMXL/D53 strigolactone signalling repressors in the model of shoot branching regulation in Pisum sativum. Plant J. 2021, 107, 1756–1770. [Google Scholar] [CrossRef]
- Ji, X.; Van den Ende, W.; Van Laere, A.; Cheng, S.; Bennett, J. Structure, evolution, and expression of the two invertase gene families of rice. J. Mol. Evol. 2005, 60, 615–634. [Google Scholar] [CrossRef]
- Zhou, Q.Y.; Zheng, Y.R.; Lai, L.M.; Du, H. Observations on sexual reproduction in Xanthoceras sorbifolium (Sapindaceae). Acta Bot. Boreal-Occident Sin. 2017, 37, 14–22. [Google Scholar]
- López-Ráez, J.A.; Bouwmeester, H. Fine-tuning regulation of strigolactone biosynthesis under phosphate starvation. Plant Signal Behav. 2008, 3, 963–965. [Google Scholar] [CrossRef]
- Aquino, B.; Bradley, J.M.; Lumba, S. On the outside looking in: Roles of endogenous and exogenous strigolactones. Plant J. 2021, 105, 322–334. [Google Scholar] [CrossRef]
- Lee, H.Y.; Yoon, G.M. Strigolactone elevates ethylene biosynthesis in etiolated Arabidopsis seedlings. Plant Signal Behav. 2020, 15, e1805232. [Google Scholar] [CrossRef]
- Matusova, R.; Rani, K.; Verstappen, F.W.A.; Franssen, M.C.R.; Beale, M.H.; Bouwmeester, H.J. The strigolactone germination stimulants of the plant-parasitic Striga and Orobanche spp. are derived from the carotenoid pathway. Plant Physiol. 2005, 139, 920–934. [Google Scholar] [CrossRef]
- Alder, A.; Jamil, M.; Marzorati, M.; Bruno, M.; Vermathen, M.; Bigler, P.; Ghisla, S.; Bouwmeester, H.; Beyer, P.; Al-Babili, S. The path from beta-carotene to carlactone, a strigolactone-like plant hormone. Science 2012, 335, 1348–1351. [Google Scholar] [CrossRef] [PubMed]
- Chevalier, F.; Nieminen, K.; Sánchez-Ferrero, J.C.; Rodríguez, M.L.; Chagoyen, M.; Hardtke, C.S.; Cubas, P. Strigolactone promotes degradation of DWARF14, an α/β hydrolase essential for strigolactone signaling in Arabidopsis. Plant Cell 2014, 26, 1134–1150. [Google Scholar] [CrossRef]
- Stanic, M.; Hickerson, N.M.N.; Arunraj, R.; Samuel, M.A. Gene-editing of the strigolactone receptor BnD14 confers promising shoot architectural changes in Brassica napus (canola). Plant Biotechnol. J. 2021, 19, 639–641. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.Y.; Cai, Q. The superoxide dismutase genes might be required for appropriate development of the ovule after fertilization in Xanthoceras sorbifolium. Plant Cell Rep. 2018, 37, 727–739. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).