Transcription Factor MYB as Therapeutic Target: Current Developments
Abstract
1. The Discovery of the MYB Gene
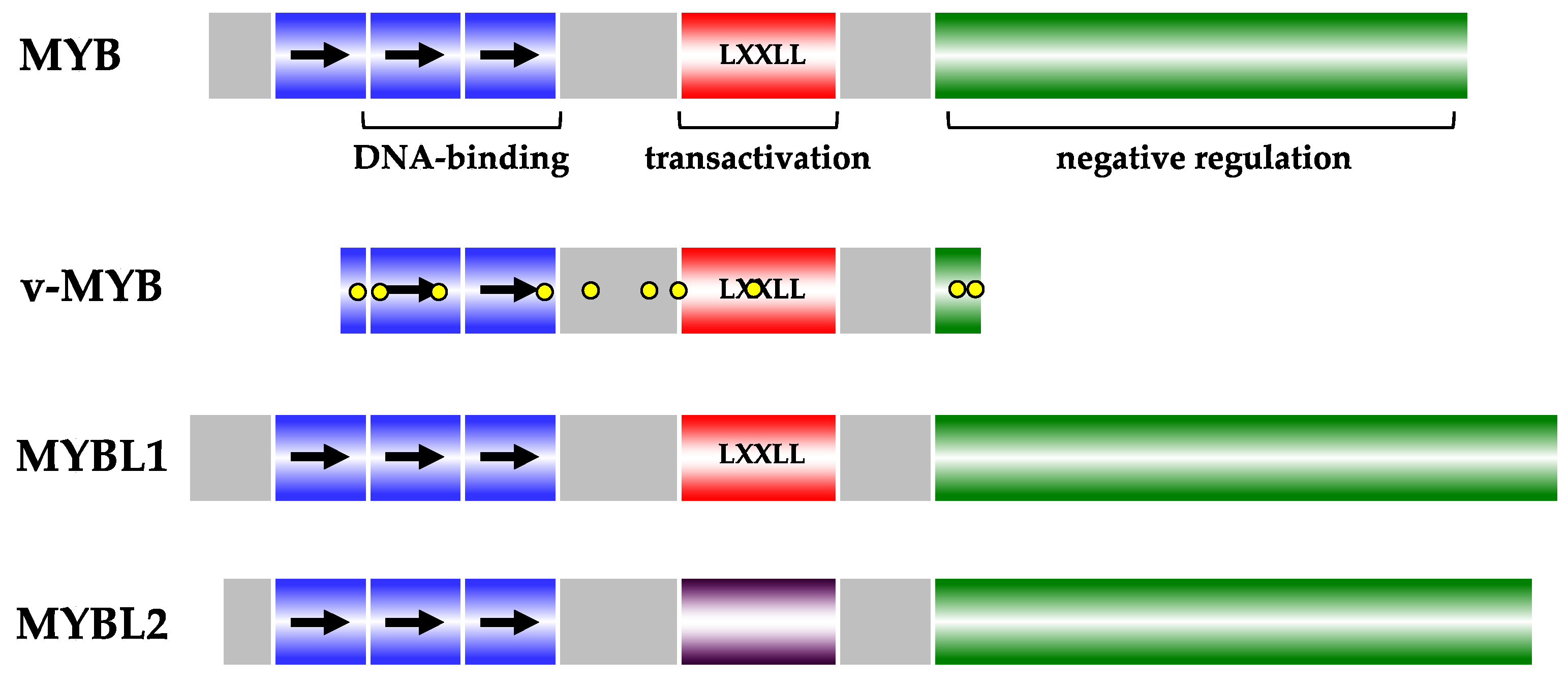
2. The MYB Protein
3. The Role of MYB in Human Cancer
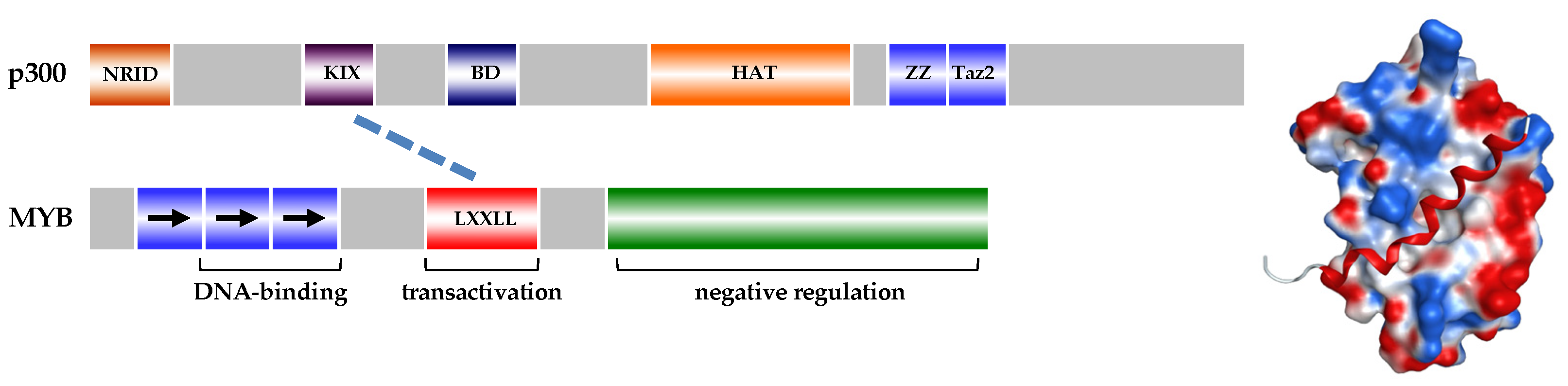
4. MYB as a Therapeutic Target
5. Concluding Remarks
Funding
Conflicts of Interest
References
- Lipsick, J.S. One billion years of Myb. Oncogene 1996, 13, 223–235. [Google Scholar] [PubMed]
- Stracke, R.; Werber, M.; Weisshaar, B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001, 4, 447–456. [Google Scholar] [CrossRef] [PubMed]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Ganter, B.; Lipsick, J.S. Myb and oncogenesis. Adv. Cancer Res. 1999, 76, 21–60. [Google Scholar] [CrossRef]
- Lipsick, J.S.; Wang, D.M. Transformation by v-Myb. Oncogene 1999, 18, 3047–3055. [Google Scholar] [CrossRef]
- Mucenski, M.L.; McLain, K.; Kier, A.B.; Swerdlow, S.H.; Schreiner, C.M.; Miller, T.A.; Pietryga, D.W.; Scott, W.J., Jr.; Potter, S.S. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell 1991, 65, 677–689. [Google Scholar] [CrossRef] [PubMed]
- Ramsay, R.G.; Gonda, T.J. MYB function in normal and cancer cells. Nat. Rev. Cancer 2008, 8, 523–534. [Google Scholar] [CrossRef]
- Ness, S.A. Myb binding proteins: Regulators and cohorts in transformation. Oncogene 1999, 18, 3039–3046. [Google Scholar] [CrossRef]
- Zhou, Y.; Ness, S.A. Myb proteins: Angels and demons in normal and transformed cells. Front. Biosci. 2011, 16, 1109–1131. [Google Scholar] [CrossRef]
- George, O.L.; Ness, S.A. Situational awareness: Regulation of the myb transcription factor in differentiation, the cell cycle and oncogenesis. Cancers 2014, 6, 2049–2071. [Google Scholar] [CrossRef]
- Biedenkapp, H.; Borgmeyer, U.; Sippel, A.E.; Klempnauer, K.-H. Viral myb oncogene encodes a sequence-specific DNA-binding activity. Nature 1988, 335, 835–837. [Google Scholar] [CrossRef] [PubMed]
- Weston, K.; Bishop, J.M. Transcriptional activation by the v-myb oncogene and its cellular progenitor, c-myb. Cell 1989, 58, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Ness, S.A.; Marknell, A.; Graf, T. The v-myb oncogene product binds to and activates the promyelocyte-specific mim-1 gene. Cell 1989, 59, 1115–1125. [Google Scholar] [CrossRef] [PubMed]
- Lang, G.; White, J.R.; Argent-Katwala, M.J.; Allinson, C.G.; Weston, K. Myb proteins regulate the expression of diverse target genes. Oncogene 2005, 24, 1375–1384. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Lei, W.; O’Rourke, J.P.; Ness, S.A. Oncogenic mutations cause dramatic, qualitative changes in the transcriptional activity of c-Myb. Oncogene 2006, 25, 795–805. [Google Scholar] [CrossRef] [PubMed]
- Quintana, A.M.; Liu, F.; O’Rourke, J.P.; Ness, S.A. Identification and regulation of c-Myb target genes in MCF-7 cells. BMC Cancers 2011, 11, 30. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Glazov, E.A.; Pattabiraman, D.R.; Al-Owaidi, F.; Zhang, P.; Brown, M.A.; Leo, P.J.; Gonda, T.J. Integrated genome-wide chromatin occupancy and expression analyses identify key myeloid pro-differentiation transcription factors repressed by Myb. Nucleic Acids Res. 2011, 39, 4664–4679. [Google Scholar] [CrossRef]
- Roe, J.S.; Mercan, F.; Rivera, K.; Pappin, D.J.; Vakoc, C.R. BET Bromodomain Inhibition Suppresses the Function of Hematopoietic Transcription Factors in Acute Myeloid Leukemia. Mol. Cell 2015, 58, 1028–1039. [Google Scholar] [CrossRef]
- Bengtsen, M.; Klepper, K.; Gundersen, S.; Cuervo, I.; Drabløs, F.; Hovig, E.; Sandve, G.K.; Gabrielsen, O.S.; Eskeland, R. c-Myb Binding Sites in Haematopoietic Chromatin Landscapes. PLoS ONE 2015, 10, e0133280. [Google Scholar] [CrossRef]
- Lemma, R.B.; Ledsaak, M.; Fuglerud, B.M.; Rodríguez-Castañeda, F.; Eskeland, R.; Gabrielsen, O.S. MYB regulates the SUMO protease SENP1 and its novel interaction partner UXT, modulating MYB target genes and the SUMO landscape. J. Biol. Chem. 2023, 299, 105062. [Google Scholar] [CrossRef]
- Takao, S.; Forbes, L.; Uni, M.; Cheng, S.; Pineda, J.M.B.; Tarumoto, Y.; Cifani, P.; Minuesa, G.; Chen, C.; Kharas, M.G.; et al. Convergent organization of aberrant MYB complex controls oncogenic gene expression in acute myeloid leukemia. Elife 2021, 10, e65905. [Google Scholar] [CrossRef]
- Dai, P.; Akimaru, H.; Tanaka, Y.; Hou, D.X.; Yasukawa, T.; Kanei-Ishii, C.; Takahashi, T.; Ishii, S. CBP as a transcriptional coactivator of c-Myb. Genes Dev. 1996, 10, 528–540. [Google Scholar] [CrossRef]
- Oelgeschläger, M.; Janknecht, R.; Krieg, J.; Schreek, S.; Lüscher, B. Interaction of the co-activator CBP with Myb proteins: Effects on Myb-specific transactivation and on the cooperativity with NF-M. EMBO J. 1996, 15, 2771–2780. [Google Scholar] [CrossRef]
- Blobel, G.A. CREB-binding protein and p300: Molecular integrators of hematopoietic transcription. Blood 2000, 95, 745–755. [Google Scholar] [CrossRef]
- Vo, N.; Goodman, R.H. CREB-binding protein and p300 in transcriptional regulation. J. Biol. Chem. 2001, 276, 13505–13508. [Google Scholar] [CrossRef]
- Zor, T.; De Guzman, R.N.; Dyson, H.J.; Wright, P.E. Solution structure of the KIX domain of CBP bound to the transactivation domain of c-Myb. J. Mol. Biol. 2004, 337, 521–534. [Google Scholar] [CrossRef]
- Rebel, V.I.; Kung, A.L.; Tanner, E.A.; Yang, H.; Bronson, R.T.; Livingston, D.M. Distinct roles for CREB-binding protein and p300 in hematopoietic stem cell self-renewal. Proc. Natl. Acad. Sci. USA 2002, 99, 14789–14794. [Google Scholar] [CrossRef] [PubMed]
- Pattabiraman, D.R.; Sun, J.; Dowhan, D.H.; Ishii, S.; Gonda, T.J. Mutations in multiple domains of c-Myb disrupt interaction with CBP/p300 and abrogate myeloid transforming ability. Mol. Cancer Res. 2009, 7, 1477–1486. [Google Scholar] [CrossRef] [PubMed]
- Pattabiraman, D.R.; McGirr, C.; Shakhbazov, K.; Barbier, V.; Krishnan, K.; Mukhopadhyay, P.; Hawthorne, P.; Trezise, A.; Ding, J.; Grimmond, S.M.; et al. Interaction of c-Myb with p300 is required for the induction of acute myeloid leukemia (AML) by human AML oncogenes. Blood 2014, 123, 2682–2690. [Google Scholar] [CrossRef] [PubMed]
- Mink, S.; Haenig, B.; Klempnauer, K.-H. Interaction and functional collaboration of p300 and C/EBPbeta. Mol. Cell Biol. 1997, 17, 6609–6617. [Google Scholar] [CrossRef] [PubMed]
- Ness, S.A.; Kowenz-Leutz, E.; Casini, T.; Graf, T.; Leutz, A. Myb and NF-M: Combinatorial activators of myeloid genes in heterologous cell types. Genes Dev. 1993, 7, 749–759. [Google Scholar] [CrossRef]
- Burk, O.; Mink, S.; Ringwald, M.; Klempnauer, K.-H. Synergistic activation of the chicken mim-1 gene by v-myb and C/EBP transcription factors. EMBO J. 1993, 12, 2027–2038. [Google Scholar] [CrossRef] [PubMed]
- Uttarkar, S.; Dassé, E.; Coulibaly, A.; Steinmann, S.; Jakobs, A.; Schomburg, C.; Trentmann, A.; Jose, J.; Schlenke, P.; Berdel, W.E.; et al. Targeting acute myeloid leukemia with a small molecule inhibitor of the Myb/p300 interaction. Blood 2016, 127, 1173–1182. [Google Scholar] [CrossRef][Green Version]
- Lahortiga, I.; De Keersmaecker, K.; Van Vlierberghe, P.; Graux, C.; Cauwelier, B.; Lambert, F.; Mentens, N.; Beverloo, H.B.; Pieters, R.; Speleman, F.; et al. Duplication of the MYB oncogene in T cell acute lymphoblastic leukemia. Nat. Genet. 2007, 39, 593–595. [Google Scholar] [CrossRef] [PubMed]
- Clappier, E.; Cuccuini, W.; Kalota, A.; Crinquette, A.; Cayuela, J.M.; Dik, W.A.; Langerak, A.W.; Montpellier, B.; Nadel, B.; Walrafen, P.; et al. The c-MYB locus is involved in chromosomal translocation and genomic duplications in human T-cell acute leukemia (T-ALL), the translocation defining a new T-ALL subtype in very young children. Blood 2007, 110, 1251–1261. [Google Scholar] [CrossRef]
- O’Neil, J.; Tchinda, J.; Gutierrez, A.; Moreau, L.; Maser, R.S.; Wong, K.K.; Li, W.; McKenna, K.; Liu, X.S.; Feng, B.; et al. Alu elements mediate MYB gene tandem duplication in human T-ALL. J. Exp. Med. 2007, 204, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Quelen, C.; Lippert, E.; Struski, S.; Demur, C.; Soler, G.; Prade, N.; Delabesse, E.; Broccardo, C.; Dastugue, N.; Mahon, F.X.; et al. Identification of a transforming MYB-GATA1 fusion gene in acute basophilic leukemia: A new entity in male infants. Blood 2011, 117, 5719–5722. [Google Scholar] [CrossRef]
- Belloni, E.; Shing, D.; Tapinassi, C.; Viale, A.; Mancuso, P.; Malazzi, O.; Gerbino, E.; Dall’Olio, V.; Egurbide, I.; Odero, M.D.; et al. In vivo expression of an aberrant MYB-GATA1 fusion induces leukemia in the presence of GATA1 reduced levels. Leukemia 2011, 25, 733–736. [Google Scholar] [CrossRef]
- Mansour, M.R.; Abraham, B.J.; Anders, L.; Berezovskaya, A.; Gutierrez, A.; Durbin, A.D.; Etchin, J.; Lawton, L.; Sallan, S.E.; Silverman, L.B.; et al. Oncogene regulation. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science 2014, 346, 1373–1377. [Google Scholar] [CrossRef]
- Rahman, S.; Magnussen, M.; León, T.E.; Farah, N.; Li, Z.; Abraham, B.J.; Alapi, K.Z.; Mitchell, R.J.; Naughton, T.; Fielding, A.K.; et al. Activation of the LMO2 oncogene through a somatically acquired neomorphic promoter in T-cell acute lymphoblastic leukemia. Blood 2017, 129, 3221–3226. [Google Scholar] [CrossRef]
- Smith, C.; Goyal, A.; Weichenhan, D.; Allemand, E.; Mayakonda, A.; Toprak, U.; Riedel, A.; Balducci, E.; Manojkumar, M.; Pejkovska, A.; et al. TAL1 activation in T-cell acute lymphoblastic leukemia: A novel oncogenic 3’ neo-enhancer. Haematologica 2023, 108, 1259–1271. [Google Scholar] [CrossRef]
- Song, H.; Liu, Y.; Tan, Y.; Zhang, Y.; Jin, W.; Chen, L.; Wu, S.; Yan, J.; Li, J.; Chen, Z.; et al. Recurrent noncoding somatic and germline WT1 variants converge to disrupt MYB binding in acute promyelocytic leukemia. Blood 2022, 140, 1132–1144. [Google Scholar] [CrossRef]
- Hess, J.L.; Bittner, C.B.; Zeisig, D.T.; Bach, C.; Fuchs, U.; Borkhardt, A.; Frampton, J.; Slany, R.K. c-Myb is an essential downstream target for homeobox-mediated transformation of hematopoietic cells. Blood 2006, 108, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Somervaille, T.C.; Matheny, C.J.; Spencer, G.J.; Iwasaki, M.; Rinn, J.L.; Witten, D.M.; Chang, H.Y.; Shurtleff, S.A.; Downing, J.R.; Cleary, M.L. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell 2009, 4, 129–140. [Google Scholar] [CrossRef] [PubMed]
- Zuber, J.; Rappaport, A.R.; Luo, W.; Wang, E.; Chen, C.; Vaseva, A.V.; Shi, J.; Weissmueller, S.; Fellmann, C.; Taylor, M.J.; et al. An integrated approach to dissecting oncogene addiction implicates a Myb-coordinated self-renewal program as essential for leukemia maintenance. Genes Dev. 2011, 25, 1628–1640. [Google Scholar] [CrossRef] [PubMed]
- Anfossi, G.; Gewirtz, A.M.; Calabretta, B. An oligomer complementary to c-myb-encoded mRNA inhibits proliferation of human myeloid leukemia cell lines. Proc. Natl. Acad. Sci. USA 1989, 86, 3379–3383. [Google Scholar] [CrossRef] [PubMed]
- Calabretta, B.; Gewirtz, A.M. Functional requirements of c-myb during normal and leukemic hematopoiesis. Crit. Rev. Oncog. 1991, 2, 187–194. [Google Scholar] [PubMed]
- Persson, M.; Andrén, Y.; Mark, J.; Horlings, H.M.; Persson, F.; Stenman, G. Recurrent fusion of MYB and NFIB transcription factor genes in carcinomas of the breast and head and neck. Proc. Natl. Acad. Sci. USA 2009, 106, 18740–18744. [Google Scholar] [CrossRef]
- Wagner, V.P.; Bingle, C.D.; Bingle, L. MYB-NFIB fusion transcript in adenoid cystic carcinoma: Current state of knowledge and future directions. Crit. Rev. Oncol. Hematol. 2022, 176, 103745. [Google Scholar] [CrossRef]
- Brayer, K.J.; Frerich, C.A.; Kang, H.; Ness, S.A. Recurrent Fusions in MYB and MYBL1 Define a Common, Transcription Factor-Driven Oncogenic Pathway in Salivary Gland Adenoid Cystic Carcinoma. Cancer Discov. 2016, 6, 176–187. [Google Scholar] [CrossRef]
- Persson, M.; Andrén, Y.; Moskaluk, C.A.; Frierson, H.F., Jr.; Cooke, S.L.; Futreal, P.A.; Kling, T.; Nelander, S.; Nordkvist, A.; Persson, F.; et al. Clinically significant copy number alterations and complex rearrangements of MYB and NFIB in head and neck adenoid cystic carcinoma. Genes Chromosomes Cancer 2012, 51, 805–817. [Google Scholar] [CrossRef] [PubMed]
- Drier, Y.; Cotton, M.J.; Williamson, K.E.; Gillespie, S.M.; Ryan, R.J.; Kluk, M.J.; Carey, C.D.; Rodig, S.J.; Sholl, L.M.; Afrogheh, A.H.; et al. An oncogenic MYB feedback loop drives alternate cell fates in adenoid cystic carcinoma. Nat. Genet. 2016, 48, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Tatevossian, R.G.; Tang, B.; Dalton, J.; Forshew, T.; Lawson, A.R.; Ma, J.; Neale, G.; Shurtleff, S.A.; Bailey, S.; Gajjar, A.; et al. MYB upregulation and genetic aberrations in a subset of pediatric low-grade gliomas. Acta Neuropathol. 2010, 120, 731–743. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wu, G.; Miller, C.P.; Tatevossian, R.G.; Dalton, J.D.; Tang, B.; Orisme, W.; Punchihewa, C.; Parker, M.; Qaddoumi, I.; et al. Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat. Genet. 2013, 45, 602–612. [Google Scholar] [CrossRef]
- Bandopadhayay, P.; Ramkissoon, L.A.; Jain, P.; Bergthold, G.; Wala, J.; Zeid, R.; Schumacher, S.E.; Urbanski, L.; O’Rourke, R.; Gibson, W.J.; et al. MYB-QKI rearrangements in angiocentric glioma drive tumorigenicity through a tripartite mechanism. Nat. Genet. 2016, 48, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Guérin, M.; Sheng, Z.M.; Andrieu, N.; Riou, G. Strong association between c-myb and oestrogen-receptor expression in human breast cancer. Oncogene 1990, 5, 131–135. [Google Scholar]
- Drabsch, Y.; Hugo, H.; Zhang, R.; Dowhan, D.H.; Miao, Y.R.; Gewirtz, A.M.; Barry, S.C.; Ramsay, R.G.; Gonda, T.J. Mechanism of and requirement for estrogen-regulated MYB expression in estrogen-receptor-positive breast cancer cells. Proc. Natl. Acad. Sci. USA 2007, 104, 13762–13767. [Google Scholar] [CrossRef] [PubMed]
- Drabsch, Y.; Robert, R.G.; Gonda, T.J. MYB suppresses differentiation and apoptosis of human breast cancer cells. Breast Cancer Res. 2010, 12, R55. [Google Scholar] [CrossRef]
- Miao, R.Y.; Drabsch, Y.; Cross, R.S.; Cheasley, D.; Carpinteri, S.; Pereira, L.; Malaterre, J.; Gonda, T.J.; Anderson, R.L.; Ramsay, R.G. MYB is essential for mammary tumorigenesis. Cancer Res. 2011, 71, 7029–7037. [Google Scholar] [CrossRef]
- Yang, R.M.; Nanayakkara, D.; Kalimutho, M.; Mitra, P.; Khanna, K.K.; Dray, E.; Gonda, T.J. MYB regulates the DNA damage response and components of the homology-directed repair pathway in human estrogen receptor-positive breast cancer cells. Oncogene 2019, 38, 5239–5249. [Google Scholar] [CrossRef]
- Ramsay, R.G.; Thompson, M.A.; Hayman, J.A.; Reid, G.; Gonda, T.J.; Whitehead, R.H. Myb expression is higher in malignant human colonic carcinoma and premalignant adenomatous polyps than in normal mucosa. Cell Growth Differ. 1992, 3, 723–730. [Google Scholar] [PubMed]
- Biroccio, A.; Benassi, B.; D’Agnano, I.; D’Angelo, C.; Buglioni, S.; Mottolese, M.; Ricciotti, A.; Citro, G.; Cosimelli, M.; Ramsay, R.G.; et al. c-Myb and Bcl-x overexpression predicts poor prognosis in colorectal cancer: Clinical and experimental findings. Am. J. Pathol. 2001, 158, 1289–1299. [Google Scholar] [CrossRef] [PubMed]
- Hugo, H.; Cures, A.; Suraweera, N.; Drabsch, Y.; Purcell, D.; Mantamadiotis, T.; Phillips, W.; Dobrovic, A.; Zupi, G.; Gonda, T.J.; et al. Mutations in the MYB intron I regulatory sequence increase transcription in colon cancers. Genes Chromosomes Cancer 2006, 45, 1143–1154. [Google Scholar] [CrossRef] [PubMed]
- Malaterre, J.; Pereira, L.; Putoczki, T.; Millen, R.; Paquet-Fifield, S.; Germann, M.; Liu, J.; Cheasley, D.; Sampurno, S.; Stacker, S.A.; et al. Intestinal-specific activatable Myb initiates colon tumorigenesis in mice. Oncogene 2016, 35, 2475–2484. [Google Scholar] [CrossRef] [PubMed]
- Wallrapp, C.; Müller-Pillasch, F.; Solinas-Toldo, S.; Lichter, P.; Friess, H.; Büchler, M.; Fink, T.; Adler, G.; Gress, T.M. Characterization of a high copy number amplification at 6q24 in pancreatic cancer identifies c-myb as a candidate oncogene. Cancer Res. 1997, 57, 3135–3139. [Google Scholar] [PubMed]
- Srivastava, S.K.; Bhardwaj, A.; Arora, S.; Singh, S.; Azim, S.; Tyagi, N.; Carter, J.E.; Wang, B.; Singh, A.P. MYB is a novel regulator of pancreatic tumour growth and metastasis. Br. J. Cancer 2015, 113, 1694–1703. [Google Scholar] [CrossRef] [PubMed]
- Anand, S.; Khan, M.A.; Zubair, H.; Sudan, S.K.; Vikramdeo, K.S.; Deshmukh, S.K.; Azim, S.; Srivastava, S.K.; Singh, S.; Singh, A.P. MYB sustains hypoxic survival of pancreatic cancer cells by facilitating metabolic reprogramming. EMBO Rep. 2023, 24, e55643. [Google Scholar] [CrossRef]
- Edwards, J.; Krishna, N.S.; Witton, C.J.; Bartlett, J.M. Gene amplifications associated with the development of hormone-resistant prostate cancer. Clin. Cancer Res. 2003, 9, 5271–5281. [Google Scholar]
- Srivastava, S.K.; Bhardwaj, A.; Singh, S.; Arora, S.; McClellan, S.; Grizzle, W.E.; Reed, E.; Singh, A.P. Myb overexpression overrides androgen depletion-induced cell cycle arrest and apoptosis in prostate cancer cells and confers aggressive malignant traits: Potential role in castration resistance. Carcinogenesis 2012, 33, 1149–1157. [Google Scholar] [CrossRef]
- Srivastava, S.K.; Khan, M.A.; Anand, S.; Zubair, H.; Deshmukh, S.K.; Patel, G.K.; Singh, S.; Andrews, J.; Wang, B.; Carter, J.E.; et al. MYB interacts with androgen receptor, sustains its ligand-independent activation and promotes castration resistance in prostate cancer. Br. J. Cancer 2022, 126, 1205–1214. [Google Scholar] [CrossRef]
- Ratajczak, M.Z.; Hijiya, N.; Catani, L.; DeRiel, K.; Luger, S.M.; McGlave, P.; Gewirtz, A.M. Acute- and chronic-phase chronic myelogenous leukemia colony-forming units are highly sensitive to the growth inhibitory effects of c-myb antisense oligodeoxynucleotides. Blood 1992, 79, 1956–1961. [Google Scholar] [CrossRef]
- Jarvis, T.C.; Wincott, F.E.; Alby, L.J.; McSwiggen, J.A.; Beigelman, L.; Gustofson, J.; DiRenzo, A.; Levy, K.; Arthur, M.; Matulic-Adamic, J.; et al. Optimizing the cell efficacy of synthetic ribozymes. Site selection and chemical modifications of ribozymes targeting the proto-oncogene c-myb. J. Biol. Chem. 1996, 271, 29107–29112. [Google Scholar] [CrossRef]
- Kasper, L.H.; Boussouar, F.; Ney, P.A.; Jackson, C.W.; Rehg, J.; van Deursen, J.M.; Brindle, P.K. A transcription-factor-binding surface of coactivator p300 is required for haematopoiesis. Nature 2002, 419, 738–743. [Google Scholar] [CrossRef]
- Sandberg, M.L.; Sutton, S.E.; Pletcher, M.T.; Wiltshire, T.; Tarantino, L.M.; Hogenesch, J.B.; Cooke, M.P. c-Myb and p300 regulate hematopoietic stem cell proliferation and differentiation. Dev. Cell 2005, 8, 153–166. [Google Scholar] [CrossRef]
- Papathanasiou, P.; Tunningley, R.; Pattabiraman, D.R.; Ye, P.; Gonda, T.J.; Whittle, B.; Hamilton, A.E.; Cridland, S.O.; Lourie, R.; Perkins, A.C. A recessive screen for genes regulating hematopoietic stem cells. Blood 2010, 116, 5849–5858. [Google Scholar] [CrossRef] [PubMed]
- Best, J.L.; Amezcua, C.A.; Mayr, B.; Flechner, L.; Murawsky, C.M.; Emerson, B.; Zor, T.; Gardner, K.H.; Montminy, M. Identification of small-molecule antagonists that inhibit an activator: Coactivator interaction. Proc. Natl. Acad. Sci. USA 2004, 101, 17622–17627. [Google Scholar] [CrossRef] [PubMed]
- Uttarkar, S.; Dukare, S.; Bopp, B.; Goblirsch, M.; Jose, J.; Klempnauer, K.-H. Naphthol AS-E Phosphate Inhibits the Activity of the Transcription Factor Myb by Blocking the Interaction with the KIX Domain of the Coactivator p300. Mol. Cancer Ther. 2015, 14, 1276–1285. [Google Scholar] [CrossRef] [PubMed]
- Uttarkar, S.; Piontek, T.; Dukare, S.; Schomburg, C.; Schlenke, P.; Berdel, W.E.; Müller-Tidow, C.; Schmidt, T.J.; Klempnauer, K.-H. Small-Molecule Disruption of the Myb/p300 Cooperation Targets Acute Myeloid Leukemia Cells. Mol. Cancer Ther. 2016, 15, 2905–2915. [Google Scholar] [CrossRef] [PubMed]
- Ramaswamy, K.; Forbes, L.; Minuesa, G.; Gindin, T.; Brown, F.; Kharas, M.G.; Krivtsov, A.V.; Armstrong, S.A.; Still, E.; de Stanchina, E.; et al. Peptidomimetic blockade of MYB in acute myeloid leukemia. Nat. Commun. 2018, 9, 110. [Google Scholar] [CrossRef] [PubMed]
- Joy, S.T.; Henley, M.J.; De Salle, S.N.; Beyersdorf, M.S.; Vock, I.W.; Huldin, A.J.L.; Mapp, A.K. A Dual-Site Inhibitor of CBP/p300 KIX is a Selective and Effective Modulator of Myb. J. Am. Chem. Soc. 2021, 143, 15056–15062. [Google Scholar] [CrossRef] [PubMed]
- Suetaka, S.; Oka, Y.; Kunihara, T.; Hayashi, Y.; Arai, M. Rational design of a helical peptide inhibitor targeting c-Myb-KIX interaction. Sci. Rep. 2022, 12, 816. [Google Scholar] [CrossRef]
- Sato, N.; Suetaka, S.; Hayashi, Y.; Arai, M. Rational peptide design for inhibition of the KIX-MLL interaction. Sci. Rep. 2023, 13, 6330. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.; Grosche, P.; Floersheimer, A.; André, J.; Gattlen, R.; Oser, D.; Tinchant, J.; Wille, R.; Chie-Leon, B.; Gerspacher, M.; et al. Design and Biochemical Characterization of Peptidic Inhibitors of the Myb/p300 Interaction. Biochemistry 2023, 62, 1321–1329. [Google Scholar] [CrossRef]
- Yusenko, M.V.; Trentmann, A.; Casolari, D.A.; Abdel Ghani, L.; Lenz, M.; Horn, M.; Dörner, W.; Klempnauer, S.; Mootz, H.D.; Arteaga, M.F.; et al. C/EBPβ is a MYB- and p300-cooperating pro-leukemogenic factor and promising drug target in acute myeloid leukemia. Oncogene 2021, 40, 4746–4758. [Google Scholar] [CrossRef]
- Abdel Ghani, L.; Yusenko, M.V.; Frank, D.; Moorthy, R.; Widen, J.C.; Dörner, W.; Khandanpour, C.; Harki, D.A.; Klempnauer, K.-H. A synthetic covalent ligand of the C/EBPβ transactivation domain inhibits acute myeloid leukemia cells. Cancer Lett. 2022, 530, 170–180. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Milazzo, J.P.; Somerville, T.D.D.; Tarumoto, Y.; Huang, Y.H.; Ostrander, E.L.; Wilkinson, J.E.; Challen, G.A.; Vakoc, C.R. A TFIID-SAGA Perturbation that Targets MYB and Suppresses Acute Myeloid Leukemia. Cancer Cell 2018, 33, 13–28. [Google Scholar] [CrossRef] [PubMed]
- Yusenko, M.; Jakobs, A.; Klempnauer, K.-H. A novel cell-based screening assay for small-molecule MYB inhibitors identifies podophyllotoxins teniposide and etoposide as inhibitors of MYB activity. Sci. Rep. 2018, 8, 13159. [Google Scholar] [CrossRef] [PubMed]
- Yusenko, M.V.; Trentmann, A.; Andersson, M.K.; Ghani, L.A.; Jakobs, A.; Arteaga Paz, M.F.; Mikesch, J.H.; Peter von Kries, J.; Stenman, G.; Klempnauer, K.-H. Monensin, a novel potent MYB inhibitor, suppresses proliferation of acute myeloid leukemia and adenoid cystic carcinoma cells. Cancer Lett. 2020, 479, 61–70. [Google Scholar] [CrossRef]
- Yusenko, M.V.; Biyanee, A.; Andersson, M.K.; Radetzki, S.; von Kries, J.P.; Stenman, G.; Klempnauer, K.-H. Proteasome inhibitors suppress MYB oncogenic activity in a p300-dependent manner. Cancer Lett. 2021, 520, 132–142. [Google Scholar] [CrossRef]
- Yusenko, M.V.; Klempnauer, K.-H. Characterization of the MYB-inhibitory potential of the Pan-HDAC inhibitor LAQ824. BBA Adv. 2021, 2, 100034. [Google Scholar] [CrossRef]
- Yusenko, M.V.; Biyanee, A.; Frank, D.; Köhler, L.H.F.; Andersson, M.K.; Khandanpour, C.; Schobert, R.; Stenman, G.; Biersack, B.; Klempnauer, K.-H. Bcr-TMP, a Novel Nanomolar-Active Compound That Exhibits Both MYB- and Microtubule-Inhibitory Activity. Cancers 2021, 14, 43. [Google Scholar] [CrossRef]
- Biyanee, A.; Yusenko, M.V.; Klempnauer, K.-H. Src-Family Protein Kinase Inhibitors Suppress MYB Activity in a p300-Dependent Manner. Cells 2022, 11, 1162. [Google Scholar] [CrossRef]
- Mandelbaum, J.; Shestopalov, I.A.; Henderson, R.E.; Chau, N.G.; Knoechel, B.; Wick, M.J.; Zon, L.I. Zebrafish blastomere screen identifies retinoic acid suppression of MYB in adenoid cystic carcinoma. J. Exp. Med. 2018, 215, 2673–2685. [Google Scholar] [CrossRef]
- Hanna, G.J.; ONeill, A.; Cutler, J.M.; Flynn, M.; Vijaykumar, T.; Clark, J.R.; Wirth, L.J.; Lorch, J.H.; Park, J.C.; Mito, J.K.; et al. A phase II trial of all-trans retinoic acid (ATRA) in advanced adenoid cystic carcinoma. Oral Oncol. 2021, 119, 105366. [Google Scholar] [CrossRef]
- Walf-Vorderwülbecke, V.; Pearce, K.; Brooks, T.; Hubank, M.; van den Heuvel-Eibrink, M.M.; Zwaan, C.M.; Adams, S.; Edwards, D.; Bartram, J.; Samarasinghe, S.; et al. Targeting acute myeloid leukemia by drug-induced c-MYB degradation. Leukemia 2018, 32, 882–889. [Google Scholar] [CrossRef]
- Clesham, K.; Walf-Vorderwülbecke, V.; Gasparoli, L.; Virely, C.; Cantilena, S.; Tsakaneli, A.; Inglott, S.; Adams, S.; Samarasinghe, S.; Bartram, J.; et al. Identification of a c-MYB-directed therapeutic for acute myeloid leukemia. Leukemia 2022, 36, 1541–1549. [Google Scholar] [CrossRef] [PubMed]
- Falkenberg, K.D.; Jakobs, A.; Matern, J.C.; Dörner, W.; Uttarkar, S.; Trentmann, A.; Steinmann, S.; Coulibaly, A.; Schomburg, C.; Mootz, H.D.; et al. Withaferin A, a natural compound with anti-tumor activity, is a potent inhibitor of transcription factor C/EBPβ. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 1349–1358. [Google Scholar] [CrossRef] [PubMed]
- Tejera Nevado, P.; Tešan Tomić, T.; Atefyekta, A.; Fehr, A.; Stenman, G.; Andersson, M.K. Synthetic oleanane triterpenoids suppress MYB oncogene activity and sensitize T-cell acute lymphoblastic leukemia cells to chemotherapy. Front. Oncol. 2023, 13, 1126354. [Google Scholar] [CrossRef]
- Konopleva, M.; Tsao, T.; Ruvolo, P.; Stiouf, I.; Estrov, Z.; Leysath, C.E.; Zhao, S.; Harris, D.; Chang, S.; Jackson, C.E.; et al. Novel triterpenoid CDDO-Me is a potent inducer of apoptosis and differentiation in acute myelogenous leukemia. Blood 2002, 99, 326–335. [Google Scholar] [CrossRef] [PubMed]
- Suh, W.S.; Kim, Y.S.; Schimmer, A.D.; Kitada, S.; Minden, M.; Andreeff, M.; Suh, N.; Sporn, M.; Reed, J.C. Synthetic triterpenoids activate a pathway for apoptosis in AML cells involving downregulation of FLIP and sensitization to TRAIL. Leukemia 2003, 17, 2122–2129. [Google Scholar] [CrossRef] [PubMed]
- Liby, K.T.; Sporn, M.B. Synthetic oleanane triterpenoids: Multifunctional drugs with a broad range of applications for prevention and treatment of chronic disease. Pharmacol. Rev. 2012, 64, 972–1003. [Google Scholar] [CrossRef] [PubMed]
- Andersson, M.K.; Mangiapane, G.; Nevado, P.T.; Tsakaneli, A.; Carlsson, T.; Corda, G.; Nieddu, V.; Abrahamian, C.; Chayka, O.; Rai, L.; et al. ATR is a MYB regulated gene and potential therapeutic target in adenoid cystic carcinoma. Oncogenesis 2020, 9, 5. [Google Scholar] [CrossRef] [PubMed]
- Cicirò, Y.; Ragusa, D.; Nevado, P.T.; Lattanzio, R.; Sala, G.; DesRochers, T.; Millard, M.; Andersson, M.K.; Stenman, G.; Sala, A. The mitotic checkpoint kinase BUB1 is a direct and actionable target of MYB in adenoid cystic carcinoma. FEBS Lett. 2024, 598, 252–265. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Klempnauer, K.-H. Transcription Factor MYB as Therapeutic Target: Current Developments. Int. J. Mol. Sci. 2024, 25, 3231. https://doi.org/10.3390/ijms25063231
Klempnauer K-H. Transcription Factor MYB as Therapeutic Target: Current Developments. International Journal of Molecular Sciences. 2024; 25(6):3231. https://doi.org/10.3390/ijms25063231
Chicago/Turabian StyleKlempnauer, Karl-Heinz. 2024. "Transcription Factor MYB as Therapeutic Target: Current Developments" International Journal of Molecular Sciences 25, no. 6: 3231. https://doi.org/10.3390/ijms25063231
APA StyleKlempnauer, K.-H. (2024). Transcription Factor MYB as Therapeutic Target: Current Developments. International Journal of Molecular Sciences, 25(6), 3231. https://doi.org/10.3390/ijms25063231





