SARS-CoV-2 Infection Alters the Phenotype and Gene Expression of Adipocytes
Abstract
1. Introduction
2. Results
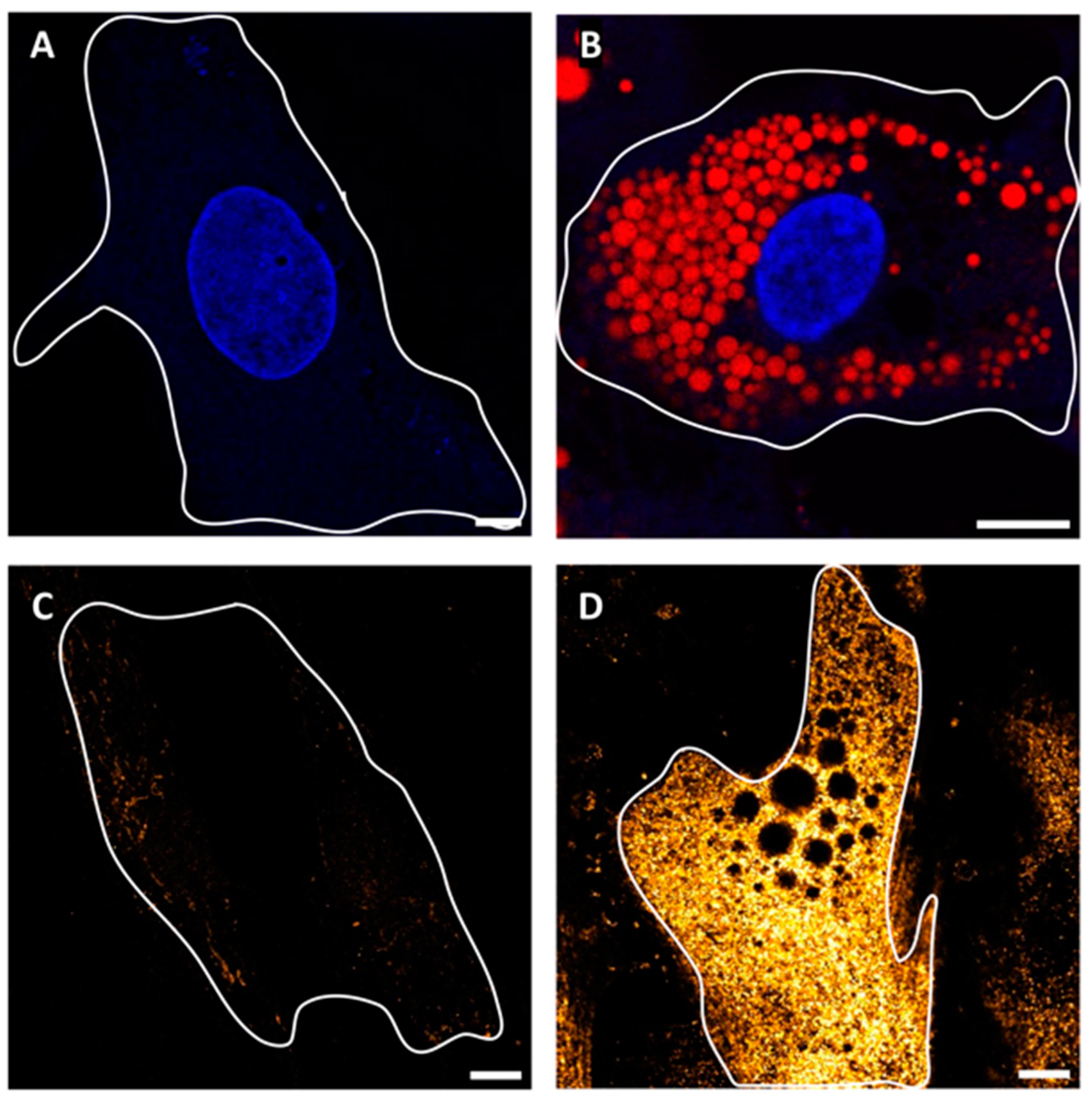
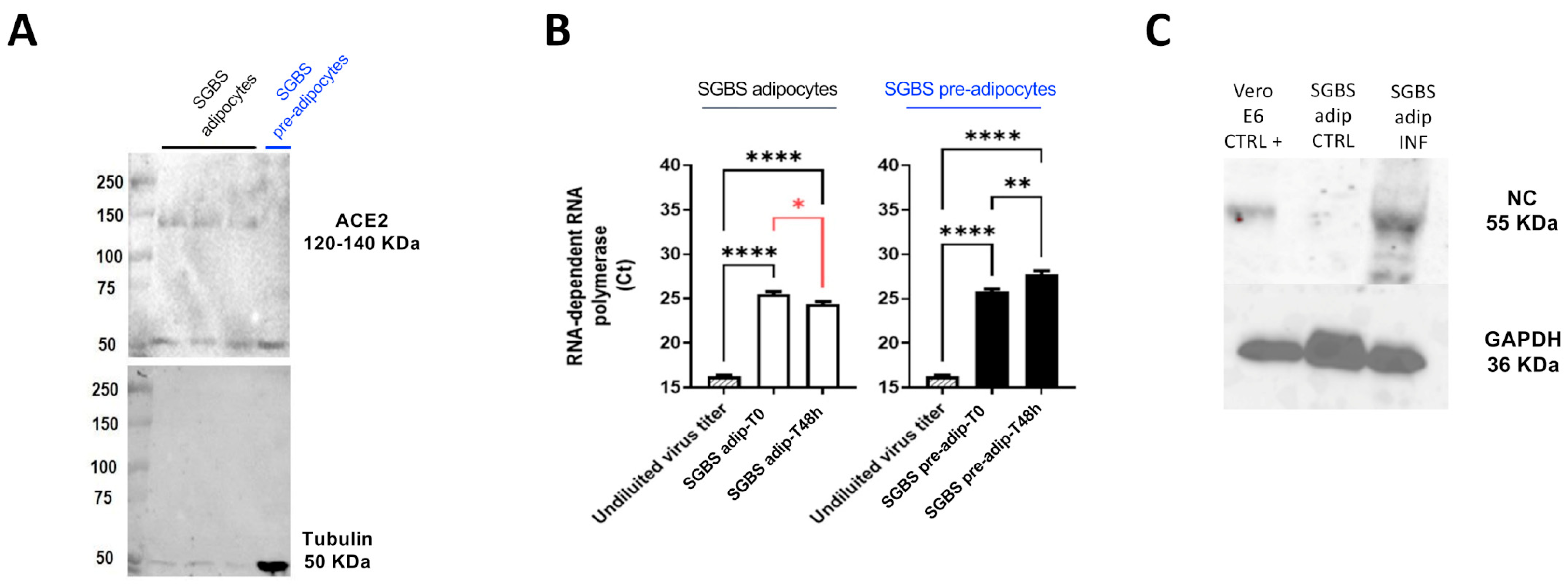
2.1. SGBS Cells Are Permissive to SARS-CoV-2 Infection
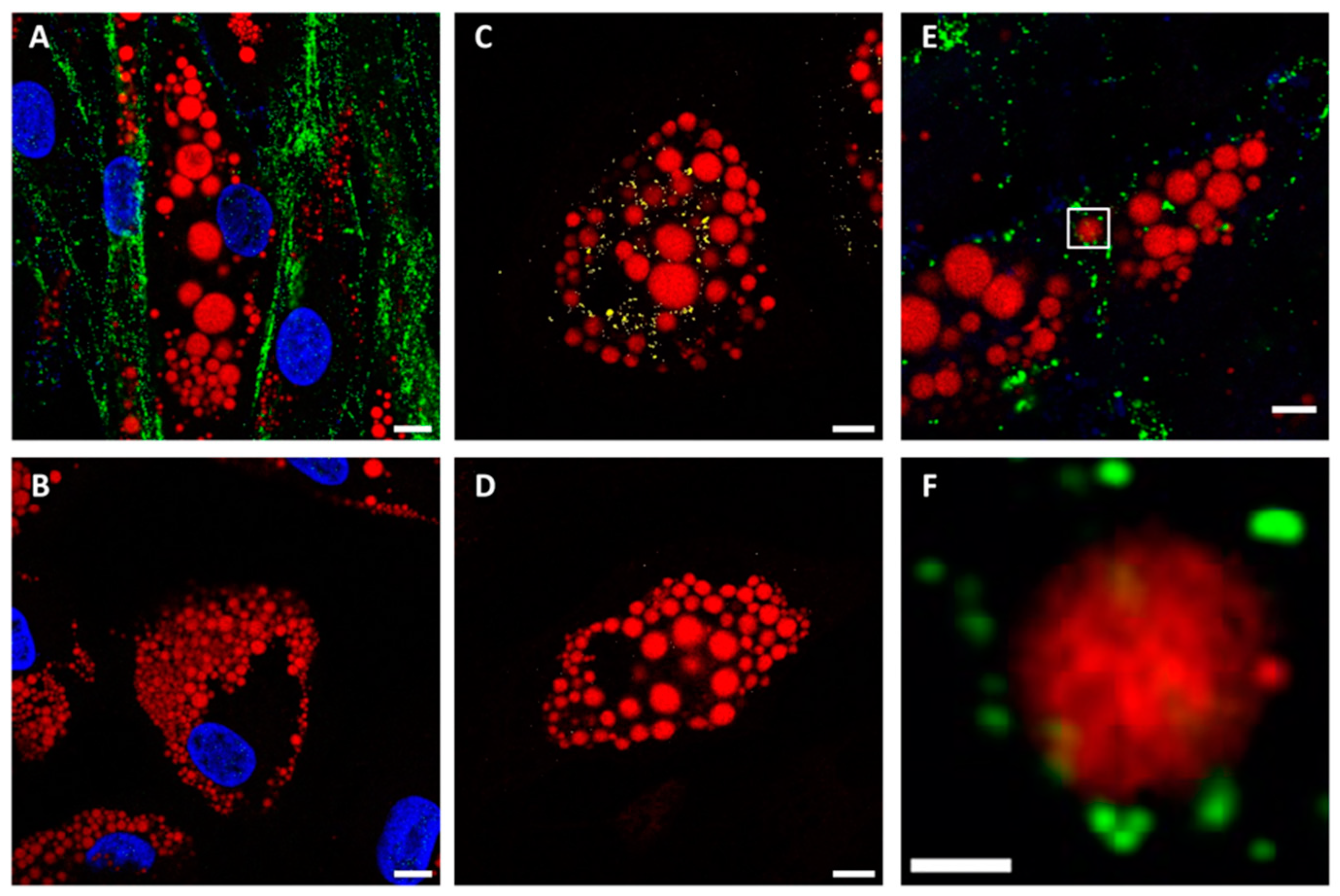
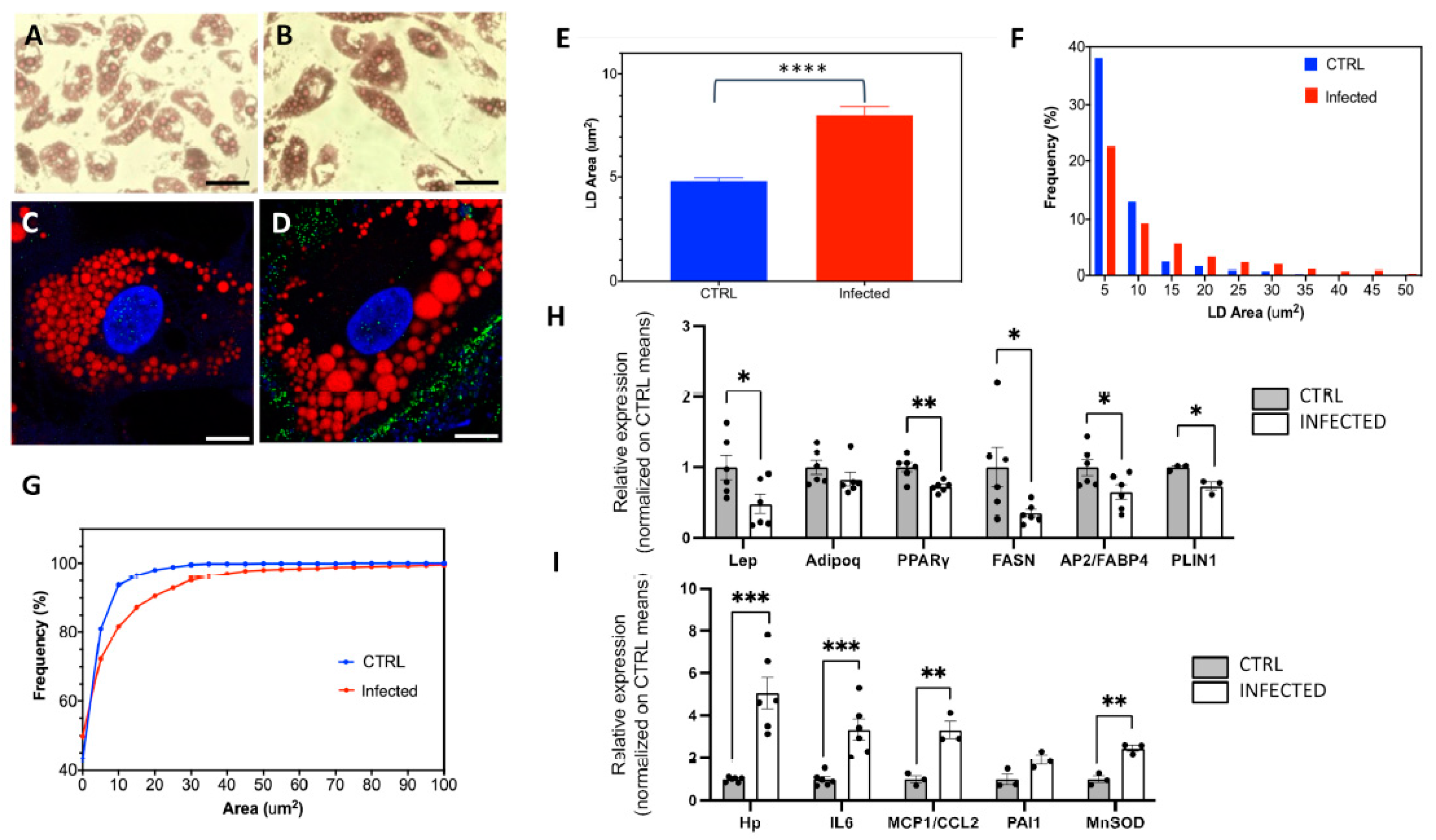
2.2. The Effects of SARS-CoV-2 Infection on SGBS Morphology and Gene Expression
2.3. The Role of Spike Protein in LD Enlargement
3. Discussion
4. Materials and Methods
4.1. Reagents, Plasmids and Antibodies
- -
- Anti-NC rabbit polyclonal antibody (GTX135357, GeneTex, Irvine, CA, USA), dilution: 1:1000.
- -
- Anti-GAPDH mouse monoclonal antibody (ab8245, Abcam, Cambridge, UK), dilution: 1:10,000.
- -
- Anti-mouse and anti-rabbit HRP-conjugated antibody (#170-6516 and #170-6515, BioRad, Hercules, CA, USA), dilution: 1:3000.
- -
- Anti-S IgG rabbit monoclonal antibody (40592-V05H, Sino Biological, Beijing, China), dilution: 1:200.
- -
- Anti-N IgG mouse monoclonal antibody (#33717, Cell Signaling, Danvers, MA, USA), dilution: 1:6400.
- -
- Anti-ACE2 IgG rabbit monoclonal antibody (ab15348, Abcam), dilution: 1:200.
- -
- αr488: donkey anti-rabbit monoclonal IgG conjugated to AlexaFluor488 (a21206, Thermo Fisher, Milan, Italy). Immunolabeling dilution: 1/400.
4.2. Cell Lines and Virus
4.3. SGBS Human Preadipocytes Culture and Differentiation
4.4. Transfection of SGBS Differentiating Cells
4.5. ORO Staining
4.6. Infection of SGBS Cells with SARS-CoV-2
- -
- in Ripa buffer 1× (Millipore 20-188, Burlington, MA, USA) 50 µL/well added with protease inhibitor cocktail 1× (Sigma-Aldrich, P8340) and phosphatase inhibitor cocktail 1× (Sigma-Aldrich, P0044) and stored at −20 °C, for Western blot analysis.
- -
- in QIAzol Lysis Reagent (Qiagen, Hilden, Germany, 79306) 50 µL/well and stored at −80 °C, for gene expression analysis.
4.7. qPCR and ddPCR
- -
- Hp, IL-6, mcp1/CCL2, ap2/FABP4, Adipoq, Lep, TBP, FAM-conjugated (Applied Biosystems, USA)
- -
- PAI-1, PPARgamma, FASN, PLIN, MnSOD, HEX-conjugated (Bio-Rad Laboratories, USA).
4.8. Western Blot
4.9. Immunostaining of Cells
4.10. Confocal and ISM Microscopy
- -
- Blue (Hoechst 33342): λex = 405 λem = 420–500 nm
- -
- Green (Alexa488): λex = 488, λem = 500–560 nm
- -
- Far-red (Oil red): λex = 640, λem = 650–700 nm
4.11. Imaging Analysis of Lipid Droplets
- A.
- Gaussian blur of the blue channel image (sigma: 1 pixel).
- B.
- Background subtraction in the blue channel image (rolling ball radius: 25 pixels).
- C.
- Thresholding (Method: Huang).
- D.
- Filling holes and watershed separation of the thresholded image.
- E.
- Particle analysis was performed by setting minimum area size as 4–16 pixels and minimum circularity as 0.05.
4.12. Graphics and Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gupta, A.; Madhavan, M.V.; Sehgal, K.; Nair, N.; Mahajan, S.; Sehrawat, T.S.; Bikdeli, B.; Ahluwalia, N.; Ausiello, J.C.; Wan, E.Y.; et al. Extrapulmonary Manifestations of COVID-19. Nat. Med. 2020, 26, 1017–1032. [Google Scholar] [CrossRef]
- Hoebel, J.; Kuntz, B.; Kroll, L.E.; Schienkiewitz, A.; Finger, J.D.; Lange, C.; Lampert, T. Socioeconomic Inequalities in the Rise of Adult Obesity: A Time-Trend Analysis of National Examination Data from Germany, 1990–2011. Obes. Facts 2019, 12, 344–356. [Google Scholar] [CrossRef]
- Petrilli, C.M.; Jones, S.A.; Yang, J.; Rajagopalan, H.; O’Donnell, L.; Chernyak, Y.; Tobin, K.A.; Cerfolio, R.J.; Francois, F.; Horwitz, L.I. Factors Associated with Hospital Admission and Critical Illness among 5279 People with Coronavirus Disease 2019 in New York City: Prospective Cohort Study. BMJ 2020, 369, m1966. [Google Scholar] [CrossRef] [PubMed]
- Simonnet, A.; Chetboun, M.; Poissy, J.; Raverdy, V.; Noulette, J.; Duhamel, A.; Labreuche, J.; Mathieu, D.; Pattou, F.; Jourdain, M.; et al. High Prevalence of Obesity in Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) Requiring Invasive Mechanical Ventilation. Obesity 2020, 28, 1195–1199. [Google Scholar] [CrossRef]
- Lopez-Jimenez, F.; Almahmeed, W.; Bays, H.; Cuevas, A.; Di Angelantonio, E.; le Roux, C.W.; Sattar, N.; Sun, M.C.; Wittert, G.; Pinto, F.J.; et al. Obesity and Cardiovascular Disease: Mechanistic Insights and Management Strategies. A Joint Position Paper by the World Heart Federation and World Obesity Federation. Eur. J. Prev. Cardiol. 2022, 29, 2218–2237. [Google Scholar] [CrossRef]
- Tartof, S.Y.; Qian, L.; Hong, V.; Wei, R.; Nadjafi, R.F.; Fischer, H.; Li, Z.; Shaw, S.F.; Caparosa, S.L.; Nau, C.L.; et al. Obesity and Mortality Among Patients Diagnosed With COVID-19: Results from an Integrated Health Care Organization. Ann. Intern. Med. 2020, 173, 773–781. [Google Scholar] [CrossRef] [PubMed]
- Moriconi, D.; Masi, S.; Rebelos, E.; Virdis, A.; Manca, M.L.; De Marco, S.; Taddei, S.; Nannipieri, M. Obesity Prolongs the Hospital Stay in Patients Affected by COVID-19, and May Impact on SARS-COV-2 Shedding. Obes. Res. Clin. Pract. 2020, 14, 205–209. [Google Scholar] [CrossRef] [PubMed]
- Heinonen, S.; Saarinen, L.; Naukkarinen, J.; Rodríguez, A.; Frühbeck, G.; Hakkarainen, A.; Lundbom, J.; Lundbom, N.; Vuolteenaho, K.; Moilanen, E.; et al. Adipocyte Morphology and Implications for Metabolic Derangements in Acquired Obesity. Int. J. Obes. 2014, 38, 1423–1431. [Google Scholar] [CrossRef]
- Maffei, M.; Barone, I.; Scabia, G.; Santini, F. The Multifaceted Haptoglobin in the Context of Adipose Tissue and Metabolism. Endocr. Rev. 2016, 37, 403–416. [Google Scholar] [CrossRef]
- Poma, A.M.; Basolo, A.; Alì, G.; Bonuccelli, D.; Di Stefano, I.; Conti, M.; Mazzetti, P.; Sparavelli, R.; Vignali, P.; Macerola, E.; et al. SARS-CoV-2 Spread to Endocrine Organs Is Associated with Obesity: An Autopsy Study of COVID-19 Cases. Endocrine 2023, 83, 110–117. [Google Scholar] [CrossRef]
- Basolo, A.; Poma, A.M.; Bonuccelli, D.; Proietti, A.; Macerola, E.; Ugolini, C.; Torregrossa, L.; Giannini, R.; Vignali, P.; Basolo, F.; et al. Adipose Tissue in COVID-19: Detection of SARS-CoV-2 in Adipocytes and Activation of the Interferon-Alpha Response. J. Endocrinol. Investig. 2022, 45, 1021–1029. [Google Scholar] [CrossRef]
- Martínez-Colón, G.J.; Ratnasiri, K.; Chen, H.; Jiang, S.; Zanley, E.; Rustagi, A.; Verma, R.; Chen, H.; Andrews, J.R.; Mertz, K.D.; et al. SARS-CoV-2 Infection Drives an Inflammatory Response in Human Adipose Tissue through Infection of Adipocytes and Macrophages. Sci. Transl. Med. 2022, 14, eabm9151. [Google Scholar] [CrossRef]
- Colleluori, G.; Graciotti, L.; Pesaresi, M.; Di Vincenzo, A.; Perugini, J.; Di Mercurio, E.; Caucci, S.; Bagnarelli, P.; Zingaretti, C.M.; Nisoli, E.; et al. Visceral Fat Inflammation and Fat Embolism Are Associated with Lung’s Lipidic Hyaline Membranes in Subjects with COVID-19. Int. J. Obes. 2022, 46, 1009–1017. [Google Scholar] [CrossRef]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 Entry into Cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef]
- Xia, S.; Liu, Z.; Jiang, S. High-Throughput Screening of Mutations Affecting SARS-CoV-2 Spike Functions. Trends Immunol. 2023, 44, 321–323. [Google Scholar] [CrossRef] [PubMed]
- Wabitsch, M.; Brenner, R.; Melzner, I.; Braun, M.; Möller, P.; Heinze, E.; Debatin, K.-M.; Hauner, H. Characterization of a Human Preadipocyte Cell Strain with High Capacity for Adipose Differentiation. Int. J. Obes. 2001, 25, 8–15. [Google Scholar] [CrossRef]
- Fischer-Posovszky, P.; Newell, F.S.; Wabitsch, M.; Tornqvist, H.E. Human SGBS Cells–a Unique Tool for Studies of Human Fat Cell Biology. Obes. Facts 2008, 1, 184–189. [Google Scholar] [CrossRef] [PubMed]
- Frese, N.; Schmerer, P.; Wortmann, M.; Schürmann, M.; König, M.; Westphal, M.; Weber, F.; Sudhoff, H.; Gölzhäuser, A. Imaging of SARS-CoV-2 Infected Vero E6 Cells by Helium Ion Microscopy. Beilstein J. Nanotechnol. 2021, 12, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Pires De Souza, G.A.; Le Bideau, M.; Boschi, C.; Wurtz, N.; Colson, P.; Aherfi, S.; Devaux, C.; La Scola, B. Choosing a Cellular Model to Study SARS-CoV-2. Front. Cell. Infect. Microbiol. 2022, 12, 1003608. [Google Scholar] [CrossRef] [PubMed]
- Nepita, I.; Piazza, S.; Ruglioni, M.; Cristiani, S.; Bosurgi, E.; Salvadori, T.; Vicidomini, G.; Diaspro, A.; Castello, M.; Bianchini, P.; et al. Image Scanning Microscopy to Investigate Polycomb Protein Colocalization onto Chromatin. Appl. Sci. 2023, 13, 1556. [Google Scholar] [CrossRef]
- Storti, B.; Quaranta, P.; Di Primio, C.; Clementi, N.; Mancini, N.; Criscuolo, E.; Spezia, P.G.; Carnicelli, V.; Lottini, G.; Paolini, E.; et al. A Spatial Multi-Scale Fluorescence Microscopy Toolbox Discloses Entry Checkpoints of SARS-CoV-2 Variants in Vero E6 Cells. Comput. Struct. Biotechnol. J. 2021, 19, 6140–6156. [Google Scholar] [CrossRef]
- Braga, L.; Ali, H.; Secco, I.; Chiavacci, E.; Neves, G.; Goldhill, D.; Penn, R.; Jimenez-Guardeño, J.M.; Ortega-Prieto, A.M.; Bussani, R.; et al. Drugs That Inhibit TMEM16 Proteins Block SARS-CoV-2 Spike-Induced Syncytia. Nature 2021, 594, 88–93. [Google Scholar] [CrossRef]
- Buchrieser, J.; Dufloo, J.; Hubert, M.; Monel, B.; Planas, D.; Rajah, M.M.; Planchais, C.; Porrot, F.; Guivel-Benhassine, F.; Van der Werf, S.; et al. Syncytia Formation by SARS-CoV-2-infected Cells. EMBO J. 2020, 39, e106267. [Google Scholar] [CrossRef]
- Martínez-Mármol, R.; Giordano-Santini, R.; Kaulich, E.; Cho, A.-N.; Przybyla, M.; Riyadh, M.A.; Robinson, E.; Chew, K.Y.; Amor, R.; Meunier, F.A.; et al. SARS-CoV-2 Infection and Viral Fusogens Cause Neuronal and Glial Fusion That Compromises Neuronal Activity. Sci. Adv. 2023, 9, eadg2248. [Google Scholar] [CrossRef] [PubMed]
- Frühbeck, G.; Catalán, V.; Valentí, V.; Moncada, R.; Gómez-Ambrosi, J.; Becerril, S.; Silva, C.; Portincasa, P.; Escalada, J.; Rodríguez, A. FNDC4 and FNDC5 Reduce SARS-CoV-2 Entry Points and Spike Glycoprotein S1-Induced Pyroptosis, Apoptosis, and Necroptosis in Human Adipocytes. Cell. Mol. Immunol. 2021, 18, 2457–2459. [Google Scholar] [CrossRef] [PubMed]
- Reiterer, M.; Rajan, M.; Gómez-Banoy, N.; Lau, J.D.; Gomez-Escobar, L.G.; Ma, L.; Gilani, A.; Alvarez-Mulett, S.; Sholle, E.T.; Chandar, V.; et al. Hyperglycemia in Acute COVID-19 Is Characterized by Insulin Resistance and Adipose Tissue Infectivity by SARS-CoV-2. Cell Metab. 2021, 33, 2484. [Google Scholar] [CrossRef] [PubMed]
- Oudshoorn, D.; Rijs, K.; Limpens, R.W.A.L.; Groen, K.; Koster, A.J.; Snijder, E.J.; Kikkert, M.; Bárcena, M. Expression and Cleavage of Middle East Respiratory Syndrome Coronavirus Nsp3-4 Polyprotein Induce the Formation of Double-Membrane Vesicles That Mimic Those Associated with Coronaviral RNA Replication. mBio 2017, 8, e01658-17. [Google Scholar] [CrossRef] [PubMed]
- Klein, S.; Cortese, M.; Winter, S.L.; Wachsmuth-Melm, M.; Neufeldt, C.J.; Cerikan, B.; Stanifer, M.L.; Boulant, S.; Bartenschlager, R.; Chlanda, P. SARS-CoV-2 Structure and Replication Characterized by in Situ Cryo-Electron Tomography. Nat. Commun. 2020, 11, 5885. [Google Scholar] [CrossRef] [PubMed]
- Cottier, S.; Schneiter, R. Lipid Droplets Form a Network Interconnected by the Endoplasmic Reticulum through Which Their Proteins Equilibrate. J. Cell Sci. 2022, 135, jcs258819. [Google Scholar] [CrossRef] [PubMed]
- Maffei, M.; Funicello, M.; Vottari, T.; Gamucci, O.; Costa, M.; Lisi, S.; Viegi, A.; Ciampi, O.; Bardi, G.; Vitti, P.; et al. The Obesity and Inflammatory Marker Haptoglobin Attracts Monocytes via Interaction with Chemokine (C-C Motif) Receptor 2 (CCR2). BMC Biol. 2009, 7, 87. [Google Scholar] [CrossRef] [PubMed]
- Lisi, S.; Gamucci, O.; Vottari, T.; Scabia, G.; Funicello, M.; Marchi, M.; Galli, G.; Arisi, I.; Brandi, R.; D’Onofrio, M.; et al. Obesity-Associated Hepatosteatosis and Impairment of Glucose Homeostasis Are Attenuated by Haptoglobin Deficiency. Diabetes 2011, 60, 2496–2505. [Google Scholar] [CrossRef]
- Kaji, H. Adipose Tissue-Derived Plasminogen Activator Inhibitor-1 Function and Regulation. In Comprehensive Physiology; Wiley: Hoboken, NJ, USA, 2016; pp. 1873–1896. [Google Scholar]
- Zagotta, I.; Dimova, E.Y.; Funcke, J.-B.; Wabitsch, M.; Kietzmann, T.; Fischer-Posovszky, P. Resveratrol Suppresses PAI-1 Gene Expression in a Human In Vitro Model of Inflamed Adipose Tissue. Oxidative Med. Cell. Longev. 2013, 2013, 793525. [Google Scholar] [CrossRef]
- Candas, D.; Li, J.J. MnSOD in Oxidative Stress Response-Potential Regulation via Mitochondrial Protein Influx. Antioxid. Redox Signal. 2014, 20, 1599–1617. [Google Scholar] [CrossRef] [PubMed]
- Dalen, K.T.; Schoonjans, K.; Ulven, S.M.; Weedon-Fekjaer, M.S.; Bentzen, T.G.; Koutnikova, H.; Auwerx, J.; Nebb, H.I. Adipose Tissue Expression of the Lipid Droplet–Associating Proteins S3-12 and Perilipin Is Controlled by Peroxisome Proliferator–Activated Receptor-γ. Diabetes 2004, 53, 1243–1252. [Google Scholar] [CrossRef]
- Medina-Gomez, G.; Gray, S.; Vidal-Puig, A. Adipogenesis and Lipotoxicity: Role of Peroxisome Proliferator-Activated Receptor γ (PPARγ) and PPARγcoactivator-1 (PGC1). Public Health Nutr. 2007, 10, 1132–1137. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xu, Y.; Zhang, X.; Wang, S.; Peng, Z.; Guo, J.; Jiang, H.; Liu, J.; Xie, Y.; Wang, J.; et al. Leptin Correlates with Monocytes Activation and Severe Condition in COVID-19 Patients. J. Leukoc. Biol. 2021, 110, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Larsson, A.; Lipcsey, M.; Hultström, M.; Frithiof, R.; Eriksson, M. Plasma Leptin Is Increased in Intensive Care Patients with COVID-19—An Investigation Performed in the PronMed-Cohort. Biomedicines 2021, 10, 4. [Google Scholar] [CrossRef] [PubMed]
- Maffei, M.; Giordano, A. Leptin, the Brain and Energy Homeostasis: From an Apparently Simple to a Highly Complex Neuronal System. Rev. Endocr. Metab. Disord. 2022, 23, 87–101. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Ambrosi, J.; Salvador, J.; Páramo, J.A.; Orbe, J.; de Irala, J.; Diez-Caballero, A.; Gil, M.J.; Cienfuegos, J.A.; Frühbeck, G. Involvement of Leptin in the Association between Percentage of Body Fat and Cardiovascular Risk Factors. Clin. Biochem. 2002, 35, 315–320. [Google Scholar] [CrossRef] [PubMed]
- Popkin, B.M.; Du, S.; Green, W.D.; Beck, M.A.; Algaith, T.; Herbst, C.H.; Alsukait, R.F.; Alluhidan, M.; Alazemi, N.; Shekar, M. Individuals with Obesity and COVID-19: A Global Perspective on the Epidemiology and Biological Relationships. Obes. Rev. 2020, 21, e13128. [Google Scholar] [CrossRef]
- Maurya, R.; Bhattacharya, P.; Dey, R.; Nakhasi, H.L. Leptin Functions in Infectious Diseases. Front. Immunol. 2018, 9, 2741. [Google Scholar] [CrossRef]
- Matarese, G.; Moschos, S.; Mantzoros, C.S. Leptin in Immunology. J. Immunol. 2005, 174, 3137–3142. [Google Scholar] [CrossRef] [PubMed]
- Prompetchara, E.; Ketloy, C.; Palaga, T. Immune Responses in COVID-19 and Potential Vaccines: Lessons Learned from SARS and MERS Epidemic. Asian Pac. J. Allergy Immunol. 2020, 38, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Honecker, J.; Ruschke, S.; Seeliger, C.; Laber, S.; Strobel, S.; Pröll, P.; Nellaker, C.; Lindgren, C.M.; Kulozik, U.; Ecker, J.; et al. Transcriptome and Fatty-Acid Signatures of Adipocyte Hypertrophy and Its Non-Invasive MR-Based Characterization in Human Adipose Tissue. EBioMedicine 2022, 79, 104020. [Google Scholar] [CrossRef] [PubMed]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.-L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM Structure of the 2019-nCoV Spike in the Prefusion Conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef]
- Rajah, M.M.; Bernier, A.; Buchrieser, J.; Schwartz, O. The Mechanism and Consequences of SARS-CoV-2 Spike-Mediated Fusion and Syncytia Formation. J. Mol. Biol. 2022, 434, 167280. [Google Scholar] [CrossRef] [PubMed]
- Dias, S.S.G.; Soares, V.C.; Ferreira, A.C.; Sacramento, C.Q.; Fintelman-Rodrigues, N.; Temerozo, J.R.; Teixeira, L.; Nunes da Silva, M.A.; Barreto, E.; Mattos, M.; et al. Lipid Droplets Fuel SARS-CoV-2 Replication and Production of Inflammatory Mediators. PLoS Pathog. 2020, 16, e1009127. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Moro Huitron, L.; De Jesús-González, L.A.; Martínez-Castillo, M.; Ulloa-Aguilar, J.M.; Cabello-Gutierrez, C.; Helguera-Repetto, C.; Garcia-Cordero, J.; León Juárez, M. Multifaceted Nature of Lipid Droplets in Viral Interactions and Pathogenesis. Microorganisms 2023, 11, 1851. [Google Scholar] [CrossRef]
- Yu, Y.; Maguire, T.G.; Alwine, J.C. Human Cytomegalovirus Infection Induces Adipocyte-Like Lipogenesis through Activation of Sterol Regulatory Element Binding Protein 1. J. Virol. 2012, 86, 2942–2949. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Quaranta, P.; Scabia, G.; Storti, B.; Dattilo, A.; Quintino, L.; Perrera, P.; Di Primio, C.; Costa, M.; Pistello, M.; Bizzarri, R.; et al. SARS-CoV-2 Infection Alters the Phenotype and Gene Expression of Adipocytes. Int. J. Mol. Sci. 2024, 25, 2086. https://doi.org/10.3390/ijms25042086
Quaranta P, Scabia G, Storti B, Dattilo A, Quintino L, Perrera P, Di Primio C, Costa M, Pistello M, Bizzarri R, et al. SARS-CoV-2 Infection Alters the Phenotype and Gene Expression of Adipocytes. International Journal of Molecular Sciences. 2024; 25(4):2086. https://doi.org/10.3390/ijms25042086
Chicago/Turabian StyleQuaranta, Paola, Gaia Scabia, Barbara Storti, Alessia Dattilo, Lara Quintino, Paola Perrera, Cristina Di Primio, Mario Costa, Mauro Pistello, Ranieri Bizzarri, and et al. 2024. "SARS-CoV-2 Infection Alters the Phenotype and Gene Expression of Adipocytes" International Journal of Molecular Sciences 25, no. 4: 2086. https://doi.org/10.3390/ijms25042086
APA StyleQuaranta, P., Scabia, G., Storti, B., Dattilo, A., Quintino, L., Perrera, P., Di Primio, C., Costa, M., Pistello, M., Bizzarri, R., & Maffei, M. (2024). SARS-CoV-2 Infection Alters the Phenotype and Gene Expression of Adipocytes. International Journal of Molecular Sciences, 25(4), 2086. https://doi.org/10.3390/ijms25042086






