Abstract
Abiotic stress affects the growth and development of maize (Zea mays). The regulator of chromosome condensation 1 (RCC1)-containing proteins (RCPs) plays crucial roles in plant growth and development and response to abiotic stresses. However, a comprehensive analysis of the maize RCP family has not been reported in detail. This study presents a systematic bioinformatics analysis of the ZmRCP family, identifying a total of 30 members distributed across nine chromosomes. The physicochemical properties and cis-acting elements in the promoters of ZmRCP members are predicted. The results of subcellular localization showed that ZmRCP3 and ZmRCP10 are targeted to mitochondria and ZmRCP2 is localized in the nucleus. A heatmap of expression levels among family members under abiotic stress conditions revealed varying degrees of induced expression, and the expression levels of 10 ZmRCP members were quantified using RT-qPCR under abiotic stress and plant hormone treatments. The results showed that ZmRCP members exhibit induced or inhibited responses to these abiotic stresses and plant hormones. These results contribute to a better understanding of the evolutionary history and potential role of the ZmRCP family in mediating responses to abiotic stress in maize.
1. Introduction
Maize is an important cereal crop that often suffers from abiotic stresses such as drought, high temperature, low temperature, and salinity during its growth and development, leading to severe yield reduction [1]. During long periods of evolution, organisms have evolved a range of mechanisms to cope with a variety of stresses, such as UV protection and osmo-protection mechanisms [2,3]. Regulator of chromosome condensation 1 (RCC1)-containing proteins (RCPs) have been reported to mediate diverse biological processes, including these abiotic stress responses [4,5,6].
Proteins that contain one or more RCC1/RCC1-like domains/repeats are classified as RCC1 proteins/RCPs [7]. Each repeat contains 51-68 amino acid residues and adopts a seven-bladed β-propeller fold [4,8]. RCP members are commonly found in eukaryotic cells. In humans, RCC1 is a key cell cycle regulatory factor involved in chromosome binding, nuclear cytoplasmic transport, and cell cycle control [8]. In animals, RCC1 is involved in many biological processes coupled with Ran in the nucleus, where RanGDP can produce RanGTP under the action of RCC1 [9,10].
In plants, RCPs have been identified in several species, including rice (Oryza sativa), maize (Zea mays), cotton (Gossypium Hirsutum), Populus euphratica, and Arabidopsis thaliana [5,6,11,12,13,14]. In Arabidopsis, 24 putative RCP family members have been identified. Six AtRCP proteins have been characterized, including UVR8 (ultraviolet receptor 8), tolerant to chilling and freezing (TCF1), RCC1/UVR8/GEF like 1 (RUG1), RUG2, RUG3, and sensitive to ABA 1 (SAB1) [5,11,12,15,16,17,18]. Under UV-B irradiation, UVR8 can dissociate from homologous dimers into monomers, which can interact with COP1 and continue to accumulate in the nucleus, triggering the UV-B cascade reaction, so that the plant activates the UV protection mechanism [3,19]. RUG1 encodes bilirubin deaminase (PBGD), which is an enzyme in the tetrapyrrole biosynthesis pathway that produces chlorophyll, heme, and so on. Deficiency of RUG1 has been shown to lead to leaf damage, abnormal plant development, and down-regulated expression of auxin-related genes [20]. RUG2 encodes a dual localization protein, which localizes in both mitochondria and chloroplasts, and its mutation exhibits pale green leaves, growth retardation, and a reduced number of mesophyll cells [16]. During seed germination and post-germination growth, mutation of RUG3 enhances the responses to ABA, suggesting that RUG3 as a negative regulatory factor mediates the ABA signaling pathway in Arabidopsis [17]. Arabidopsis SAB1 is also an RCP member as a negative regulator of ABI5 (abscisic acid-insensitive 5); it functions upstream of ABI5 and regulates promoter binding activity, its stability, and histone-methylation-mediated gene silencing of ABI5 post-germination [11]. Moreover, Arabidopsis TCF1 encodes a glycosylphosphatidylinositol anchored protein that mediates a plant-specific freezing-resistant transcription program by regulating lignin biosynthesis independently of the CBF pathway [5]. These cases indicate that RCP members are involved in the response to abiotic stresses in Arabidopsis.
In addition, rice Root Length Regulator 4 (OsRLR4) encodes an RCP protein that affects root elongation by negatively regulating RAM (root apical meristem) activity [13]. Overexpression of Populus euphratica UVR8 (PeUVR8) in the Arabidopsis uvr8 mutant rescued its phenotype and overexpression of PeUVR8 in wild-type Arabidopsis enhanced anthocyanin accumulation and inhibited hypocotyl elongation, indicating that PeUVR8 functions in controlling plant photomorphogenesis [14]. In maize, several RCP family members have been reported. For example, DEK47 is responsible for splicing four introns of mitochondrial nad2 in maize; therefore, it is critical for the assembly and activity of mitochondrial complex I and seed development [21]. The down-regulated expression of ZmRCC1-1 causes cytological changes, such as altering grain orientation and affecting cell cycle progression. ZmRCC1-2 and ZmRCC1-3 neutralize the changes caused by ZmRCC1-1 down-regulation, returning chromosome function to a normal state [22]. Although RCP members have been studied for over thirty years, there is still a lack of systematic identification and an analysis of the RCP family in maize.
In this study, we identified a total of 30 RCP members in maize through a systematic bioinformatics analysis. A comprehensive analysis of the ZmRCP family, including physicochemical properties, a phylogenetic tree, and subcellular localization, is performed. The expression analysis of ZmRCP genes were determined by RT-qPCR under abiotic stresses and plant hormones treatments. These data provide theoretical support and optional gene resources for future research on the biological functions of the ZmRCP family and the cultivation of stress-resistant maize varieties.
2. Results
2.1. Identification and Prediction of Protein Physicochemical Properties
To identify the RCP family members in maize, we operated the homologous alignment using the Arabidopsis RCP proteins using bidirectional BLASTp from NCBI. After domain validation, a total of 30 maize RCP family members were identified (Table 1, Figure 1). The number of amino acid residues (aa) in the ZmRCP family ranges from 181 to 1092. The molecular weight of ZmRCP family proteins ranges from 19,246.71 Da to 117,959.71 Da. The isoelectric point is between 4.95 and 9.84; most are alkaline proteins. The instability index of ZmRCP family proteins ranges from 6.81 to 49.16, and 19 members have an instability index lower than 40, indicating their stability. The hydrophilicity index ranges from −0.609 to −0.01.

Table 1.
Prediction of physicochemical properties of the maize RCP family members.
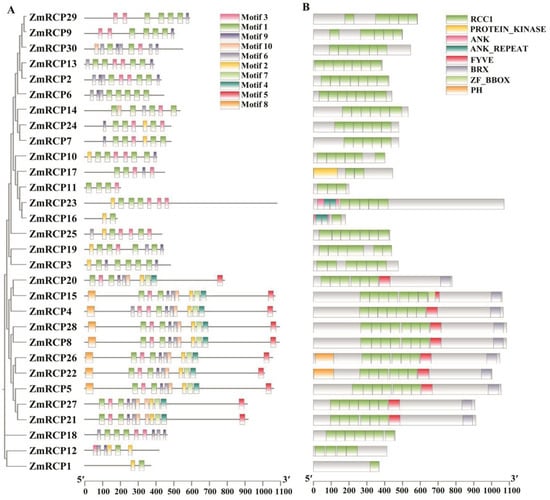
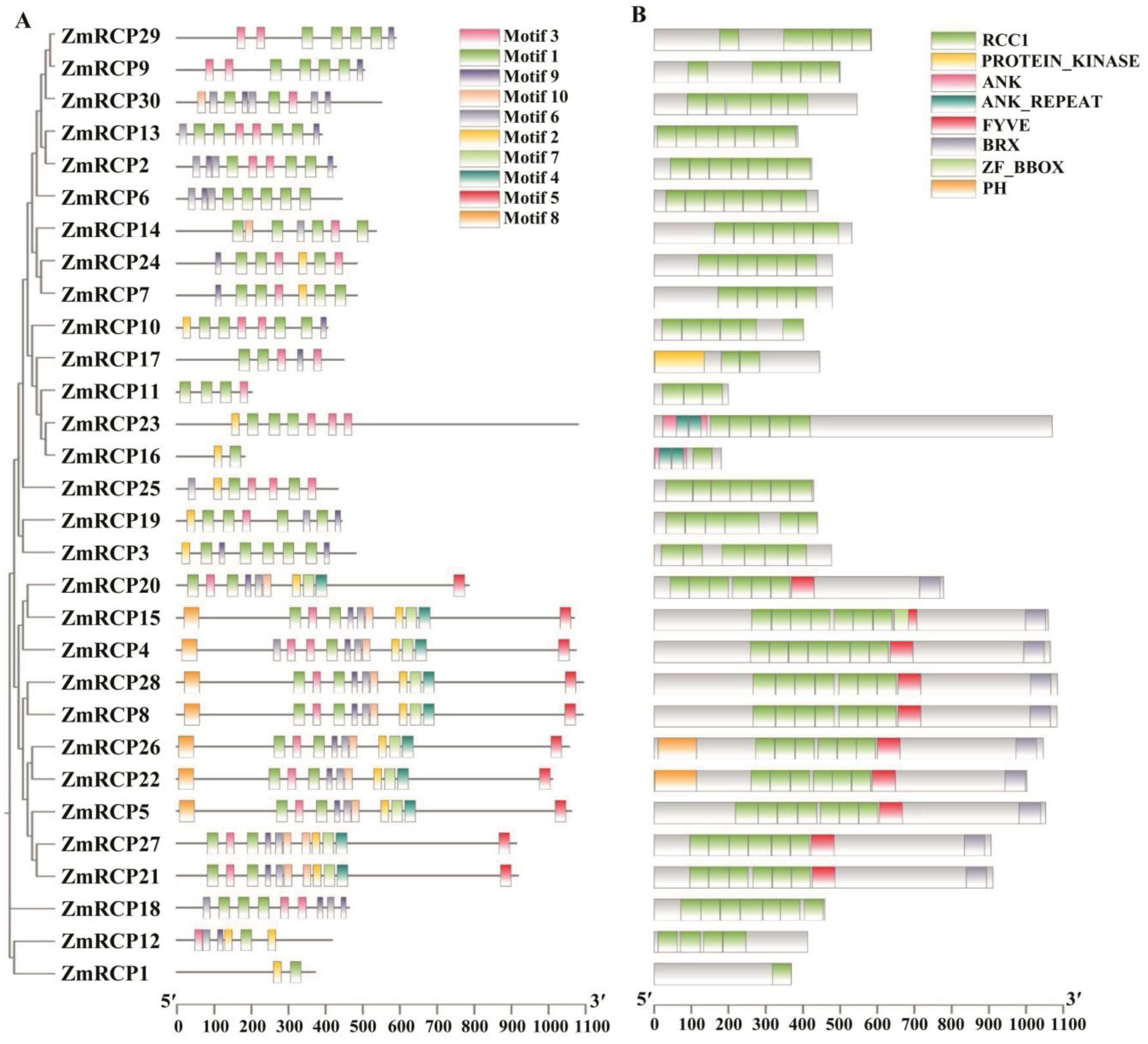
Figure 1.
Prediction of conserved motifs and domains of ZmRCP family members in maize. (A) The phylogenetic relationship (left panel) and conserved motifs (right panel) of ZmRCP members are listed. (B) Prediction of conserved domains of ZmRCP members.
2.2. Phylogenetic Relationship and Collinearity Analysis
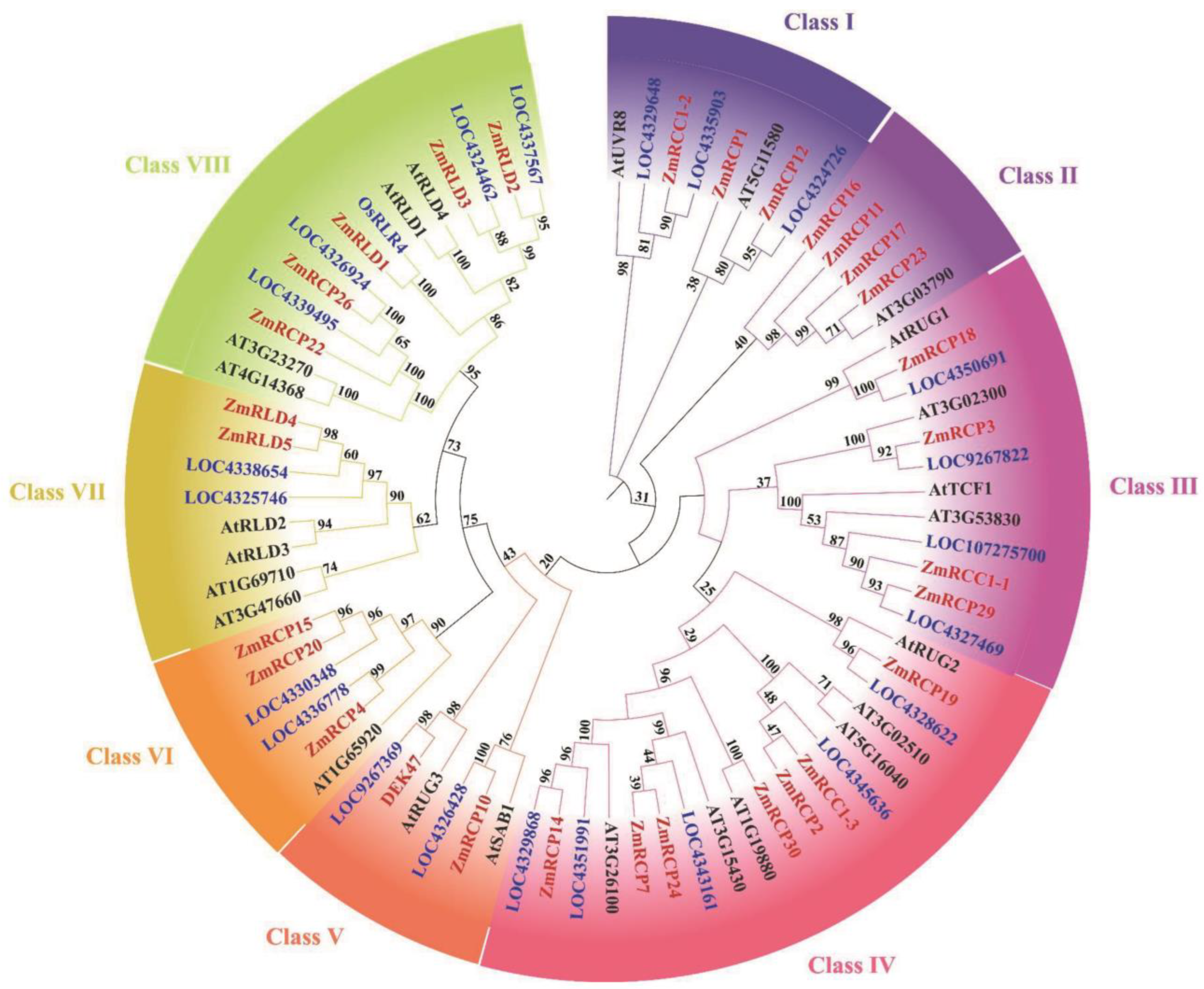
In order to explore the specific phylogenetic relationship of RCP members in maize and other species, we constructed a phylogenetic tree of RCP proteins in three species, including Zea mays, Arabidopsis thaliana, and Oryza sativa. After domain validation of the rice protein sequence downloaded from NCBI, it was found that there are 22 members of the same domain in rice. There are 30 members in maize, 24 members in Arabidopsis, and 23 members in rice, indicating that the number of RCP family members is relatively conservative with no significant expansion (Figure 2). After constructing the tree using the adjacency method, all RCP family members in these three species were clustered into eight classes based on their genetic relationships, from Class I to VIII. In maize, there are two members in Class V, three members in Class I, VI, and VII, four members in Class II and III, five members in Class VIII, and seven members in Class IV (Figure 2).
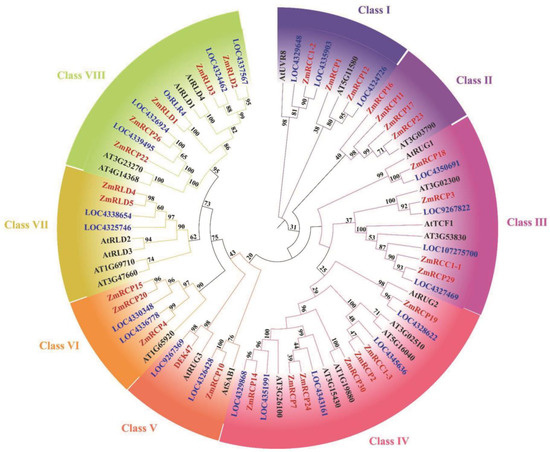
Figure 2.
Phylogenetic tree of RCP family members among maize, rice, and Arabidopsis. Bootstrap values are marked at the branches. Different groups are marked with different colors.
To further understand the evolutionary relationship of the RCP gene family in different species, we selected three plants for a species collinearity analysis, Zea mays, Oryza sativa, and Arabidopsis thaliana. The results showed that there are two RCP genes in Zea mays that are collinear with Arabidopsis thaliana (Figure S1). However, there are 22 RCP genes in maize that are collinear with Oryza sativa. Among them, ZmRCP9 and ZmRCP29 are collinear with both Arabidopsis thaliana and Oryza sativa. Compared with dicotyledonous plants like Arabidopsis thaliana, more RCP genes in Zea mays show collinearity with monocotyledonous plants like Oryza sativa, indicating that Zea mays and Oryza sativa, both of which are monocotyledonous plants, have a higher homology of RCP members. Most RCP genes originated from the ancestors of monocotyledonous plants, while a small number of genes may originate from dicotyledonous plants.
Collinearity analysis is a technique used to identify homologous protein sequences in the genome that are arranged in almost the same order at different positions in order to further reveal the genetic relationships between or within species of these proteins. The results of an intra-species collinearity analysis in maize showed that there were eight fragment duplication events on chromosomes 1, 2, 3, 4, 5, 7, and 8, involving a total of 12 genes (Figure S2). Based on the results of the evolutionary tree, we found that two genes with collinearity are classified in the same class (Figure 2). For example, ZmRCP2 and ZmRCP13/ZmRCC1-3 both belong to the Class IV subfamily, indicating that their gene structures and conserved domains are similar, which further supports the rationality of evolutionary tree grouping. The members of the RCP gene family have a certain degree of conservation and many gene duplications, which not only play an important role in the survival process of maize but also provide possibilities for the diversification of gene functions and gene evolution.
2.3. Conserved Motifs, Domains, and Gene Structure Analysis
In order to study the genetic structural changes of the RCP family genes in maize, their gene structures were analyzed. The diagram of gene structure shows that all members of the maize RCP family contain different quantities of introns, and most members contain untranslated regions at the 5′ and/or 3′ ends except for three members (Figure S3). The numbers of introns of RCP members contained are from 2 to 15.
A motif is commonly referred to as a super secondary structure. This structure is formed by the aggregation of secondary structural units of proteins, and the motif plays an important role in the function and structure of proteins. The prediction of motifs of RCP family members showed that the ZmRCP gene family in maize contains a total of 10 motifs, with ZmRCP1 and ZmRCP16 having only two motifs (Figure 1A). The vast majority of members contain motif 1 and motif 3, except for ZmRCP1, ZmRCP3, ZmRCP6, and ZmRCP16 (Figure 1A).
The conserved domains of the maize RCP gene family were analyzed using the MEME tool of Tbtools. The prediction results of conserved domains showed that all the ZmRCP members have the RCC1 domain, and the amounts of the RCC1 domain are from 1 to 7 (Figure 1B). In addition, there are the PROTEIN_KINASE, ANK (Ankyrin), ANK-REAPEAT, FYVE, BRX (BREVIS RADIX domain), ZF_BBOX (B-box type zinc finger), and PH (Pleckstrin Homology) domains. Among them, there are members that are relatively close in evolutionary relationships, such as ZmRCP29 and ZmRCP9 and ZmRCP13 and ZmRCP2 (Figure 1B).
2.4. Distribution of ZmRCP Family Members on Chromosomes
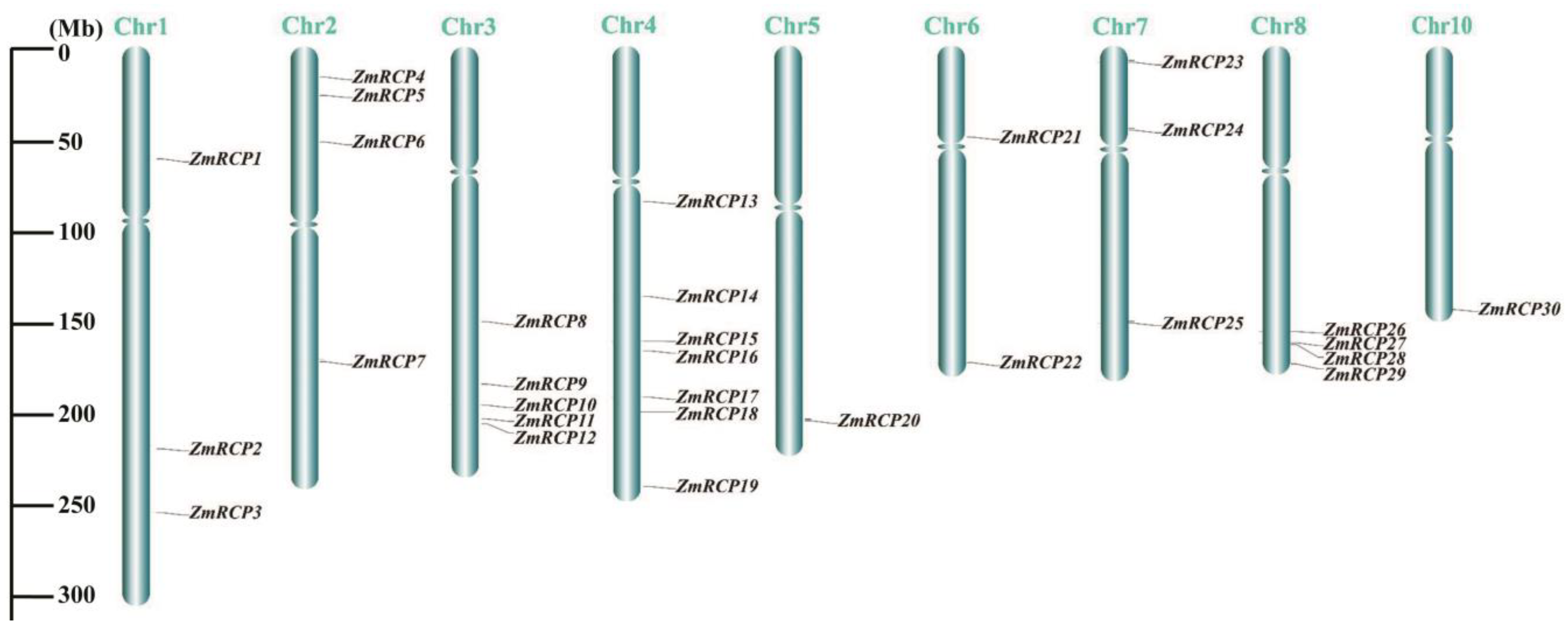
After obtaining the chromosomal location information of ZmRCP family members from the NCBI and MaizeGDB databases, TBtools software II was used to visualize it. The diagram showed that the 30 members of the ZmRCP family are distributed on nine chromosomes (Chr), with 1 member on Chr5 and Chr10; 2 members on Chr6; 3 members on each of Chr1 and Chr7; 4 members on each of Chr2 and Chr8; 5 members on Chr3; and 7 members on Chr4 (Figure 3). Most of the genes are distributed at the ends of the chromosome.
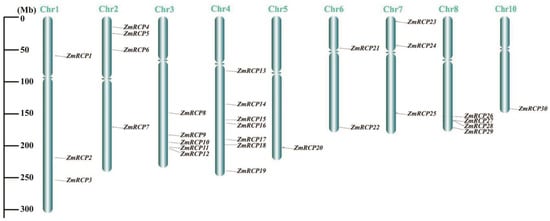
Figure 3.
Distribution of ZmRCP family members on chromosomes (Chr). Gray lines indicate the location of the genes on the chromosomes, and the length of the chromosomes is shown on the left scale.
2.5. Subcellular Localization of ZmRCP Family Members
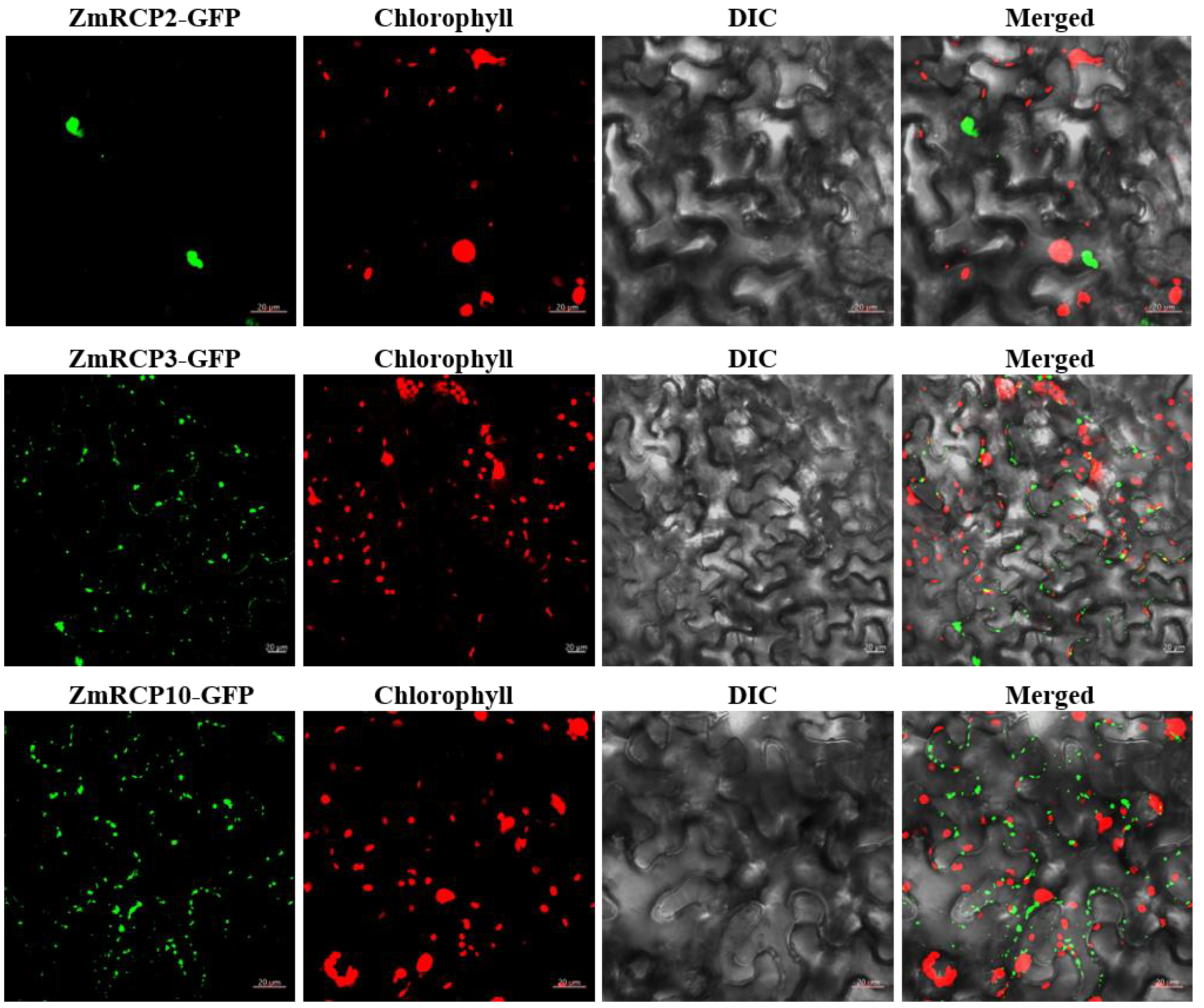
In order to understand the subcellular compartments where RCP family members are located, we conducted a subcellular localization prediction (Table 1). The results indicate that there are 22 ZmRCPs localized in the nucleus and four members localized in both the cell wall and nucleus. To verify the accuracy of subcellular localization prediction, we selected three family members for subcellular localization observation, ZmRCP2, 3, and 10. The resulting constructs ZmRCP2-GFP, ZmRCP3-GFP, and ZmRCP10-GFP were transfected to Nicotiana benthamiana leaves via Agrobacterium-mediated methods. Fluorescence signals were imaged using fluorescence confocal microscopy. The results showed that the green fluorescence signals of ZmRCP2-GFP were observed in the nucleus, whereas ZmRCP3-GFP and ZmRCP10-GFP were detected in dots that were not merged with the chloroplast autofluorescence signals (Figure 4), indicating that ZmRCP3-GFP and ZmRCP10 are targeted to mitochondria.
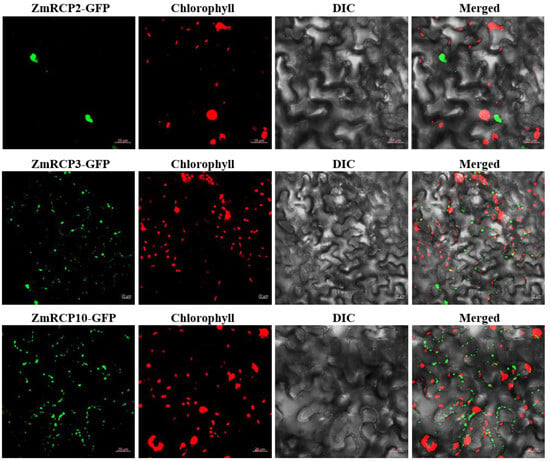
Figure 4.
The subcellular localization of ZmRCP2, ZmRCP3, and ZmRCP10 in tobacco leaf epidermal cells. Fluorescence signals from GFP (green) and chlorophyll (red) were imaged under a confocal microscope. Scale bars = 20 µm.
2.6. Prediction of Cis-Elements in the Promoters
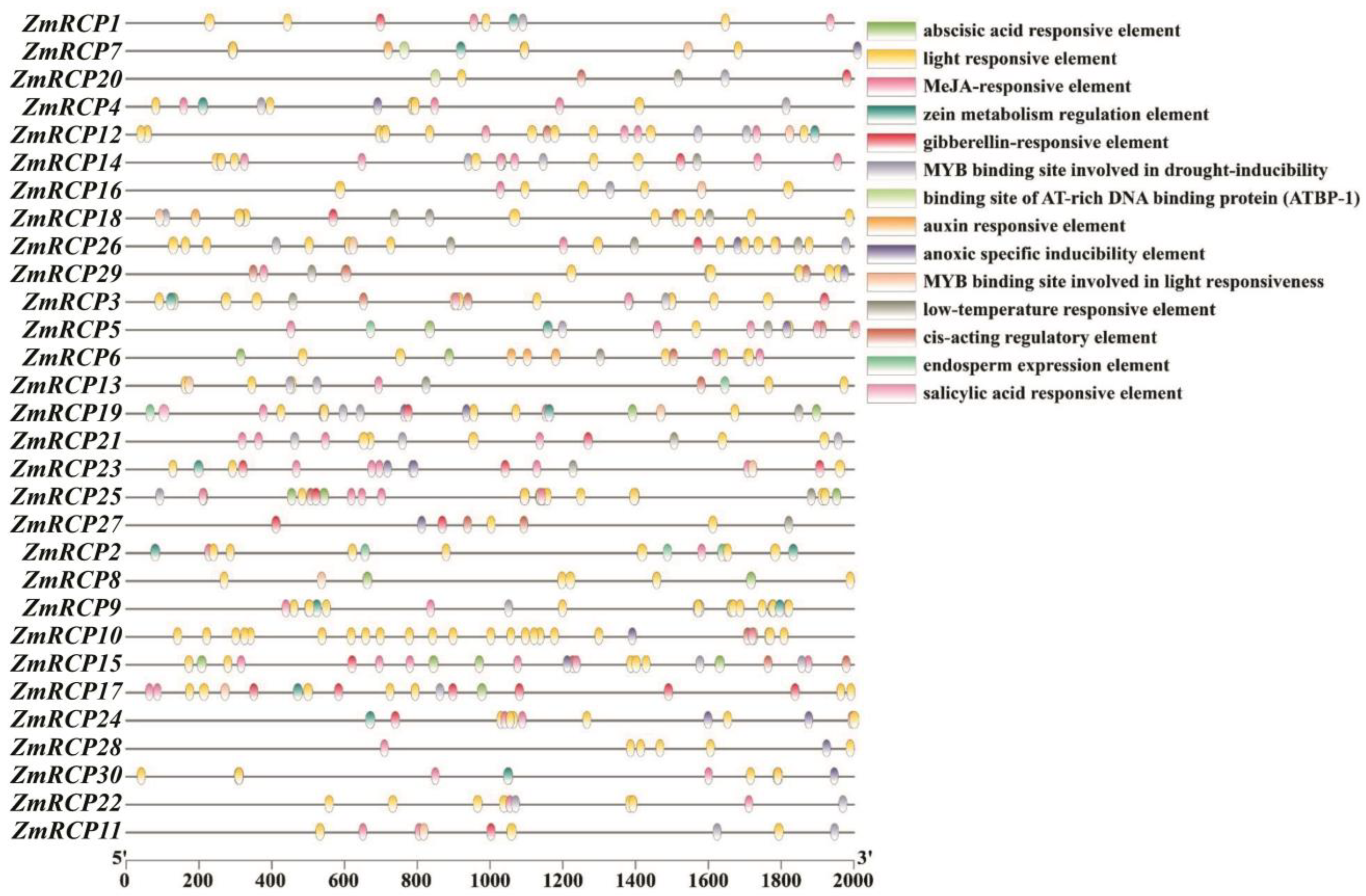
The 2000 bp sequences upstream of the start codons of 30 ZmRCP genes were extracted as the promoter regions used for cis-element prediction (Figure 5). The predicted cis-acting elements can be roughly divided into three categories: abiotic stress, plant hormones, and the growth and development process of maize. Abiotic-stress-responsive elements include drought, low-temperature, anoxic, and light-responsive elements. Plant-hormone-responsive elements include abscisic acid, MeJA, gibberellin, salicylic acid, and auxin-inductive or -responsive elements. The cis-acting elements during the growth and development of maize include the zein metabolism regulation element and endosperm expression element. All ZmRCP family members’ promoter regions contain light-responsive elements, and the majority of ZmRCP family members’ promoters contain multiple light-responsive elements. A total of 14 ZmRCP member promoter regions contain low-temperature responsive elements, and 22 ZmRCP gene promoter regions contain a MYB binding site involved in drought-inducibility elements. These prediction results indicate that the ZmRCP family may play an important regulatory role in the growth and development of and response to abiotic stress and plant hormones. The prediction of cis-acting elements can provide ideas for future research on related genes.
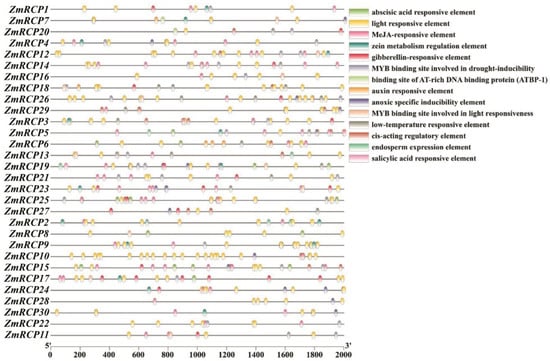
Figure 5.
Prediction of cis-elements in promoters of 30 ZmRCP family members. Various colored boxes indicate different cis-elements that correspond to each colored box shown on the right side of the picture, and promoter lengths (base pairs) are shown on the scale below.
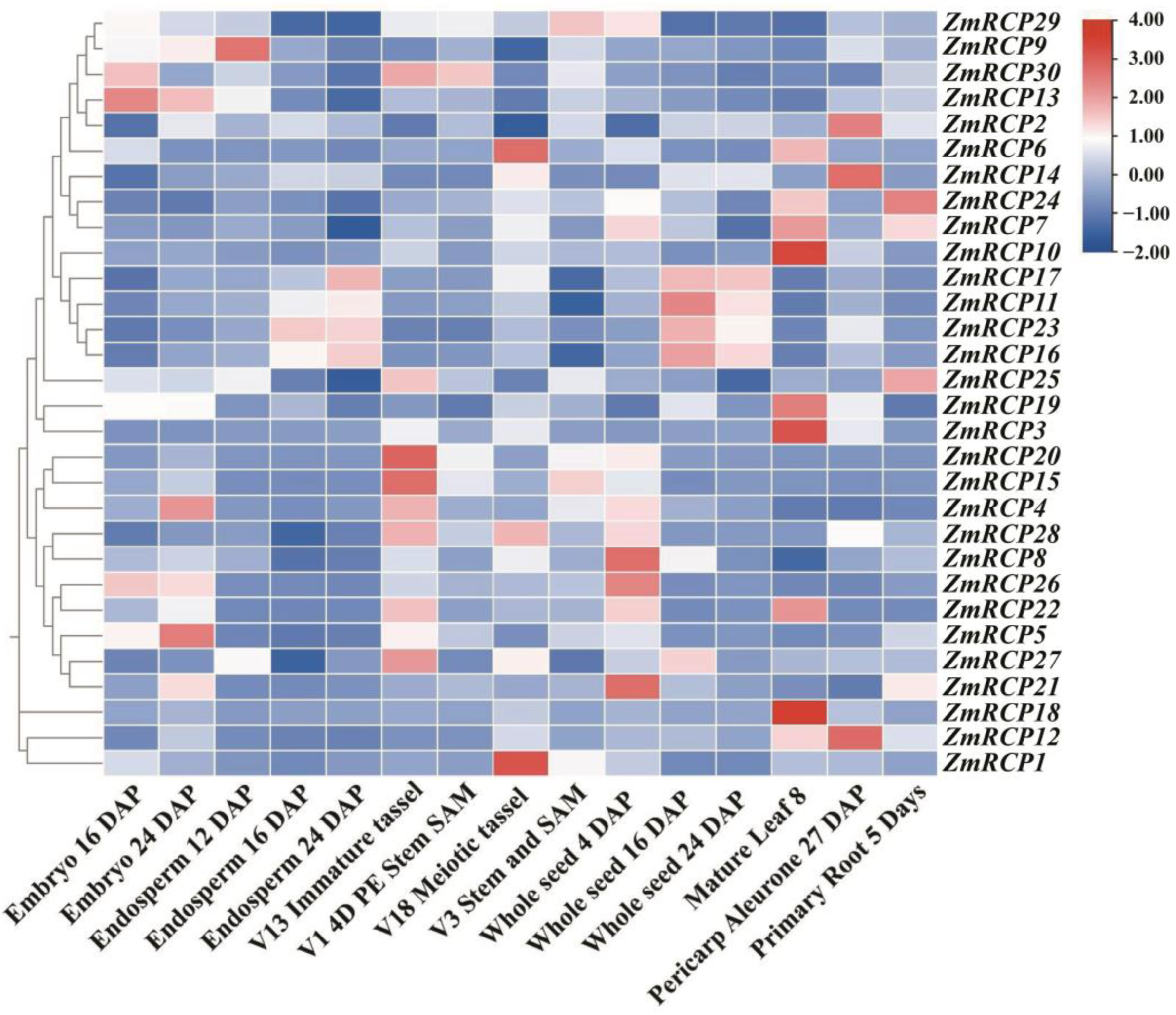
2.7. Analysis of Expression Patterns under Different Tissues and Stresses
In plants, gene expression in different tissues is often closely related to their functions. By analyzing transcriptome data from different tissues or developmental stages of plants, the expression levels of gene family members in a certain tissue or developmental stage can be determined, leading to speculation that these genes may play important roles in plant growth and development. The results of an expression pattern heatmap of ZmRCP members showed that the relative expression levels of genes in the red area are higher, while those in the blue area indicate lower levels (Figure 6). A total of 24 ZmRCP genes have relatively higher expression levels in embryos, endosperms, seeds, or the pericarp aleurone than other genes, indicating that these genes may function in the development of embryos, endosperms, seeds, or seed germination. As shown in Figure 6, ZmRCP30, 29, and 15 have higher expression levels in SAM. ZmRCP6, 24, 7, 10, 19, and 3 have higher expression levels in mature leaf 8 than others. The expression of ZmRCP24, 7, and 25 is higher than other genes in primary roots at day 5 (different days after pollination), indicating that these three genes may play a role in root development.
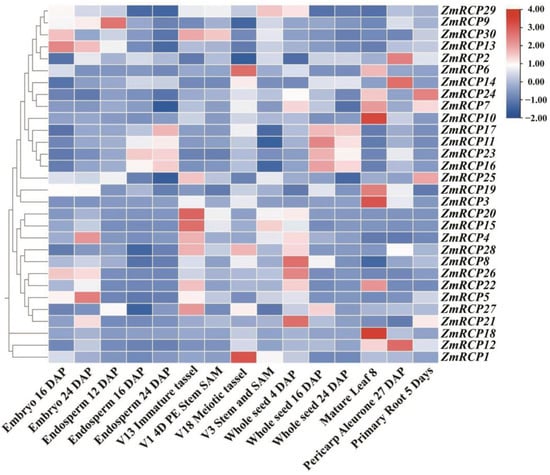
Figure 6.
Expression pattern of 30 ZmRCP family members in different developmental stages and tissues. The different colors indicate the expression levels of the ZmRCP genes in maize. The expression data were downloaded from qTeller in MaizeGDB (https://qteller.maizegdb.org/genes_by_name_B73v5.php (accessed on 27 June 2024)) and the heatmap was generated using Tbtools software.
To investigate the stress response of ZmRCP family members under abiotic stresses, we analyzed the RNA-seq data online of ZmRCP members under different abiotic stresses, including heat, cold, salt, UV, and drought stress. The results showed that the expression levels of ZmRCP29, 9, and 3 were induced under heat stress; the expression of ZmRCP30, 7, 10, 17, 11, 23, 16, and 20 was induced by cold stress; the expression of ZmRCP29, 9, 13, 25, 3, 5, 27, 18, 12, and 1 was induced by salt stress; and the expression of ZmRCP30, 13, 17, 23, 16, 25, 20, 15, 4, 28, 26, 5, 27, and 21 was induced by UV stress (Figure S4A). In addition, the expression of ZmRCP29, 20, 14, 17, 11, 23, 16, 19, 3, 20, 15, 4, 8, 27, and 18 was induced by a mild water deficit (24 h). Under a severe water deficit, the expression of ZmRCP29, 9, 14, 7, 10, 17, 11, 23, 16, 3, 20, 4, 28, 26, 22, 5, 27, 21, 18, and 12 was induced (24 h) (Figure S4B). These data suggest that ZmRCP family members possibly play important roles in the response to abiotic stresses.
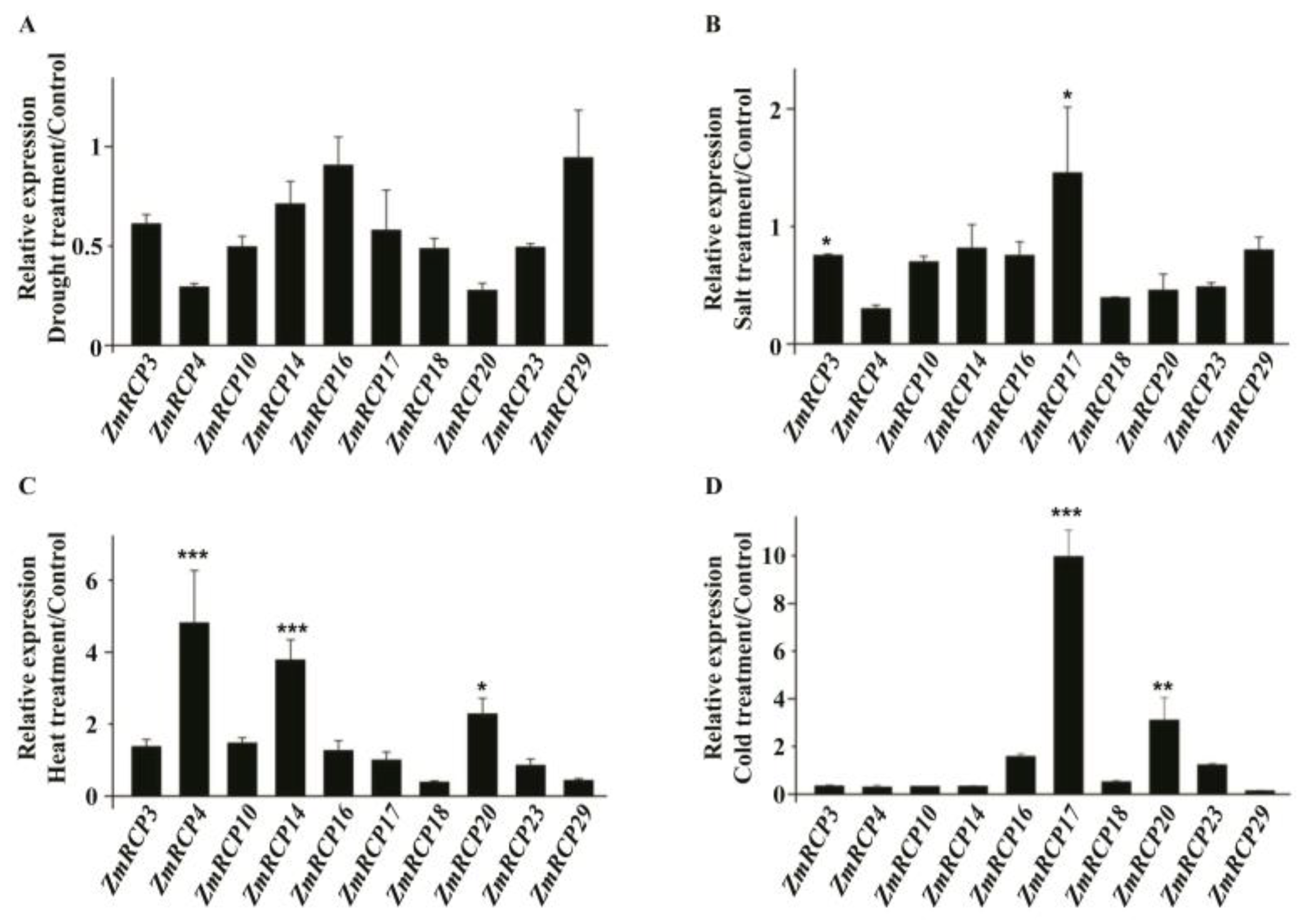
To further study the potential role of ZmRCPs, we performed RT-qPCR to detect the expression levels of 10 family members (ZmRCP3, 4, 10, 14, 16, 17, 18, 20, 23, and 29) under different abiotic stresses and plant hormone treatments, which were induced by abiotic stresses based on RNA-Seq data online (Figure S4). The results showed that the transcript of these 10 ZmRCP members was decreased under drought stress at 6 h (Figure 7A), while the transcript of ZmRCP17 was increased under salt stress (Figure 7B). ZmRCP4, 14, and 20 were remarkably induced under heat stress (Figure 7C), whereas ZmRCP17 and 20 were significantly induced under clod stress (Figure 7D). These results suggest that ZmRCP members differentially respond to drought, salt, heat, and cold treatments.
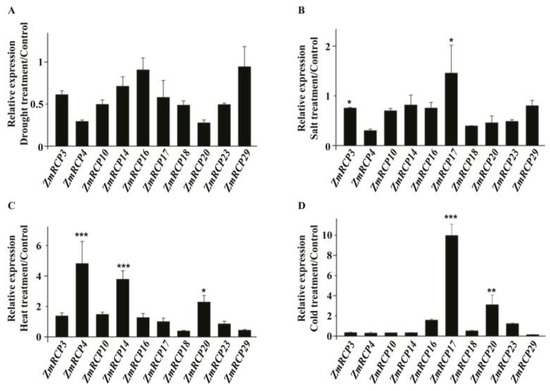
Figure 7.
Expression levels of 10 ZmRCP members under abiotic stress treatments. Expression levels of 10 ZmRCP members were detected by RT-qPCR under drought (A), salt (B), heat (C), and cold (D) treatments. ZmActin (GRMZM2G126010) was used as the reference gene. Data are means (±SE) of three biological replicates. The comparison groups of Student’s t-test are treatment and control. Asterisks indicate significant differences between means calculated with Student’s t-test. * p < 0.05; ** p < 0.01; *** p < 0.001.
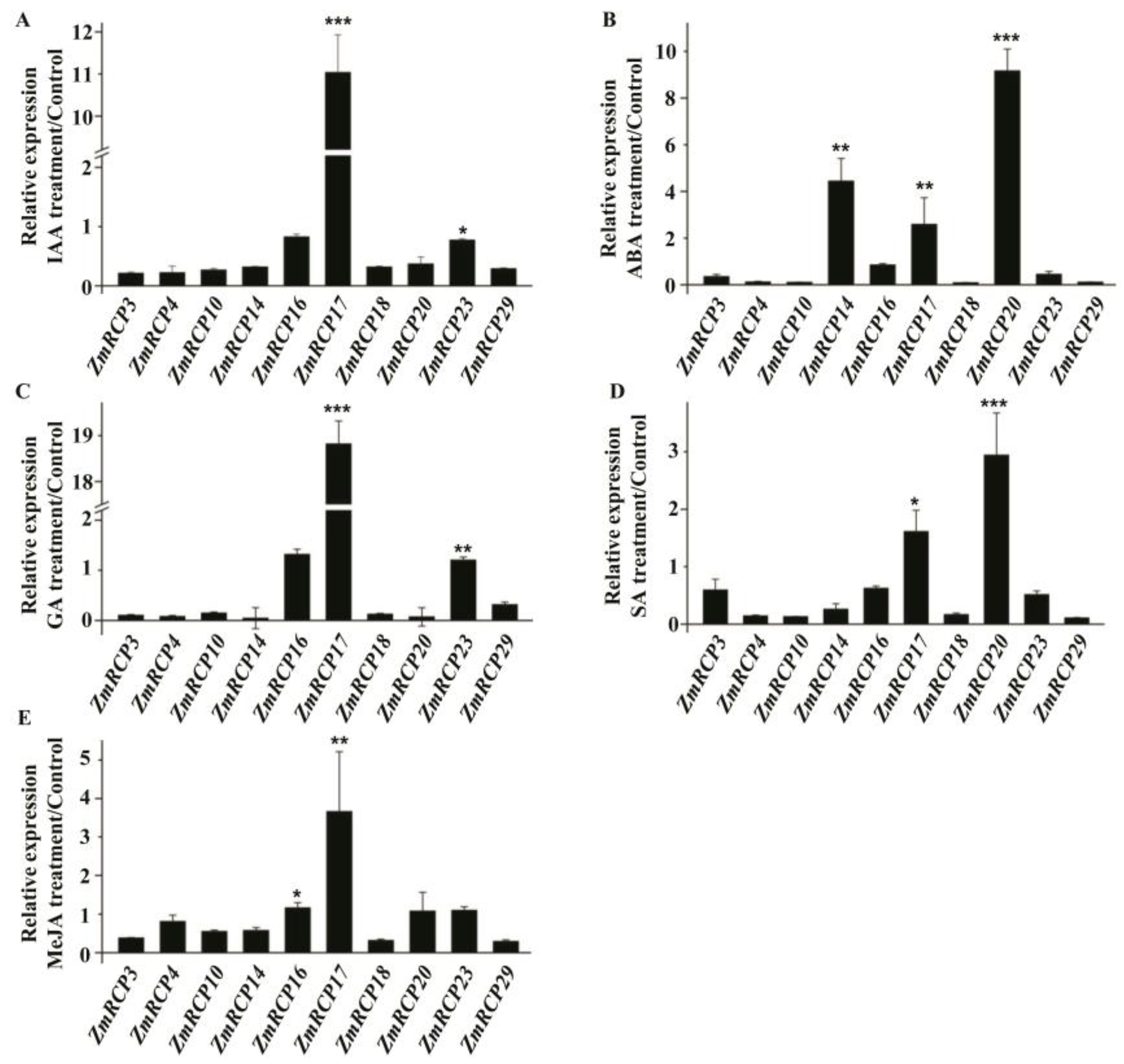
In addition, ZmRCP17 showed induced expression under IAA, GA, and MeJA treatment (Figure 8A,C,E). ZmRCP14, 17, and 20 showed induced expression under ABA treatment (Figure 8B), while ZmRCP17 and 20 showed induced expression under GA treatment (Figure 8E). These results suggest that ZmRCP members respond to different hormone treatments.
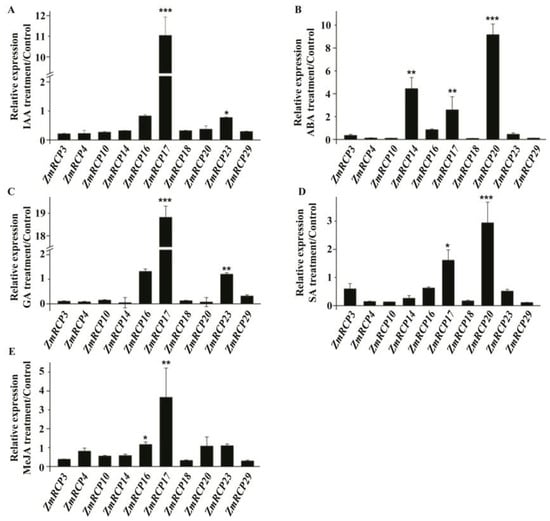
Figure 8.
Expression levels of 10 ZmRCP members under different hormone treatments. Expression levels of 10 ZmRCP members were detected by RT-qPCR under IAA (A), ABA (B), GA (C), SA (D), and MeJA (E) treatments. ZmActin (GRMZM2G126010) was used as the reference gene. Data are means (±SE) of three biological replicates. The comparison groups of Student’s t-test are treatment and control. Asterisks indicate significant differences between means calculated with Student’s t-test. * p < 0.05; ** p < 0.01; *** p < 0.001.
2.8. Prediction of Protein Interaction Network
In order to understand the potential interacting proteins of RCP gene family members, we used a website to predict the possible interacting proteins of RCP family members and then drew the diagram of the protein interaction network. There are a total of 37 proteins that possibly interact with the RCP family members, with the majority of RCP family members interacting with ATG7 (Figure S5). The ATG7 protein is an autophagy-related protein that allows cells to degrade damaged tissues through autophagy while also providing energy to help plants resist external stress and adverse factors [23,24]. In addition, other possible interacting proteins predicted include PSAH (photosystem I reaction center subunit VI) and TIDP3014 (GTP binding nucleoprotein), etc., most of which are related to stress and plant growth and development [25,26]. These results suggest that the ZmRCP family members may participate in plant growth and development processes and respond to abiotic stress through protein–protein interactions.
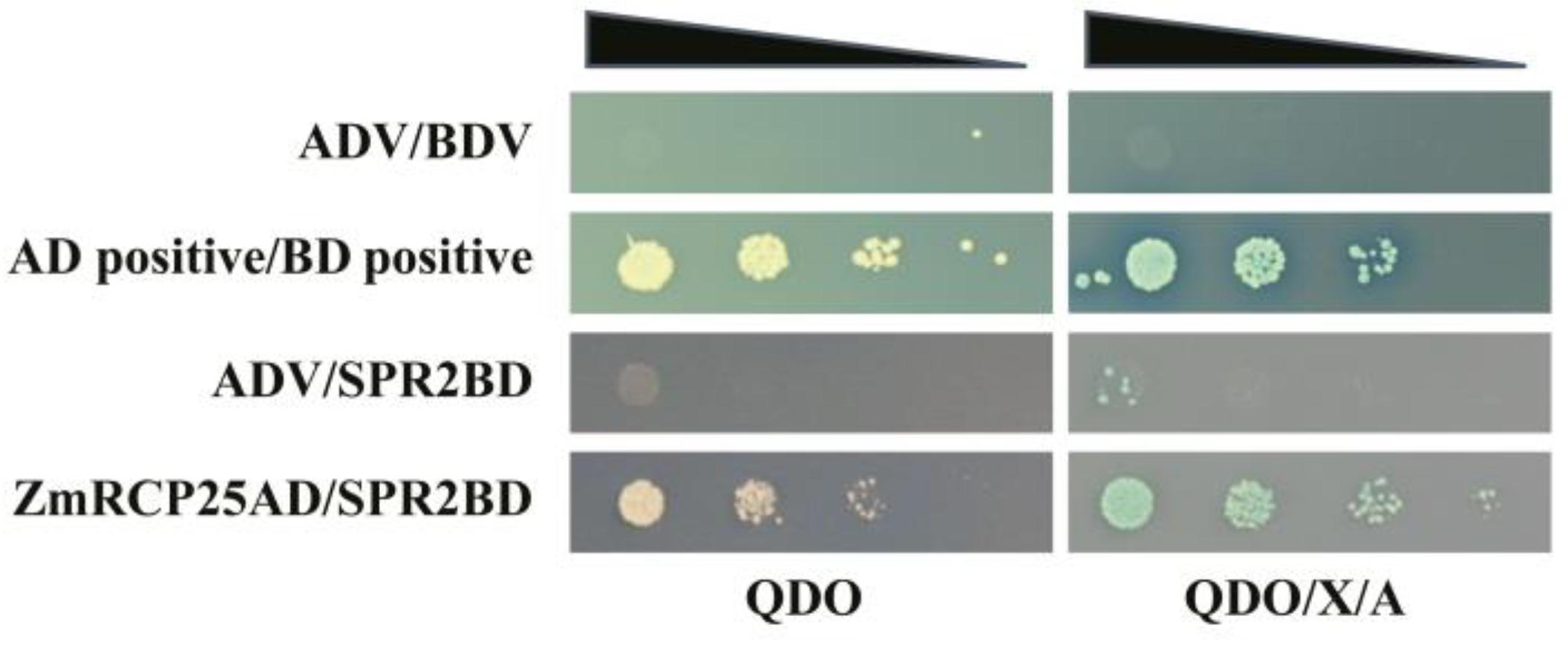
A previous study reported that ZmRCP25 (ZmDEK47) is required for the splicing of four introns of mitochondrial nad2 transcript and seed development. The small PPR protein 2 (SPR2) is required for the splicing of 15 mitochondrial group II introns in maize, including the 4 introns of nad2 [27], indicating that the functions of these two splicing factors are overlapped with each other. To test whether these two proteins interact with each other, we performed a yeast two-hybrid assay. The results showed that ZmRCP25AD/SPR2BD can grow on QDO or QDO/X/A medium (Figure 9), indicating that ZmRCP25 interacts with SPR2 in the yeast system.
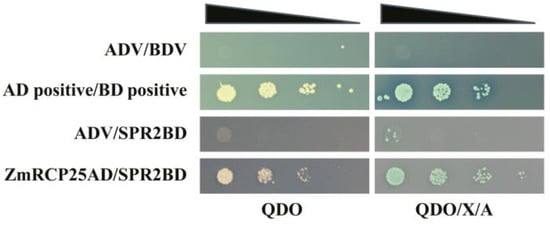
Figure 9.
ZmRCP25 interacts with SPR2 in yeast two-hybrid system. Yeast two-hybrid assay was performed to determine interactions between ZmRCP25 and SPR2. These constructs were co-transfected into Y2H Gold strain and QDO (SD/−Ade/-His/−Leu/−Trp) medium with added X-a-gal and AbA (QDO/X/A) medium to reveal protein–protein interactions. AD: GAL4 activation domain; BD: GAL4 DNA binding domain.
3. Discussion
In this study, a phylogenetic tree of RCP family members was constructed among maize, rice, and Arabidopsis (Figure 2). The results showed that there are slightly more members of the RCP family in maize than in rice and Arabidopsis, indicating that the number of members of the RCP family has slightly increased during species evolution and that its functions may be more diverse in maize. Regarding the RCC1-conserved domain, members of the RCP family typically consist of six or seven RCC1 domains [12]. Most members in maize still retained this characteristic, but some members had a reduced number of RCC1 domains, or even only one RCC1 domain remained (Figure 1B). This may be because some conservative structural domains are lost when dealing with changes in the environment or other situations. Gene duplication is important for biological evolution, as it can promote the production of new adaptive functions in plants and expand gene families [28]. In the maize RCP gene family, a total of eight fragment duplication events were identified (Figure S2). These repeated fragments may enable maize to adapt to complex environments.
Previous studies reported that RCP members function in various aspects of plant growth and development. For example, the RCC1 domain protein ZmRCP25/DEK47 is required for the splicing of mitochondrial nad2 introns and kernel development in maize [21]. The small PPR protein SPR2 is also required for the splicing of nad2 introns and kernel development in maize [27]. Interestingly, ZmRCP25 interacted with SPR2 in the yeast two-hybrid assay (Figure 9), implying that ZmRCP25/SPR2 may form a protein complex in intron splicing. The RCC1-containing protein RUG3 is involved in the splicing of mitochondrial nad2 intron 2 and 3 in Arabidopsis [12]. AtRUG3 and DEK47 cluster into a branch in Class V (Figure 2), which are both required for the intron splicing of mitochondrial nad2, indicating they have a functional homology. Similarly, ZmRCP18 is in Class III with AtRUG1, which is related to chlorophyll synthesis [20], implying that ZmRCP18 may be related to chlorophyll synthesis. Moreover, ZmRCP19, 2, 30, 24, 7, and 14 are in the Class IV, as are AtRUG2 and ZmRCC1-3 (Figure 2). AtRUG2 maintains normal transcriptional function in chloroplasts and mitochondria and ZmRCC1-3 and ZmRCC1-2 attenuate the defects of the cell cycle caused by ZmRCC1-1 dysfunction [16,22], implying that these ZmRCPs may be involved in certain functions in chloroplasts and mitochondria or play a role in cell cycle regulation. Because of the importance of chloroplast photosynthesis and mitochondrial oxidative respiration, loss of these ZmRCPs may cause severe defects in plant growth and development and even embryo lethality in maize.
Several RCP members were reported to be involved in response to abiotic stresses, such as responding to UV stress, cold stress, drought stress, heat stress, and salt stress [5,29,30]. For instance, AtUVR8 is involved in chromatin condensation and plays a role in the UV-B signaling pathway, inducing flavonoid biosynthesis [31]. AtUVR8 can activate a protection mechanism to cope with UV stress. Under drought stress, salt stress, or cold stress, the expression level of the AtUVR8 gene increased compared to the corresponding sensitive and wild-type plants [3,19,29]. The RCC1-domain protein TCF1 can improve plant cold tolerance by reducing lignin accumulation under low-temperature conditions in Arabidopsis and soybeans [5,30]. In our data, the phylogenetic tree analysis showed that ZmRCC1-1 and ZmRCP29 are clustered in the same branch as AtTCF1, and ZmRCC1-2 is clustered in the same branch as AtUVR8 (Figure 2). Meanwhile, the prediction of cis-acting elements shows that promoter regions of multiple ZmRCP members contain low-temperature-responsive elements and a MYB binding site involved in drought-inducibility elements (Figure 5). The heatmap of gene expression under abiotic stress shows that the expression levels of most ZmRCP members increase under UV stress (Figure S4A). Moreover, the expression of several ZmRCP members was induced under different abiotic stresses (Figure 6 and Figure S4A,B). These results suggest that the ZmRCP family may play important regulatory roles in the response to abiotic stresses in maize.
In previous reports, RCP family members have also been shown to respond to plant hormones such as ABA and auxin [5,11]. ABA and auxin are important plant hormones that regulate plant growth, development, and stress response. ABA plays a crucial role in various physiological processes of plants, such as stomatal closure, the accumulation of epidermal wax, leaf senescence, bud dormancy, seed germination, osmotic regulation, and growth inhibition [31]. The RCC1 protein SAB1 is involved in ABA signaling and affects the stability of ABI5 by initiating complex mechanisms, thereby negatively regulating the expression of ABI5 genes [11]. RCC1 proteins AtRLD1 and AtRLD4 change the gravitropism of roots by altering the distribution of auxin [32]. The prediction of cis-acting elements shows that promoters of many ZmRCP members contain elements responding to plant hormones, including auxin, abscisic acid, MeJA, gibberellin, etc. (Figure 5). In addition, RT-qPCR results showed that the expression of ZmRCP14, 17, and 20 was remarkably induced under different plant hormone treatments (Figure 7). These data suggest that ZmRCP members are involved in the response to plant hormones in maize.
Based on the above results, it is speculated that the ZmRCP gene may play a role in the response to abiotic stress and plant hormones, particularly in drought stress and cold stress. Although the potential functions of ZmRCP family members may be speculated based on the above analysis, whether these members truly participate in these functions still needs to be experimentally proven.
4. Materials and Methods
4.1. Materials and Treatments
Maize inbred line B73 seedlings grown to three leaves were used for abiotic stress and hormone treatments, and RNA was extracted for subsequent RT-qPCR. Seedlings were subjected to drought, salt, or control treatments with 20% (m/v) PEG-6000, 200 mM NaCl, or ddH2O for 6h, respectively. Cold, high-temperature, and control treatments were carried out in a light incubator at a low temperature of 4 °C, a high temperature of 42 °C, and a control temperature of 25 °C for 24 h, respectively. Hormone treatments were carried out with sterile water (ddH2O) or abscisic acid (ABA), auxin (IAA), salicylic acid (SA), methyl jasmonate (MeJA), and gibberellin (GA) at concentrations of 100 μmol·L−1, and the hormone solutions were used to soak roots for 4 h. Each of the above groups was planted with 3 cups of sand-cultured maize seedlings, and 5 seeds were sown in each cup.
4.2. Identification of RCP Family Members
The genome and annotation files of maize were downloaded from Ensembl Plants (http://plants.ensembl.org/info/data/ftp/index.html (accessed on 16 February 2024)). The 24 Arabidopsis thaliana RCP family members were searched, and their protein sequences were downloaded from the Arabidopsis TAIR (https://www.arabidopsis.org/ (accessed on 16 February 2024)) website. The extraction of the protein sequences of maize was carried out using TBtools software II [33,34], and the protein sequences of 24 Arabidopsis thaliana RCP family members were used as candidate family members using BLASTp bidirectional BLAST from NCBI (https://blast.ncbi.nlm.nih.gov/ (accessed on 16 February 2024)). The InterPro (https://www.ebi.ac.uk/interpro/ (accessed on 18 February 2024)) website was used to detect the RCP structural domain, and the ones that did not contain this conserved domain were removed, which finally determined the maize RCP gene family member list.
4.3. Construction of Phylogenetic Tree
The rice RCP members were obtained through a homologous blast. The RCP protein sequences of Arabidopsis thaliana, Oryza sativa, and Zea mays were imported, and a sequence alignment was performed by applying the ClustalW algorithm of MEGA7.0 software, choosing the neighbor-joining method, and setting the step value (Bootstrap) to 1000 to construct the multi-sequence phylogenetic tree [35]. The phylogenetic tree was uploaded to iTOL (https://itol.embl.de/itol.cgi (accessed on 26 February 2024)) to beautify and classify it.
4.4. Collinearity Analysis
The intra-species and inter-species collinearity analyses of maize RCP members were performed using McScanX, and the collinearity diagrams were drawn using the built-in Circos function in Tbtools software [33,34].
4.5. Analysis of Cis-Acting Elements in the Promoters
The 2000 bp promoter sequences upstream the start codon of each of the maize RCP family members were obtained from the official maize database MaizeGDB (https://maizegdb.org/ (accessed on 19 May 2024)), the cis-acting elements of each promoter of the maize RCP family members were predicted using the online tool PlantCARE (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/ (accessed on 20 May 2024)), and the drawing/diagram was completed using TBtools software [33,34].
4.6. Prediction of Protein Physicochemical Properties
The protein molecular weight, instability index, average hydropathicity, isoelectric point, and number of amino acid residues of RCP family members were analyzed by uploading each identified protein sequence to the ExPASy ProtParam official website (https://web.expasy.org/protparam/ (accessed on 11 March 2024)). The subcellular localization of each family member was predicted using CELLO (http://cello.life.nctu.edu.tw/ (accessed on 12 March 2024)), wolf psort (https://wolfpsort.hgc.jp/ (accessed on 12 March 2024)), and Cell-Ploc (http://www.csbio.sjtu.edu.cn/bioinf/Cell-PLoc-2/ (accessed on 12 March 2024)).
4.7. Gene Structure, Conserved Motif, and Structural Domain Analysis
The information about the length of open reading frames and the number of exons of maize RCP family members was obtained from the MaizeGDB (https://maizegdb.org/ (accessed on 25 March 2024)). The conserved motifs were predicted by the MEME online tool (https://memesuite.org/meme/tools/meme (accessed on 26 March 2024)), and the number of conserved motifs was set as 10. The conserved domains of the maize RCP family members were further obtained by using the CD-search tool on the official website of NCBI (https://www.ncbi.nlm.nih.gov/ (accessed on 27 March 2024)). Diagrams of gene structure, protein conserved motifs, and conserved domains were visualized using the Gene Structure View function in TBtools software [33,34].
4.8. Location on Chromosome
The diagram of chromosome localization was obtained by uploading GFF3 files and gene IDs in the Gene Location Visualize from GTF/GFF tool of TBtools [33,34].
4.9. Expression Pattern Analysis
The expression data of ZmRCP family members under abiotic stresses and in different tissues were downloaded from qTeller in MaizeGDB. The clustering analysis was performed and the expression heatmaps of maize RCP family members were generated by using Tbtools software [33,34].
4.10. Subcellular Localization
The full-length cDNAs of ZmRCP-2, 3, and 10 were cloned into the pSuper1300-GFP vector and fused to the N-terminal of GFP (Green Fluorescence Protein). The constructs were transformed into tobacco leaves through Agrobacterium EHA105-mediated transformation as described previously [27]. The subcellular localization of the target proteins was preliminarily determined where the was green fluorescence signal observed.
4.11. Analysis of Protein Interaction Network Prediction
The interaction relationships corresponding to maize RCP proteins were extracted directly from the STRING (https://string-db.org/ (accessed on 29 July 2024)) database and the PlantSPEAD database [36]. The protein interaction network was constructed by inputting the protein interaction information in Cystoscope v3.10.1 software.
4.12. RT-qPCR
Total RNA was extracted from 0.2 g of leaves from seedlings after treatment with different hormones or abiotic stresses using the RNA easy Isolation Reagent kit (Vazyme Biotech Co., Ltd., Nanjing, China). Reverse transcription was conducted after DNA digestion using a HiFi Script cDNA Synthesis Kit (Vazyme Biotech Co., Ltd., Nanjing, China) using 1 μg total RNA as the template. RT-qPCR was performed with a CFX96 Real Time PCR instrument (Bio-Rad, Hercules, CA, USA) and an SYBR Green I Master PCR kit (Roche, Basel, Switzerland) using ZmActin (GRMZM2G126010) as the internal reference gene. Three independent biological and technical replicates were performed, and the relative expression level of the gene was calculated using the formula 2-ΔΔCt [37]. The primers used in this study are in Supplementary Table S1.
4.13. Yeast Two-Hybrid Assay
The full-length cDNA of ZmRCP25 and SPR2 was cloned into pGADT7 and pGBKT7, respectively. ZmRCP25-pGADT7 and SPR2-pGBKT7 were co-transformed into the yeast strain Y2H Gold. The growth conditions of the co-transformants were imaged after being inoculated onto Minimal Media Quadruple Dropout (QDO, SD-Ade/-His/-Leu/-Trp) plates for 4 days as described previously [27].
5. Conclusions
In this study, 30 ZmRCP members were identified and analyzed in maize through a systematic bioinformatics analysis. We performed a physicochemical property analysis, a phylogenetic tree analysis, cis-acting elements prediction, and subcellular localization of ZmRCP. Several ZmRCP members exhibited induced or inhibited responses to abiotic stresses and plant hormones. These results contribute to a better understanding of the evolutionary history and potential role of the ZmRCP family in mediating responses to abiotic stresses in maize.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/ijms252111437/s1.
Author Contributions
R.L. and S.-K.C. designed the experiments. R.L., T.M., Y.L., X.L., H.J. and S.-K.C. performed the experiments and bioinformatics analysis. R.L., H.D., J.Z. and S.-K.C. analyzed the data and wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Hubei Natural Science Foundation (2022CFB183, 2024AFB301, and 2022CFA030).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article and Supplementary Materials.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Zhang, H.M.; Zhu, J.H.; Gong, Z.Z.; Zhu, J.K. Abiotic stress responses in plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Park, H.J.; Kim, W.Y.; Yun, D.J. A new insight of salt stress signaling in plant. Mol. Cells 2016, 39, 447–459. [Google Scholar] [CrossRef] [PubMed]
- Brown, B.A.; Cloix, C.; Jiang, G.H.; Kaiserli, E.; Herzyk, P.; Kliebenstein¶, D.J.; Jenkins, G.I. A UV-B-specific signaling component orchestrates plant UV protection. Proc. Natl. Acad. Sci. USA 2005, 102, 18225–18230. [Google Scholar] [CrossRef] [PubMed]
- Hadjebi, O.; Casas-Terradellas, E.; Garcia-Gonzalo, F.R.; Rosa, J.L. The RCC1 superfamily: From genes, to function, to disease. Biochim. Biophys. Acta 2008, 1783, 1467–1479. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Wang, Y.; Cloix, C.; Li, K.; Jenkins, G.I.; Wang, S.; Shang, Z.; Shi, Y.; Yang, S.; Li, X. The Arabidopsis RCC1 family protein TCF1 regulates freezing tolerance and cold acclimation through modulating lignin biosynthesis. PLoS Genet. 2015, 11, e1005471. [Google Scholar] [CrossRef]
- Liu, X.; Wu, X.; Sun, C.; Rong, J. Identification and expression profiling of the regulator of chromosome condensation 1 (RCC1) gene family in Gossypium Hirsutum L. under abiotic stress and hormone treatments. Int. J. Mol. Sci. 2019, 20, 1727. [Google Scholar] [CrossRef]
- Dasso, M. RCC1 in the cell cycle: The regulator of chromosome condensation takes on new roles. Trends Biochem. Sci. 1993, 18, 96–101. [Google Scholar] [CrossRef]
- Renault, L.; Nassar, N.; Vetter, I.; Becker, J.; Klebe, C.; Roth, M.; Wittinghofer, A. The 1.7 A crystal structure of the regulator of chromosome condensation (RCC1) reveals a seven-bladed propeller. Nature 1998, 392, 97–101. [Google Scholar] [CrossRef]
- Bischoff, F.R.; Ponstingl, H. Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature 1991, 354, 80–82. [Google Scholar] [CrossRef]
- Cekan, P.; Hasegawa, K.; Pan, Y.; Tubman, E.; Odde, D.; Chen, J.Q.; Herrmann, M.A.; Kumar, S.; Kalab, P. RCC1-dependent activation of Ran accelerates cell cycle and DNA repair, inhibiting DNA damage-induced cell senescence. Mol. Biol. Cell 2016, 27, 1346–1357. [Google Scholar] [CrossRef]
- Ji, H.; Wang, S.; Cheng, C.; Li, R.; Wang, Z.; Jenkins, G.I.; Kong, F.; Li, X. The RCC1 family protein SAB1 negatively regulates ABI5 through multidimensional mechanisms during postgermination in Arabidopsis. New Phytol. 2019, 222, 907–922. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, K.; Carrie, C.; Giraud, E.; Wang, Y.; Meyer, E.H.; Narsai, R.; des Francs-Small, C.C.; Zhang, B.; Murcha, M.W.; Whelan, J. The RCC1 family protein RUG3 is required for splicing of nad2 and complex I biogenesis in mitochondria of Arabidopsis thaliana. Plant J. 2011, 67, 1067–1080. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Li, D.; Gao, Z.; Gao, L.; Shang, L.; Wang, M.; Qiao, J.; Ding, S.; Li, C.; Geisler, M.; et al. OsRLR4 binds to the OsAUX1 promoter to negatively regulate primary root development in rice. J. Integr. Plant Biol. 2022, 64, 118–134. [Google Scholar] [CrossRef] [PubMed]
- Mao, K.; Wang, L.; Li, Y.Y.; Wu, R. Molecular cloning and functional analysis of UV RESISTANCE LOCUS 8 (PeUVR8) from Populus euphratica. PLoS ONE 2015, 10, e0132390. [Google Scholar] [CrossRef]
- Rizzini, L.; Favory, J.J.; Cloix, C.; Faggionato, D.; O’Hara, A.; Kaiserli, E.; Baumeister, R.; Schafer, E.; Nagy, F.; Ulm, R.; et al. Perception of UV-B by the Arabidopsis UVR8 protein. Science 2011, 332, 103–106. [Google Scholar] [CrossRef]
- Quesada, V.; Sarmiento-Manus, R.; Gonzalez-Bayon, R.; Hricova, A.; Perez-Marcos, R.; Gracia-Martinez, E.; Medina-Ruiz, L.; Leyva-Diaz, E.; Ponce, M.R.; Micol, J.L. Arabidopsis RUGOSA2 encodes an mTERF family member required for mitochondrion, chloroplast and leaf development. Plant J. 2011, 68, 738–753. [Google Scholar] [CrossRef]
- Su, C.; Yuan, J.; Zhao, H.; Zhao, Y.; Ji, H.; Wang, Y.; Li, X. RUG3 is a negative regulator of plant responses to ABA in Arabidopsis thaliana. Plant Signal Behav. 2017, 12, e1333217. [Google Scholar] [CrossRef]
- Su, C.; Zhao, H.; Zhao, Y.; Ji, H.; Wang, Y.; Zhi, L.; Li, X. RUG3 and ATM synergistically regulate the alternative splicing of mitochondrial nad2 and the DNA damage response in Arabidopsis thaliana. Sci. Rep. 2017, 7, 43897. [Google Scholar] [CrossRef]
- Yin, R.; Skvortsova, M.Y.; Loubery, S.; Ulm, R. COP1 is required for UV-B-induced nuclear accumulation of the UVR8 photoreceptor. Proc. Natl. Acad. Sci. USA 2016, 113, E4415–E4422. [Google Scholar] [CrossRef]
- Quesada, V.; Sarmiento-Manus, R.; Gonzalez-Bayon, R.; Hricova, A.; Ponce, M.R.; Micol, J.L. PORPHOBILINOGEN DEAMINASE deficiency alters vegetative and reproductive development and causes lesions in Arabidopsis. PLoS ONE 2013, 8, e53378. [Google Scholar] [CrossRef]
- Cao, S.-K.; Liu, R.; Sayyed, A.; Sun, F.; Song, R.; Wang, X.; Xiu, Z.; Li, X.; Tan, B.-C. Regulator of chromosome condensation 1-domain protein DEK47 functions on the intron splicing of mitochondrial nad2 and seed development in maize. Front. Plant Sci. 2021, 12, 695249. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Wu, H.; Pan, X.; Jin, W.; Li, X. Aberrant meiotic modulation partially contributes to the lower germination rate of pollen grains in maize (Zea mays L.) under low nitrogen supply. Plant Cell Physiol. 2017, 58, 342–353. [Google Scholar] [CrossRef] [PubMed]
- Minina, E.A.; Moschou, P.N.; Vetukuri, R.R.; Sanchez-Vera, V.; Cardoso, C.; Liu, Q.; Elander, P.H.; Dalman, K.; Beganovic, M.; Lindberg Yilmaz, J.; et al. Transcriptional stimulation of rate-limiting components of the autophagic pathway improves plant fitness. J. Exp. Bot. 2018, 69, 1415–1432. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Alseekh, S.; Wen, W.; Cheng, Y.; Fernie, A.R. Genome-wide association studies of Arabidopsis dark-induced senescence reveals signatures of autophagy in metabolic reprogramming. Autophagy 2022, 18, 457–458. [Google Scholar] [CrossRef] [PubMed]
- Jensen, P.E.; Bassi, R.; Boekema, E.J.; Dekker, J.P.; Jansson, S.; Leister, D.; Robinson, C.; Scheller, H.V. Structure, function and regulation of plant photosystem I. Biochim. Biophys. Acta 2007, 1767, 335–352. [Google Scholar] [CrossRef]
- Haizel, T.; Merkle, T.; Pay, A.; Fejes, E.; Nagy, F. Characterization of proteins that interact with the GTP-bound form of the regulatory GTPase Ran in Arabidopsis. Plant J. 1997, 11, 93–103. [Google Scholar] [CrossRef]
- Cao, S.K.; Liu, R.; Wang, M.; Sun, F.; Sayyed, A.; Shi, H.; Wang, X.; Tan, B.C. The small PPR protein SPR2 interacts with PPR-SMR1 to facilitate the splicing of introns in maize mitochondria. Plant Physiol. 2022, 190, 1763–1776. [Google Scholar] [CrossRef]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S.H. Evolution of Gene Duplication in Plants. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef]
- Tossi, V.E.; Regalado, J.J.; Iannicelli, J.; Laino, L.E.; Burrieza, H.P.; Escandon, A.S.; Pitta-Alvarez, S.I. Beyond Arabidopsis: Differential UV-B response mediated by UVR8 in diverse species. Front. Plant Sci. 2019, 10, 780. [Google Scholar] [CrossRef]
- Dong, Z.; Wang, H.; Li, X.; Ji, H. Enhancement of plant cold tolerance by soybean RCC1 family gene GmTCF1a. BMC Plant Biol. 2021, 21, 369. [Google Scholar] [CrossRef]
- Chen, K.; Li, G.J.; Bressan, R.A.; Song, C.P.; Zhu, J.K.; Zhao, Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef] [PubMed]
- Furutani, M.; Hirano, Y.; Nishimura, T.; Nakamura, M.; Taniguchi, M.; Suzuki, K.; Oshida, R.; Kondo, C.; Sun, S.; Kato, K.; et al. Polar recruitment of RLD by LAZY1-like protein during gravity signaling in root branch angle control. Nat. Commun. 2020, 11, 76. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Chen, M.X.; Mei, L.C.; Wang, F.; Boyagane Dewayalage, I.K.W.; Yang, J.F.; Dai, L.; Yang, G.F.; Gao, B.; Cheng, C.L.; Liu, Y.G.; et al. PlantSPEAD: A web resource towards comparatively analysing stress-responsive expression of splicing-related proteins in plant. Plant Biotechnol. J. 2021, 19, 227–229. [Google Scholar] [CrossRef]
- Song, Y.C.; Chen, M.X.; Zhang, K.L.; Reddy, A.S.N.; Cao, F.L.; Zhu, F.Y. QuantAS: A comprehensive pipeline to study alternative splicing by absolute quantification of splice isoforms. New Phytol. 2023, 240, 928–939. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).