The Overexpression of Zea mays Strigolactone Receptor Gene D14 Enhances Drought Resistance in Arabidopsis thaliana L.
Abstract
1. Introduction
2. Results
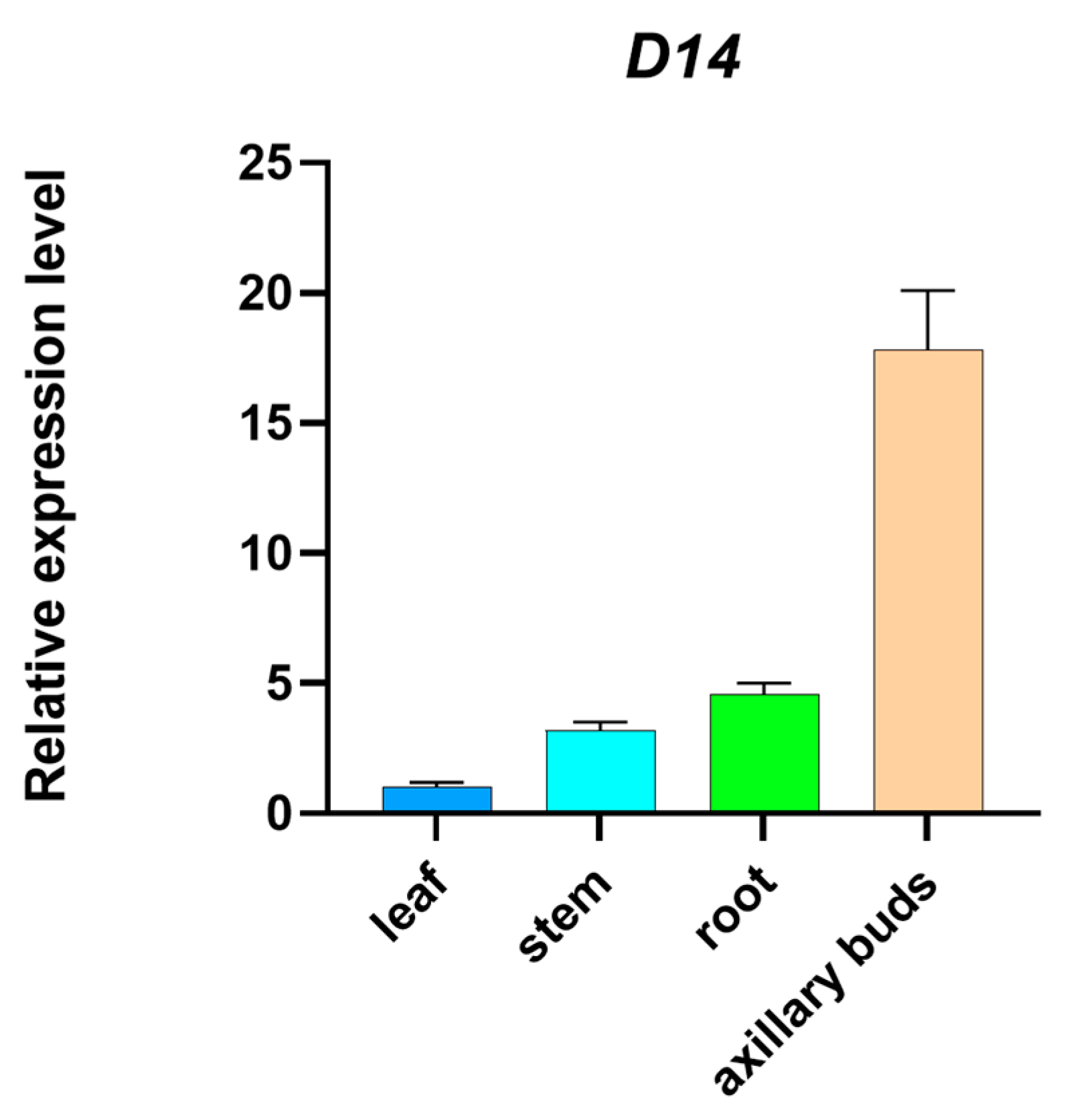
2.1. Tissue-Specific Expression of the Maize D14 Gene
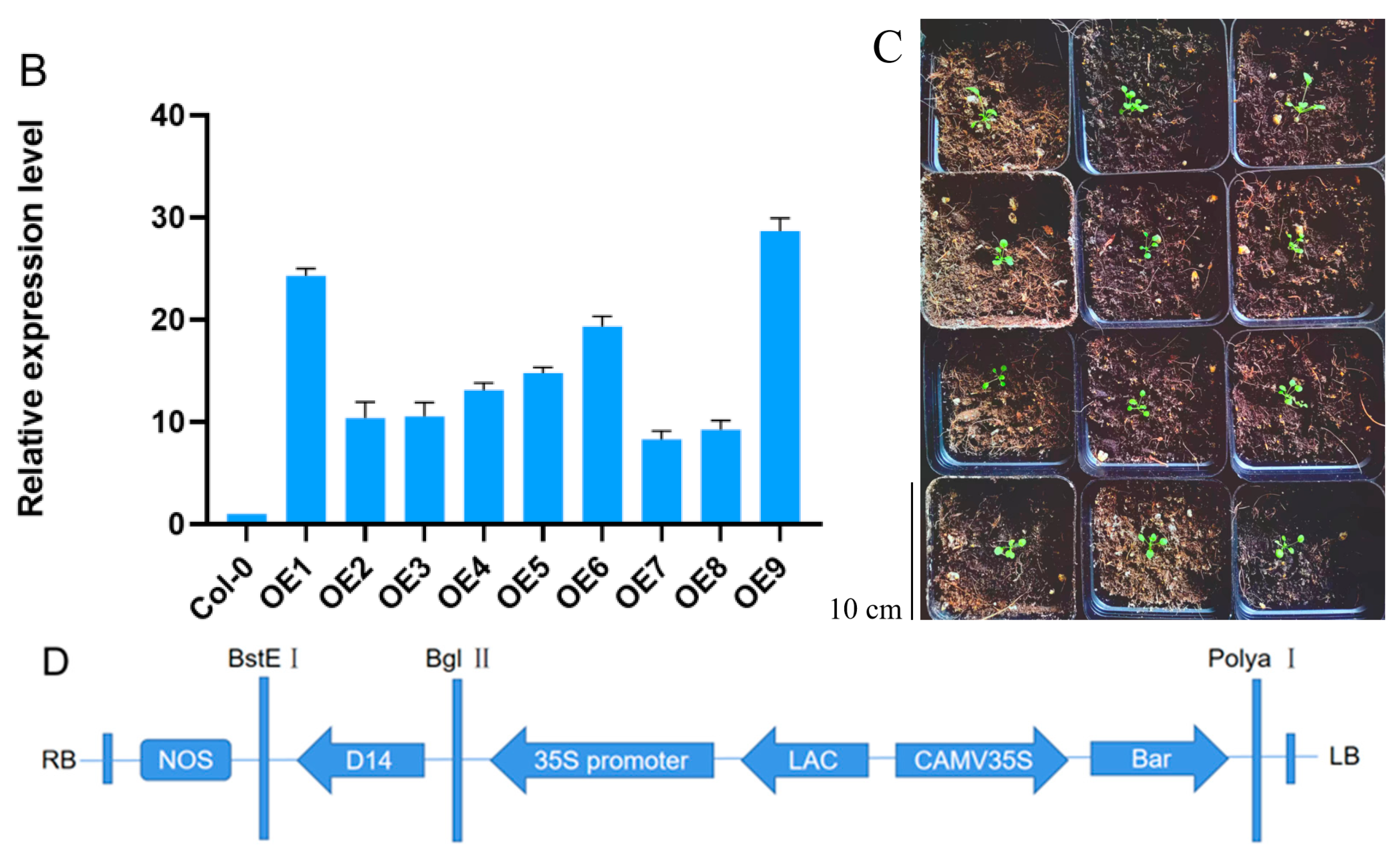
2.2. Generation and Molecular Identification of Transgenic Arabidopsis
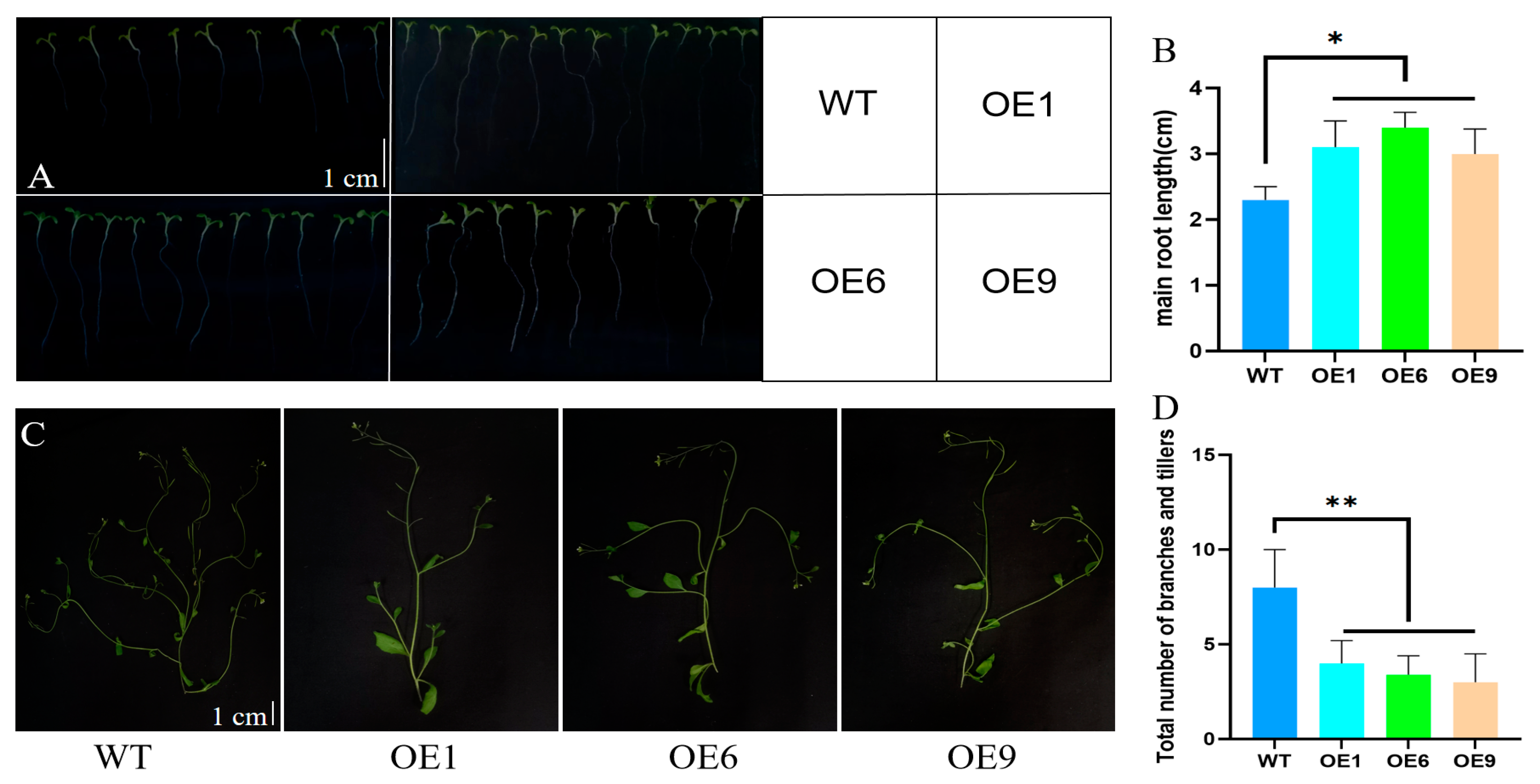
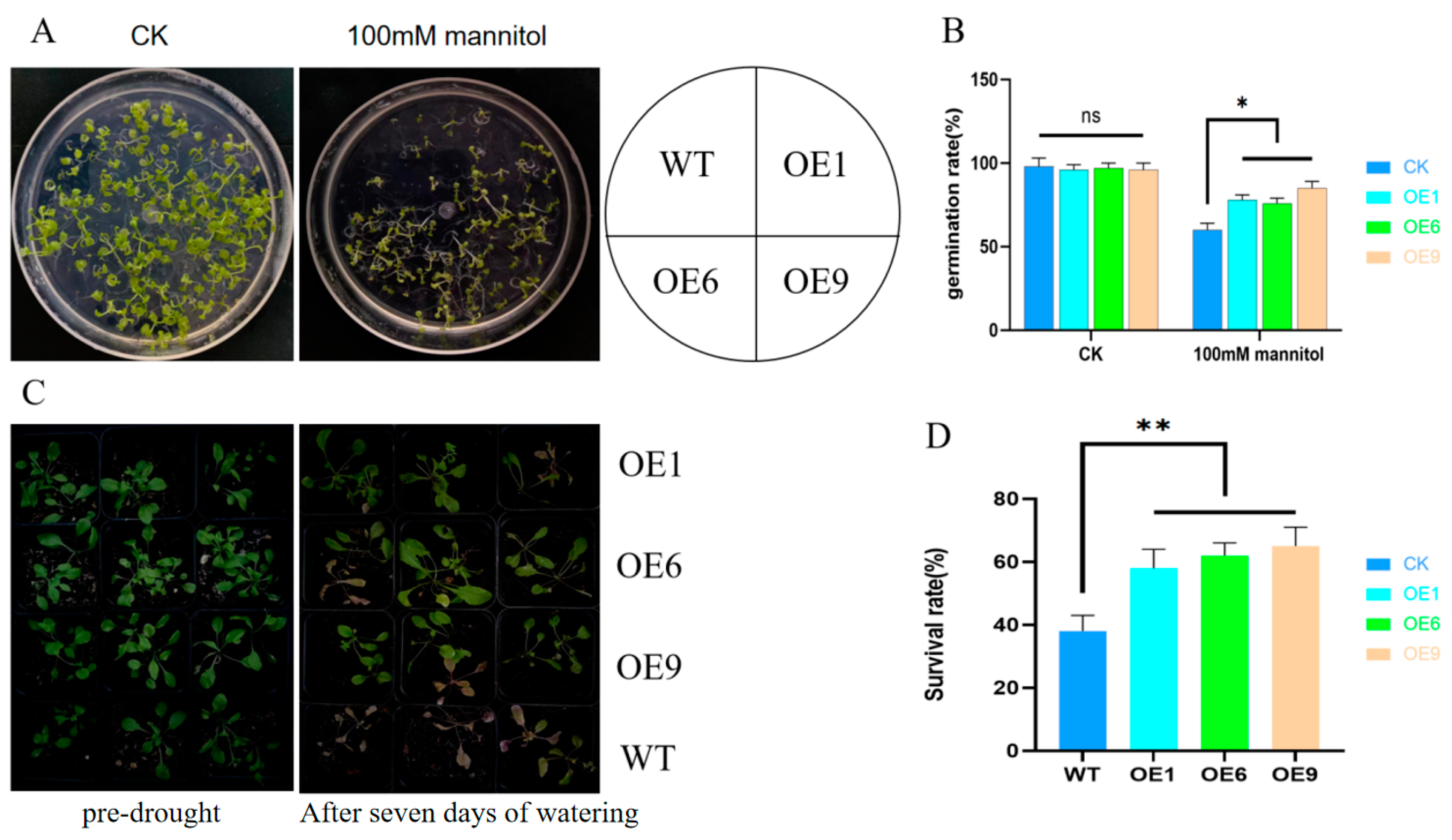
2.3. Phenotypic Analysis of Arabidopsis with D14 Gene Overexpression
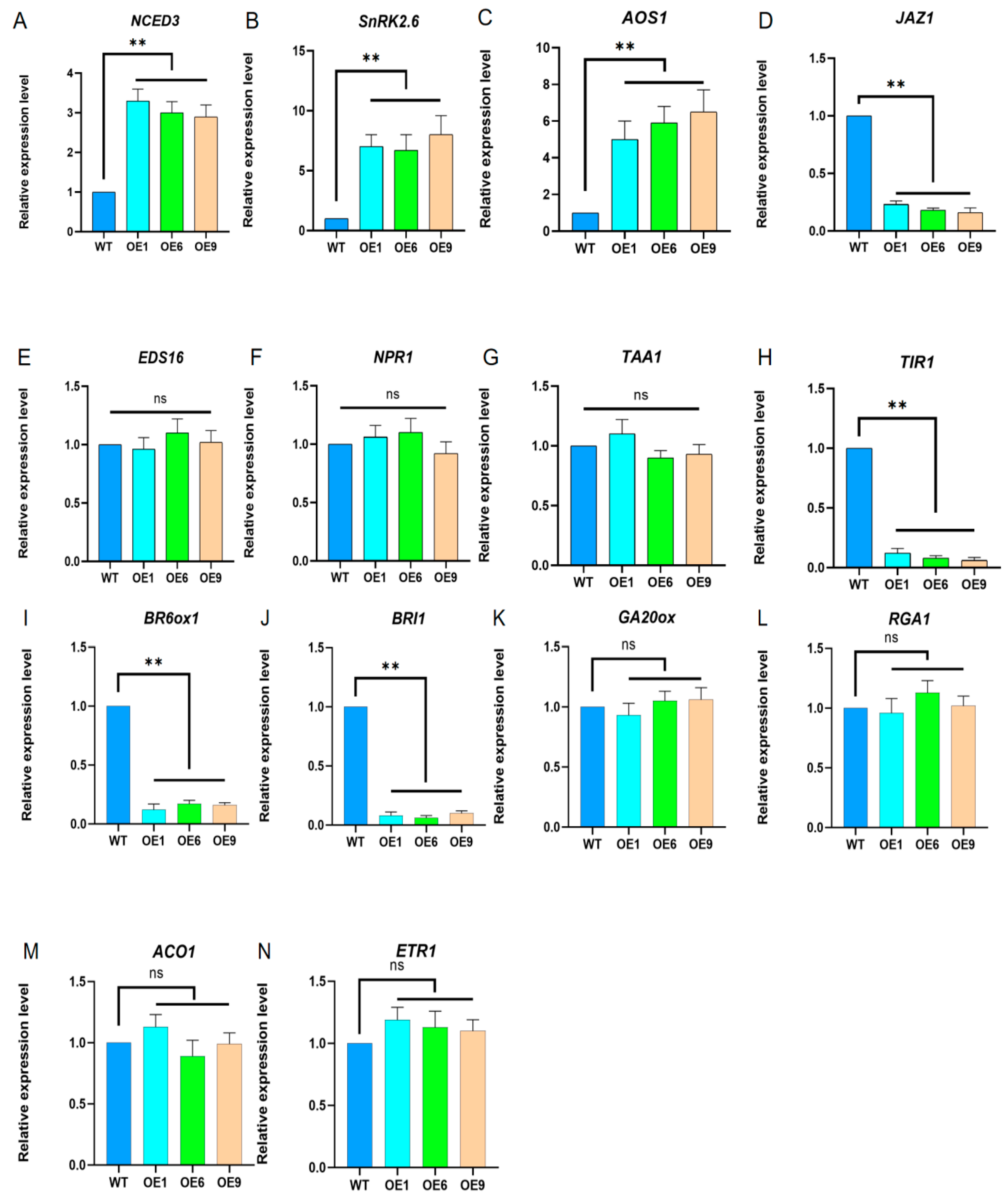
2.4. Analysis of the Expression of Various Hormones in Arabidopsis with D14 Gene Overexpression
2.5. Overexpression of Arabidopsis Improves Survival after Drought-Recovered Water
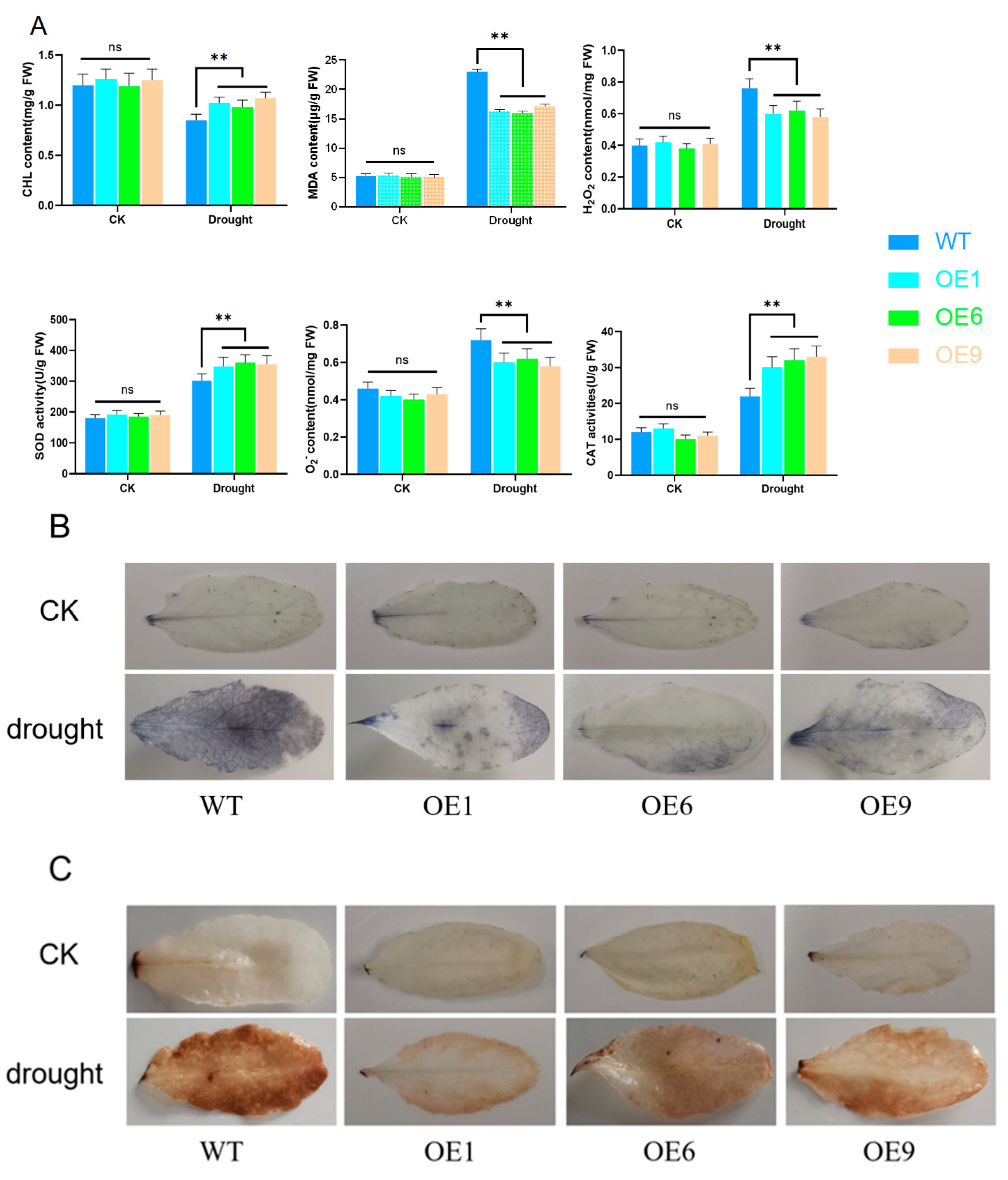
2.6. Overexpression of D14 Gene Reduced ROS Accumulation under Drought Stress Conditions in Arabidopsis
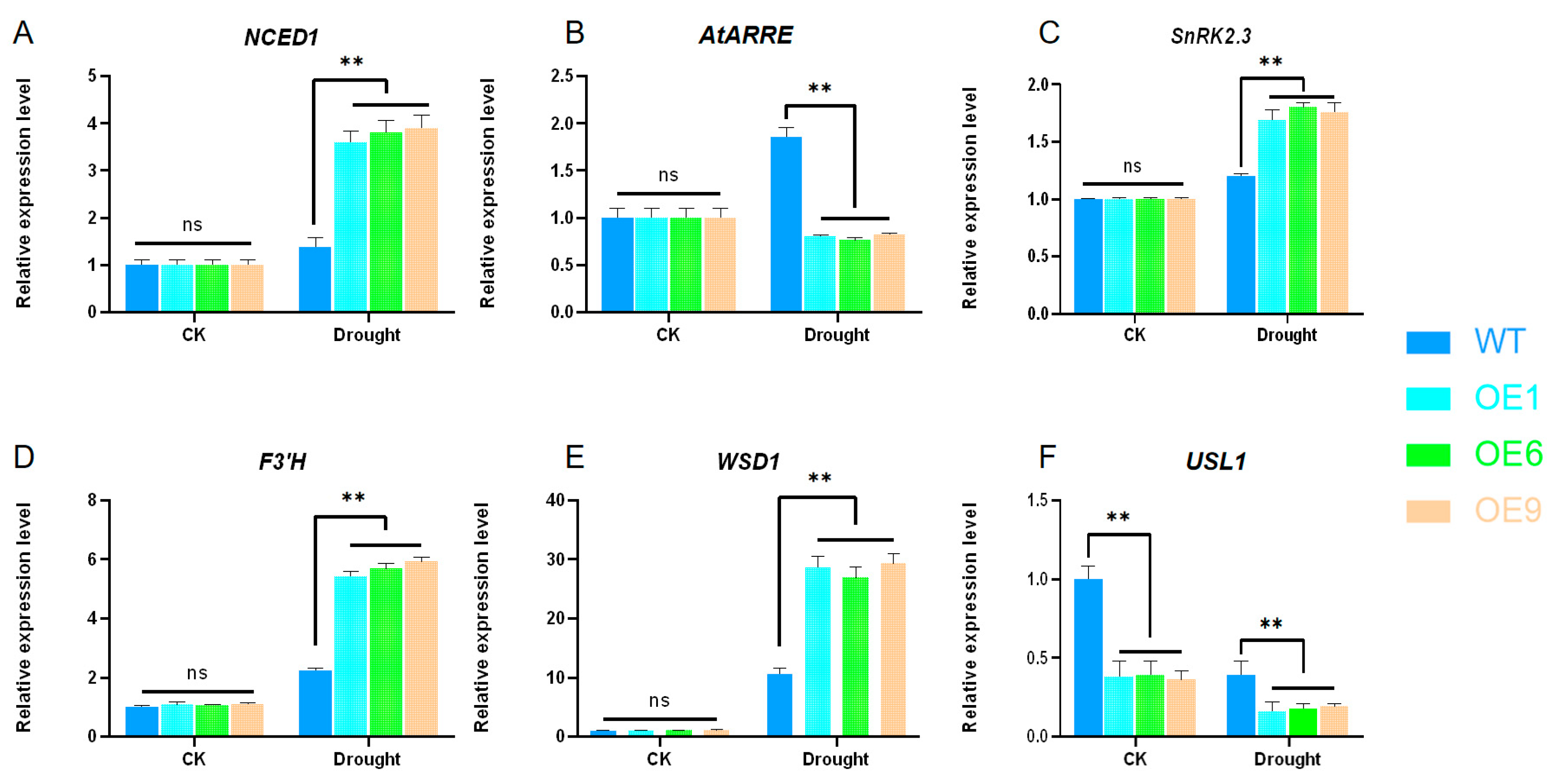
2.7. Changes in Transcription Levels of Key Genes Related to Drought Resistance Pathways in Arabidopsis
3. Discussion
4. Materials and Methods
4.1. The Growth Conditions of Zea mays
4.2. Production and Phenotypic Analyses of Transgenic Plants
4.3. Arabidopsis Abiotic Stress Treatments
4.4. Culture, Infection, and Detection of Plant Pathogens
4.5. NBT (Nitroblue Tetrazolium Chloride) Staining
4.6. DAB (3,3′-Diaminobenzidine) Staining
4.7. Determination of Physiological and Biochemical Activities
4.8. qRT-PCR
4.9. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Jiao, P.; Jiang, Z.; Wei, X.; Liu, S.; Qu, J.; Guan, S.; Ma, Y. Overexpression of the homeobox-leucine zipper protein ATHB-6 improves the drought tolerance of maize (Zea mays L.). Plant Sci. 2022, 316, 111159. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Hao, Z. Quantifying likelihoods of extreme occurrences causing maize yield reduction at the global scale. Sci. Total Environ. 2020, 704, 135250. [Google Scholar] [CrossRef] [PubMed]
- Toreti, A.; Cronie, O.; Zampieri, M. Concurrent climate extremes in the key wheat producing regions of the world. Sci. Rep. 2019, 9, 5493. [Google Scholar] [CrossRef]
- Li, Y.; Guan, K.; Schnitkey, G.D.; DeLucia, E.; Peng, B. Excessive rainfall leads to maize yield loss of a comparable magnitude to extreme drought in the United States. Glob. Chang. Biol. 2019, 25, 2325–2337. [Google Scholar] [CrossRef] [PubMed]
- Beillouin, D.; Schauberger, B.; Bastos, A.; Ciais, P.; Makowski, D. Impact of extreme weather conditions on European crop production in 2018. Philos. Trans. R. Soc. B Biol. Sci. 2020, 375, 20190510. [Google Scholar] [CrossRef]
- Li, C.; Dong, L.; Durairaj, J.; Guan, J.C.; Yoshimura, M.; Quinodoz, P.; Horber, R.; Gaus, K.; Li, J.; Setotaw, Y.B.; et al. Maize resistance to witchweed through changes in strigolactone biosynthesis. Science 2023, 379, 94–99. [Google Scholar] [CrossRef]
- Koltai, H.; Prandi, C. Strigolactones: Past, present and future. Planta 2016, 243, 1309. [Google Scholar] [CrossRef]
- Waters, M.T.; Gutjahr, C.; Bennett, T.; Nelson, D.C. Strigolactone Signaling and Evolution. Annu. Rev. Plant Biol. 2017, 68, 291–322. [Google Scholar] [CrossRef]
- Guan, J.C.; Li, C.; Flint-Garcia, S.; Suzuki, M.; Wu, S.; Saunders, J.W.; Dong, L.; Bouwmeester, H.J.; McCarty, D.R.; Koch, K.E. Maize domestication phenotypes reveal strigolactone networks coordinating grain size evolution with kernel-bearing cupule architecture. Plant Cell 2023, 35, 1013–1037. [Google Scholar] [CrossRef]
- Yao, R.; Ming, Z.; Yan, L.; Li, S.; Wang, F.; Ma, S.; Yu, C.; Yang, M.; Chen, L.; Chen, L.; et al. DWARF14 is a non-canonical hormone receptor for strigolactone. Nature 2016, 536, 469–473. [Google Scholar] [CrossRef]
- Yao, R.; Li, J.; Xie, D. Recent advances in molecular basis for strigolactone action. Sci. China Life Sci. 2018, 61, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Lin, Q.; Zhu, L.; Ren, Y.; Zhou, K.; Shabek, N.; Wu, F.; Mao, H.; Dong, W.; Gan, L.; et al. D14-SCF(D3)-dependent degradation of D53 regulates strigolactone signalling. Nature 2013, 504, 406–410. [Google Scholar] [CrossRef] [PubMed]
- Yao, R.; Wang, F.; Ming, Z.; Du, X.; Chen, L.; Wang, Y.; Zhang, W.; Deng, H.; Xie, D. ShHTL7 is a non-canonical receptor for strigolactones in root parasitic weeds. Cell Res. 2017, 27, 838–841. [Google Scholar] [CrossRef]
- Kelly, J.H.; Tucker, M.R.; Brewer, P.B. The Strigolactone Pathway Is a Target for Modifying Crop Shoot Architecture and Yield. Biology 2023, 12, 95. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Martin-Fontecha, E.S.; Khosla, A.; White, A.; Chang, S.; Cubas, P.; Nelson, D.C. The strigolactone receptor D14 targets SMAX1 for degradation in response to GR24 treatment and osmotic stress. Plant Commun. 2022, 3, 100303. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Kahmann, R. Manipulation of Phytohormone Pathways by Effectors of Filamentous Plant Pathogens. Front. Plant Sci. 2019, 10, 822. [Google Scholar] [CrossRef] [PubMed]
- de Haro, L.A.; Arellano, S.M.; Novak, O.; Feil, R.; Dumon, A.D.; Mattio, M.F.; Tarkowska, D.; Llauger, G.; Strnad, M.; Lunn, J.E.; et al. Mal de Rio Cuarto virus infection causes hormone imbalance and sugar accumulation in wheat leaves. BMC Plant Biol. 2019, 19, 112. [Google Scholar] [CrossRef]
- Agusti, J.; Herold, S.; Schwarz, M.; Sanchez, P.; Ljung, K.; Dun, E.A.; Brewer, P.B.; Beveridge, C.A.; Sieberer, T.; Sehr, E.M.; et al. Strigolactone signaling is required for auxin-dependent stimulation of secondary growth in plants. Proc. Natl. Acad. Sci. USA 2011, 108, 20242–20247. [Google Scholar] [CrossRef]
- Sedaghat, M.; Emam, Y.; Mokhtassi-Bidgoli, A.; Hazrati, S.; Lovisolo, C.; Visentin, I.; Cardinale, F.; Tahmasebi-Sarvestani, Z. The Potential of the Synthetic Strigolactone Analogue GR24 for the Maintenance of Photosynthesis and Yield in Winter Wheat under Drought: Investigations on the Mechanisms of Action and Delivery Modes. Plants 2021, 10, 1223. [Google Scholar] [CrossRef]
- Li, Y.; Li, S.; Feng, Q.; Zhang, J.; Han, X.; Zhang, L.; Yang, F.; Zhou, J. Effects of exogenous Strigolactone on the physiological and ecological characteristics of Pennisetum purpureum Schum. Seedlings under drought stress. BMC Plant Biol. 2022, 22, 578. [Google Scholar] [CrossRef]
- Marzec, M.; Daszkowska-Golec, A.; Collin, A.; Melzer, M.; Eggert, K.; Szarejko, I. Barley strigolactone signalling mutant hvd14.d reveals the role of strigolactones in abscisic acid-dependent response to drought. Plant Cell Environ. 2020, 43, 2239–2253. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Nguyen, K.H.; Tran, C.D.; Watanabe, Y.; Tian, C.; Yin, X.; Li, K.; Yang, Y.; Guo, J.; Miao, Y.; et al. Negative Roles of Strigolactone-Related SMXL6, 7 and 8 Proteins in Drought Resistance in Arabidopsis. Biomolecules 2020, 10, 607. [Google Scholar] [CrossRef] [PubMed]
- Nasir, F.; Tian, L.; Shi, S.; Chang, C.; Ma, L.; Gao, Y.; Tian, C. Strigolactones positively regulate defense against Magnaporthe oryzae in rice (Oryza sativa). Plant Physiol. Biochem. 2019, 142, 106–116. [Google Scholar] [CrossRef] [PubMed]
- Foo, E.; Bullier, E.; Goussot, M.; Foucher, F.; Rameau, C.; Beveridge, C.A. The branching gene RAMOSUS1 mediates interactions among two novel signals and auxin in pea. Plant Cell 2005, 17, 464–474. [Google Scholar] [CrossRef] [PubMed]
- Johnson, X.; Brcich, T.; Dun, E.A.; Goussot, M.; Haurogne, K.; Beveridge, C.A.; Rameau, C. Branching genes are conserved across species. Genes controlling a novel signal in pea are coregulated by other long-distance signals. Plant Physiol. 2006, 142, 1014–1026. [Google Scholar] [CrossRef]
- Bennett, T.; Sieberer, T.; Willett, B.; Booker, J.; Luschnig, C.; Leyser, O. The Arabidopsis MAX pathway controls shoot branching by regulating auxin transport. Curr. Biol. 2006, 16, 553–563. [Google Scholar] [CrossRef]
- Lin, H.; Wang, R.; Qian, Q.; Yan, M.; Meng, X.; Fu, Z.; Yan, C.; Jiang, B.; Su, Z.; Li, J.; et al. DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. Plant Cell 2009, 21, 1512–1525. [Google Scholar] [CrossRef]
- Shinohara, N.; Taylor, C.; Leyser, O. Strigolactone can promote or inhibit shoot branching by triggering rapid depletion of the auxin efflux protein PIN1 from the plasma membrane. PLoS Biol. 2013, 11, e1001474. [Google Scholar] [CrossRef]
- Ito, S.; Yamagami, D.; Umehara, M.; Hanada, A.; Yoshida, S.; Sasaki, Y.; Yajima, S.; Kyozuka, J.; Ueguchi-Tanaka, M.; Matsuoka, M.; et al. Regulation of Strigolactone Biosynthesis by Gibberellin Signaling. Plant Physiol. 2017, 174, 1250–1259. [Google Scholar] [CrossRef]
- Luqman, M.; Shahbaz, M.; Maqsood, M.F.; Farhat, F.; Zulfiqar, U.; Siddiqui, M.H.; Masood, A.; Aqeel, M.; Haider, F.U. Effect of strigolactone on growth, photosynthetic efficiency, antioxidant activity, and osmolytes accumulation in different maize (Zea mays L.) hybrids grown under drought stress. Plant Signal. Behav. 2023, 18, 2262795. [Google Scholar] [CrossRef]
- Ton, J.; Flors, V.; Mauch-Mani, B. The multifaceted role of ABA in disease resistance. Trends Plant Sci. 2009, 14, 310–317. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Bai, B.; Lu, X.; Li, H. A gibberellin-deficient maize mutant exhibits altered plant height, stem strength and drought tolerance. Plant Cell Rep. 2023, 42, 1687–1699. [Google Scholar] [CrossRef] [PubMed]
- Cowling, C.L.; Dash, L.; Kelley, D.R. Roles of auxin pathways in maize biology. J. Exp. Bot. 2023, 74, 6989–6999. [Google Scholar] [CrossRef]
- Lu, C.; Chen, M.X.; Liu, R.; Zhang, L.; Hou, X.; Liu, S.; Ding, X.; Jiang, Y.; Xu, J.; Zhang, J.; et al. Abscisic Acid Regulates Auxin Distribution to Mediate Maize Lateral Root Development Under Salt Stress. Front. Plant Sci. 2019, 10, 716. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Zhong, J.; Sun, X.; Wang, B.; Terzaghi, W.; Dai, M. The Maize ABA Receptors ZmPYL8, 9, and 12 Facilitate Plant Drought Resistance. Front. Plant Sci. 2018, 9, 422. [Google Scholar] [CrossRef] [PubMed]
- Shohat, H.; Eliaz, N.I.; Weiss, D. Gibberellin in tomato: Metabolism, signaling and role in drought responses. Mol. Hortic. 2021, 1, 15. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, Q.; Zhang, Y.; Yang, X.; Yang, C.; Hou, D.; Liu, H.; Xu, H. Overexpression of OsPIN9 Impairs Chilling Tolerance via Disturbing ROS Homeostasis in Rice. Plants 2023, 12, 2809. [Google Scholar] [CrossRef]
- Wang, H.Q.; Zhao, X.Y.; Xuan, W.; Wang, P.; Zhao, F.J. Rice roots avoid asymmetric heavy metal and salinity stress via an RBOH-ROS-auxin signaling cascade. Mol. Plant 2023, 16, 1678–1694. [Google Scholar] [CrossRef]
- Chi, C.; Xu, X.; Wang, M.; Zhang, H.; Fang, P.; Zhou, J.; Xia, X.; Shi, K.; Zhou, Y.; Yu, J. Strigolactones positively regulate abscisic acid-dependent heat and cold tolerance in tomato. Hortic. Res. Engl. 2021, 8, 237. [Google Scholar] [CrossRef]
- Wang, X.; Li, Z.; Shi, Y.; Liu, Z.; Zhang, X.; Gong, Z.; Yang, S. Strigolactones promote plant freezing tolerance by releasing the WRKY41-mediated inhibition of CBF/DREB1 expression. EMBO J. 2023, 42, e112999. [Google Scholar] [CrossRef]
- Bilska-Kos, A.; Solecka, D.; Dziewulska, A.; Ochodzki, P.; Jonczyk, M.; Bilski, H.; Sowinski, P. Low temperature caused modifications in the arrangement of cell wall pectins due to changes of osmotic potential of cells of maize leaves (Zea mays L.). Protoplasma 2017, 254, 713–724. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Fountain, J.C.; Wang, H.; Ni, X.; Ji, P.; Lee, R.D.; Kemerait, R.C.; Scully, B.T.; Guo, B. Stress Sensitivity Is Associated with Differential Accumulation of Reactive Oxygen and Nitrogen Species in Maize Genotypes with Contrasting Levels of Drought Tolerance. Int. J. Mol. Sci. 2015, 16, 24791–24819. [Google Scholar] [CrossRef] [PubMed]
- Corpas, F.J.; Barroso, J.B.; Palma, J.M.; Rodriguez-Ruiz, M. Plant peroxisomes: A nitro-oxidative cocktail. Redox Biol. 2017, 11, 535–542. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.S.; Kang, C.H.; Park, J.H.; Lee, S.Y. Physiological Significance of Plant Peroxiredoxins and the Structure-Related and Multifunctional Biochemistry of Peroxiredoxin 1. Antioxid. Redox Signal. 2018, 28, 625–639. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R. ROS Are Good. Trends Plant Sci. 2017, 22, 11–19. [Google Scholar] [CrossRef]
- Weber, H.; Chetelat, A.; Reymond, P.; Farmer, E.E. Selective and powerful stress gene expression in Arabidopsis in response to malondialdehyde. Plant J. 2004, 37, 877–888. [Google Scholar] [CrossRef]
- Gonzalez-Villagra, J.; Rodrigues-Salvador, A.; Nunes-Nesi, A.; Cohen, J.D.; Reyes-Diaz, M.M. Age-related mechanism and its relationship with secondary metabolism and abscisic acid in Aristotelia chilensis plants subjected to drought stress. Plant Physiol. Biochem. 2018, 124, 136–145. [Google Scholar] [CrossRef]
- Zhang, F.P.; Sussmilch, F.; Nichols, D.S.; Cardoso, A.A.; Brodribb, T.J.; McAdam, S. Leaves, not roots or floral tissue, are the main site of rapid, external pressure-induced ABA biosynthesis in angiosperms. J. Exp. Bot. 2018, 69, 1261–1267. [Google Scholar] [CrossRef]
- Wang, B.; Li, C.; Kong, X.; Li, Y.; Liu, Z.; Wang, J.; Li, X.; Yang, Y. AtARRE, an E3 ubiquitin ligase, negatively regulates ABA signaling in Arabidopsis thaliana. Plant Cell Rep. 2018, 37, 1269–1278. [Google Scholar] [CrossRef]
- Cai, G.; Wang, Y.; Tu, G.; Chen, P.; Luan, S.; Lan, W. Type A2 BTB Members Decrease the ABA Response during Seed Germination by Affecting the Stability of SnRK2.3 in Arabidopsis. Int. J. Mol. Sci. 2020, 21, 3153. [Google Scholar] [CrossRef]
- Tan, W.; Zhang, D.; Zhou, H.; Zheng, T.; Yin, Y.; Lin, H. Transcription factor HAT1 is a substrate of SnRK2.3 kinase and negatively regulates ABA synthesis and signaling in Arabidopsis responding to drought. PLoS Genet. 2018, 14, e1007336. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Vimolmangkang, S.; Soria-Guerra, R.E.; Rosales-Mendoza, S.; Zheng, D.; Lygin, A.V.; Korban, S.S. Ectopic expression of apple F3’H genes contributes to anthocyanin accumulation in the Arabidopsis tt7 mutant grown under nitrogen stress. Plant Physiol. 2010, 153, 806–820. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, H.M.; Rodriguez, J.; Salacup, J.M.; Castaneda, I.S.; Schnell, D.J.; Pareek, A.; Dhankher, O.P. Increased Cuticle Waxes by Overexpression of WSD1 Improves Osmotic Stress Tolerance in Arabidopsis thaliana and Camelina sativa. Int. J. Mol. Sci. 2021, 22, 5173. [Google Scholar] [CrossRef] [PubMed]
- Schoenbohm, C.; Martens, S.; Eder, C.; Forkmann, G.; Weisshaar, B. Identification of the Arabidopsis thaliana flavonoid 3’-hydroxylase gene and functional expression of the encoded P450 enzyme. Biol. Chem. 2000, 381, 749–753. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Wu, X.; Lam, P.; Bird, D.; Zheng, H.; Samuels, L.; Jetter, R.; Kunst, L. Identification of the wax ester synthase/acyl-coenzyme A: Diacylglycerol acyltransferase WSD1 required for stem wax ester biosynthesis in Arabidopsis. Plant Physiol. 2008, 148, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Min, Z.; Li, R.; Chen, L.; Zhang, Y.; Li, Z.; Liu, M.; Ju, Y.; Fang, Y. Alleviation of drought stress in grapevine by foliar-applied strigolactones. Plant Physiol. Biochem. 2019, 135, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.N.; Min, Z.; Wu, J.R.; Liu, B.C.; Xu, X.L.; Fang, Y.L.; Ju, Y.L. Physiological and transcriptomic analysis of Cabernet Sauvginon (Vitis vinifera L.) reveals the alleviating effect of exogenous strigolactones on the response of grapevine to drought stress. Plant Physiol. Biochem. 2021, 167, 400–409. [Google Scholar] [CrossRef]
- Yu, Y.; Xu, J.; Wang, C.; Pang, Y.; Li, L.; Tang, X.; Li, B.; Sun, Q. Genome-wide analysis of the strigolactone biosynthetic and signaling genes in grapevine and their response to salt and drought stresses. PeerJ 2022, 10, e13551. [Google Scholar] [CrossRef]
- Karssen, C.M.; Brinkhorst-van, D.S.D.; Breekland, A.E.; Koornneef, M. Induction of dormancy during seed development by endogenous abscisic acid: Studies on abscisic acid deficient genotypes of Arabidopsis thaliana (L.) Heynh. Planta 1983, 157, 158–165. [Google Scholar] [CrossRef]
- Seo, P.J.; Xiang, F.; Qiao, M.; Park, J.Y.; Lee, Y.N.; Kim, S.G.; Lee, Y.H.; Park, W.J.; Park, C.M. The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol. 2009, 151, 275–289. [Google Scholar] [CrossRef]
- Korwin, K.P.; Visentin, I.; Russo, G.; Minerdi, D.; Bendahmane, A.; Schubert, A.; Cardinale, F. Transcriptome Analysis Points to BES1 as a Transducer of Strigolactone Effects on Drought Memory in Arabidopsis thaliana. Plant Cell Physiol. 2023, 63, 1873–1889. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.Q.; Yuan, J.J.; Xiao, C.C.; Li, G.X.; Yan, J.Y.; Zheng, S.J.; Ding, Z.J. RING-box proteins regulate leaf senescence and stomatal closure via repression of ABA transporter gene ABCG40. J. Integr. Plant Biol. 2022, 64, 979–994. [Google Scholar] [CrossRef] [PubMed]
- Kusajima, M.; Fujita, M.; Soudthedlath, K.; Nakamura, H.; Yoneyama, K.; Nomura, T.; Akiyama, K.; Maruyama-Nakashita, A.; Asami, T.; Nakashita, H. Strigolactones Modulate Salicylic Acid-Mediated Disease Resistance in Arabidopsis thaliana. Int. J. Mol. Sci. 2022, 23, 5246. [Google Scholar] [CrossRef] [PubMed]
- Torres-Vera, R.; Garcia, J.M.; Pozo, M.J.; Lopez-Raez, J.A. Do strigolactones contribute to plant defence? Mol. Plant Pathol. 2014, 15, 211–216. [Google Scholar] [CrossRef] [PubMed]
- Jia, K.P.; Baz, L.; Al-Babili, S. From carotenoids to strigolactones. J. Exp. Bot. 2018, 69, 2189–2204. [Google Scholar] [CrossRef] [PubMed]
- Alder, A.; Jamil, M.; Marzorati, M.; Bruno, M.; Vermathen, M.; Bigler, P.; Ghisla, S.; Bouwmeester, H.; Beyer, P.; Al-Babili, S. The path from beta-carotene to carlactone, a strigolactone-like plant hormone. Science 2012, 335, 1348–1351. [Google Scholar] [CrossRef] [PubMed]
- Yi, F.; Song, A.; Cheng, K.; Liu, J.; Wang, C.; Shao, L.; Wu, S.; Wang, P.; Zhu, J.; Liang, Z.; et al. Strigolactones positively regulate Verticillium wilt resistance in cotton via crosstalk with other hormones. Plant Physiol. 2023, 192, 945–966. [Google Scholar] [CrossRef]
- Dor, E.; Joel, D.M.; Kapulnik, Y.; Koltai, H.; Hershenhorn, J. The synthetic strigolactone GR24 influences the growth pattern of phytopathogenic fungi. Planta 2011, 234, 419–427. [Google Scholar] [CrossRef]
- Zhang, X.; Henriques, R.; Lin, S.S.; Niu, Q.W.; Chua, N.H. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 2006, 1, 641–646. [Google Scholar] [CrossRef]
- He, M.; Wang, Y.; Jahan, M.S.; Liu, W.; Raziq, A.; Sun, J.; Shu, S.; Guo, S. Characterization of SlBAG Genes from Solanum lycopersicum and Its Function in Response to Dark-Induced Leaf Senescence. Plants 2021, 10, 947. [Google Scholar] [CrossRef]
- Ozturk, M.; Turkyilmaz, U.B.; Garcia-Caparros, P.; Khursheed, A.; Gul, A.; Hasanuzzaman, M. Osmoregulation and its actions during the drought stress in plants. Physiol. Plant. 2021, 172, 1321–1335. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Chen, N.; Liu, J.; Jiao, P.; Liu, S.; Qu, J.; Guan, S.; Ma, Y. Overexpression of ZmSAG39 in maize accelerates leaf senescence in Arabidopsis thaliana. Plant Growth Regul. 2022, 98, 451–463. [Google Scholar] [CrossRef]
- Abdulaal, W.H. Purification and characterization of cysteine protease from miswak Salvadora persica. BMC Boichem. 2018, 19, 10. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, C.; Wang, F.; Jiao, P.; Liu, J.; Zhang, H.; Liu, S.; Guan, S.; Ma, Y. The Overexpression of Zea mays Strigolactone Receptor Gene D14 Enhances Drought Resistance in Arabidopsis thaliana L. Int. J. Mol. Sci. 2024, 25, 1327. https://doi.org/10.3390/ijms25021327
Zhang C, Wang F, Jiao P, Liu J, Zhang H, Liu S, Guan S, Ma Y. The Overexpression of Zea mays Strigolactone Receptor Gene D14 Enhances Drought Resistance in Arabidopsis thaliana L. International Journal of Molecular Sciences. 2024; 25(2):1327. https://doi.org/10.3390/ijms25021327
Chicago/Turabian StyleZhang, Chen, Fanhao Wang, Peng Jiao, Jiaqi Liu, Honglin Zhang, Siyan Liu, Shuyan Guan, and Yiyong Ma. 2024. "The Overexpression of Zea mays Strigolactone Receptor Gene D14 Enhances Drought Resistance in Arabidopsis thaliana L." International Journal of Molecular Sciences 25, no. 2: 1327. https://doi.org/10.3390/ijms25021327
APA StyleZhang, C., Wang, F., Jiao, P., Liu, J., Zhang, H., Liu, S., Guan, S., & Ma, Y. (2024). The Overexpression of Zea mays Strigolactone Receptor Gene D14 Enhances Drought Resistance in Arabidopsis thaliana L. International Journal of Molecular Sciences, 25(2), 1327. https://doi.org/10.3390/ijms25021327





