Principal Component Analysis (PCA) of Molecular Descriptors for Improving Permeation through the Blood–Brain Barrier of Quercetin Analogues
Abstract
:1. Introduction
2. Results
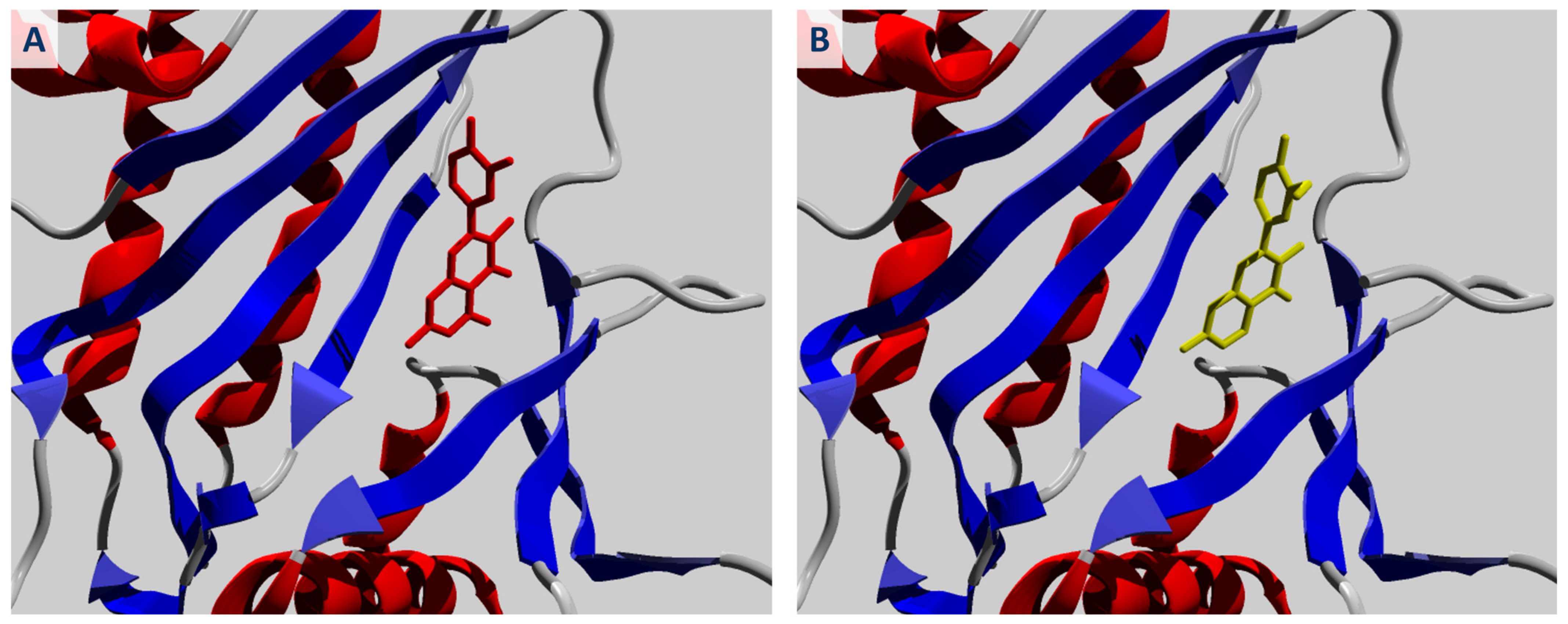
2.1. Molecular Docking of Quercetin and Its Analogues against IPMK
2.2. In Silico Prediction of CNS Distribution
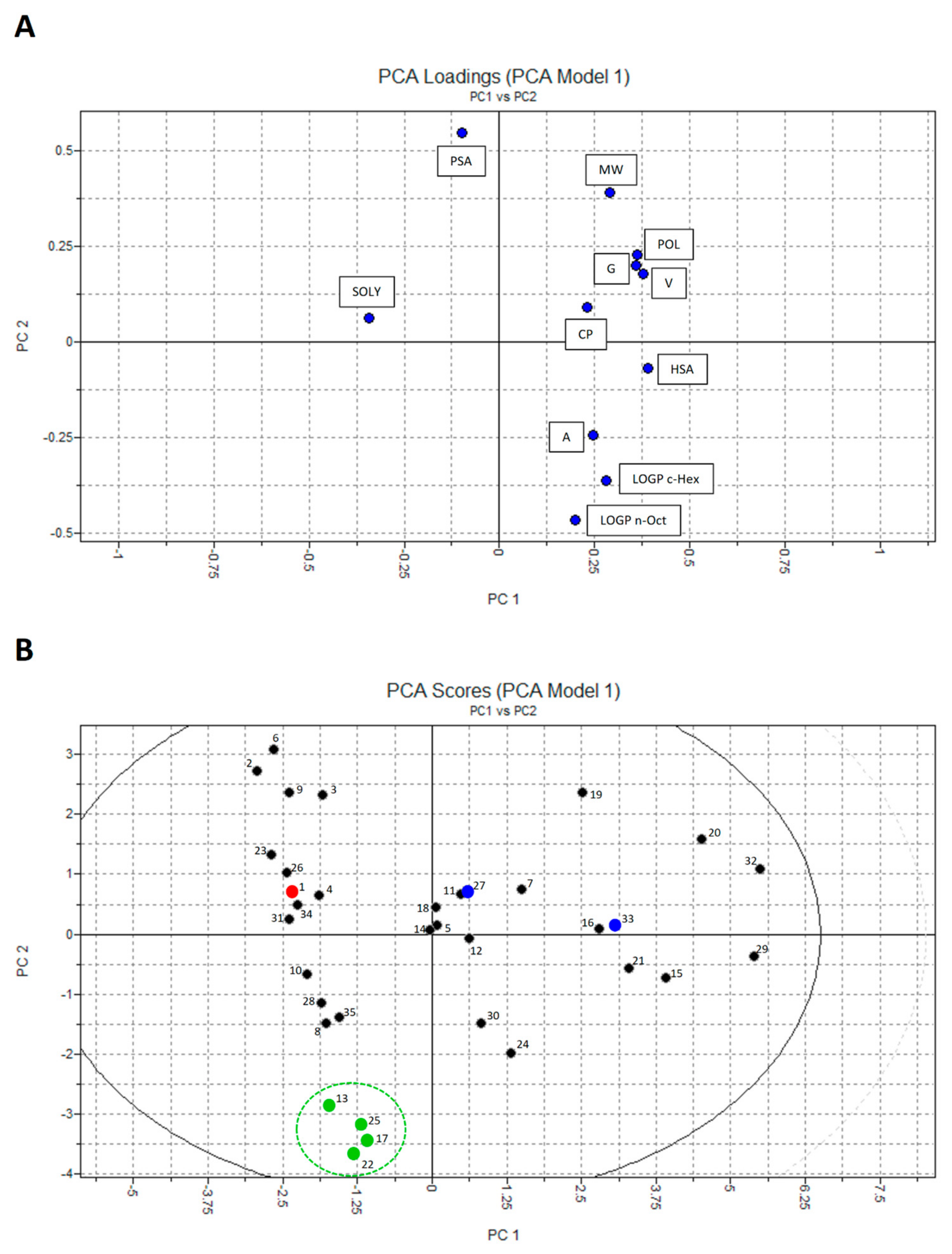
2.3. Principal Component Analysis (PCA)
3. Discussion
4. Methods and Materials
4.1. Protein and Ligand Preparation
4.2. Molecular Docking Analysis
4.3. In Silico Prediction of CNS Distribution
4.4. Principal Component Analysis (PCA)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sak, K. Site-specific anticancer effects of dietary flavonoid quercetin. Nutr. Cancer 2014, 66, 177–193. [Google Scholar] [CrossRef] [PubMed]
- Patel, R.V.; Mistry, B.M.; Shinde, S.K.; Syed, R.; Singh, V.; Shin, H.S. Therapeutic potential of quercetin as a cardiovascular agent. Eur. J. Med. Chem. 2018, 155, 889–904. [Google Scholar] [CrossRef] [PubMed]
- Carullo, G.; Cappello, A.R.; Frattaruolo, L.; Badolato, M.; Armentano, B.; Aiello, F. Quercetin and derivatives: Useful tools in inflammation and pain management. Future Med. Chem. 2017, 9, 79–93. [Google Scholar] [CrossRef] [PubMed]
- Khan, H.; Ullah, H.; Aschner, M.; Cheang, W.S.; Akkol, E.K. Neuroprotective effects of quercetin in alzheimer’s disease. Biomolecules 2020, 10, 59. [Google Scholar] [CrossRef] [PubMed]
- Rabiei, Z.; Solati, K.; Amini-Khoei, H. Phytotherapy in treatment of Parkinson’s disease: A review. Pharm. Biol. 2019, 57, 355–362. [Google Scholar] [CrossRef]
- Gu, C.; Stashko, M.A.; Puhl-Rubio, A.C.; Chakraborty, M.; Chakraborty, A.; Frye, S.V.; Pearce, K.H.; Wang, X.; Shears, S.B.; Wang, H. Inhibition of Inositol Polyphosphate Kinases by Quercetin and Related Flavonoids: A Structure-Activity Analysis. J. Med. Chem. 2019, 62, 1443–1454. [Google Scholar] [CrossRef]
- Kim, E.; Ahn, H.; Kim, M.G.; Lee, H.; Kim, S. The expanding significance of inositol polyphosphate multikinase as a signaling hub. Mol. Cells 2017, 40, 315–321. [Google Scholar] [CrossRef]
- Amanzadeh, E.; Esmaeili, A.; Rahgozar, S.; Nourbakhshnia, M. Application of quercetin in neurological disorders: From nutrition to nanomedicine. Rev. Neurosci. 2019, 30, 555–572. [Google Scholar] [CrossRef]
- El-Saber Batiha, G.; Beshbishy, A.M.; Ikram, M.; Mulla, Z.S.; Abd El-Hack, M.E.; Taha, A.E.; Algammal, A.M.; Ali Elewa, Y.H. The pharmacological activity, biochemical properties, and pharmacokinetics of the major natural polyphenolic flavonoid: Quercetin. Foods 2020, 9, 374. [Google Scholar] [CrossRef]
- Bruno, A.; Costantino, G.; Sartori, L.; Radi, M. The in silico drug discovery toolbox: Applications in lead discovery and optimization. Curr. Med. Chem. 2019, 26, 3838–3873. [Google Scholar] [CrossRef]
- Vizirianakis, I.S. Challenges in current drug delivery from the potential application of pharmacogenomics and personalized medicine in clinical practice. Curr. Drug Deliv. 2004, 1, 73–80. [Google Scholar] [CrossRef]
- Yoo, C.; Shahlaei, M. The applications of PCA in QSAR studies: A case study on CCR5 antagonists. Chem. Biol. Drug Des. 2018, 91, 137–152. [Google Scholar] [CrossRef]
- Lauria, A.; Ippolito, M.; Almerico, A.M. Principal component analysis on molecular descriptors as an alternative point of view in the search of new Hsp90 inhibitors. Comput. Biol. Chem. 2009, 33, 386–390. [Google Scholar] [CrossRef]
- Cai, X.; Fang, Z.; Dou, J.; Yu, A.; Zhai, G. Bioavailability of quercetin: Problems and promises. Curr. Med. Chem. 2013, 20, 2572–2582. [Google Scholar] [CrossRef]
- Pavlović, N.; Milošević, N.; Djanić, M.; Goločorbin-Kon, S.; Stanimirov, B.; Stankov, K.; Mikov, M. Antimetastatic Potential of Quercetin Analogues with Improved Pharmacokinetic Profile: A Pharmacoinformatic Preliminary Study. Anti-Cancer Agents Med. Chem. 2022, 22, 1407–1413. [Google Scholar] [CrossRef]
- Singh, S.P.; Konwar, B.K. Molecular docking studies of quercetin and its analogues against human inducible nitric oxide synthase. SpringerPlus 2012, 1, 69. [Google Scholar] [CrossRef]
- Pavlović, N.; Đanić, M.; Stanimirov, B.; Goločorbin-Kon, S.; Stankov, K.; Lalić-Popović, M.; Mikov, M. In silico discovery of resveratrol analogues as potential agents in treatment of metabolic disorders. Curr. Pharm. Des. 2019, 25, 3776–3783. [Google Scholar] [CrossRef]
- Viceconti, M.; Pappalardo, F.; Rodriguez, B.; Horner, M.; Bischoff, J.; Musuamba Tshinanu, F. In silico trials: Verification, validation and uncertainty quantification of predictive models used in the regulatory evaluation of biomedical products. Methods 2021, 185, 120–127. [Google Scholar] [CrossRef]
- Touil, Y.S.; Auzeil, N.; Boulinguez, F.; Saighi, H.; Regazzetti, A.; Scherman, D.; Chabot, G.G. Fisetin disposition and metabolism in mice: Identification of geraldol as an active metabolite. Biochem. Pharmacol. 2011, 82, 1731–1739. [Google Scholar] [CrossRef]
- Nabavi, S.F.; Braidy, N.; Habtemariam, S.; Sureda, A.; Manayi, A.; Nabavi, S.M. Neuroprotective effects of fisetin in alzheimer’s and parkinson’s diseases: From chemistry to medicine. Curr. Top. Med. Chem. 2016, 16, 1910–1915. [Google Scholar] [CrossRef]
- Fischer, W.; Currais, A.; Liang, Z.; Pinto, A.; Maher, P. Old age-associated phenotypic screening for Alzheimer’s disease drug candidates identifies sterubin as a potent neuroprotective compound from Yerba santa. Redox Biol. 2019, 21, 101089. [Google Scholar] [CrossRef]
- Dash, R.; Emran, T.B.; Uddin, M.M.; Islam, A.; Junaid, M. Molecular docking of fisetin with AD associated AChE, ABAD and BACE1 proteins. Bioinformation 2014, 10, 562–568. [Google Scholar] [CrossRef]
- Giuliani, A. The application of principal component analysis to drug discovery and biomedical data. Drug Discov. Today 2017, 22, 1069–1076. [Google Scholar] [CrossRef]
- Wang, J.; Skolnik, S. Permeability diagnosis model in drug discovery: A diagnostic tool to identify the most influencing properties for gastrointestinal permeability. Curr. Top. Med. Chem. 2013, 13, 1308–1316. [Google Scholar] [CrossRef]
- Namanja, A.T.; Xu, J.; Wu, H.; Sun, Q.; Upadhyay, A.K.; Sun, C.; Van Doren, S.R.; Petros, A.M. NMR-based fragment screening and lead discovery accelerated by principal component analysis. J. Biomol. NMR 2019, 73, 675–685. [Google Scholar] [CrossRef]
- Shaikh, S.; Ahmad, S.S.; Ansari, M.A.; Shakil, S.; Rizvi, S.M.D.; Shakil, S.; Tabrez, S.; Akhtar, S.; Kamal, M.A. Prediction of comparative inhibition efficiency for a novel natural ligand, galangin against human brain acetylcholinesterase, butyrylcholinesterase and 5-lipoxygenase: A neuroinformatics study. CNS Neurol. Disord.-Drug Targets 2014, 13, 452–459. [Google Scholar] [CrossRef]
- Yang, C.C.; Lin, C.C.; Hsiao, L.D.; Yang, C.M. Galangin inhibits thrombin-induced MMP-9 expression in SK-N-SH cells via protein kinase-dependent NF-κB phosphorylation. Int. J. Mol. Sci. 2018, 19, 4084. [Google Scholar] [CrossRef]
- Caliskan, M.; Mogulkoc, R.; Baltaci, A.K.; Menevse, E. The Effect of 3′,4′-Dihydroxyflavonol on Lipid Peroxidation in Rats with Cerebral Ischemia Reperfusion Injury. Neurochem. Res. 2016, 41, 1732–1740. [Google Scholar] [CrossRef]
- Oz, M.; Demir, E.A.; Caliskan, M.; Mogulkoc, R.; Baltaci, A.K.; Nurullahoglu Atalik, K.E. 3′,4′-Dihydroxyflavonol attenuates spatial learning and memory impairments in global cerebral ischemia. Nutr. Neurosci. 2017, 20, 119–126. [Google Scholar] [CrossRef]
- Kwon, S.H.; Hong, S.I.; Ma, S.X.; Lee, S.Y.; Jang, C.G. 3′,4′,7-Trihydroxyflavone prevents apoptotic cell death in neuronal cells from hydrogen peroxide-induced oxidative stress. Food Chem. Toxicol. 2015, 80, 41–51. [Google Scholar] [CrossRef]
- Thilakarathna, S.H.; Vasantha Rupasinghe, H.P. Flavonoid bioavailability and attempts for bioavailability enhancement. Nutrients 2013, 5, 3367–3387. [Google Scholar] [CrossRef]
- Wen, X.; Walle, T. Methylated flavonoids have greatly improved intestinal absorption and metabolic stability. Drug Metab. Dispos. 2006, 34, 1786–1792. [Google Scholar] [CrossRef]
- Akamine, Y.; Yasui-Furukori, N.; Ieiri, I.; Uno, T. Psychotropic drug-drug interactions involving P-glycoprotein. CNS Drugs 2012, 26, 959–973. [Google Scholar] [CrossRef]
- Choi, Y.H.; Yu, A.M. ABC transporters in multidrug resistance and pharmacokinetics, and strategies for drug development. Curr. Pharm. Des. 2014, 20, 793–807. [Google Scholar] [CrossRef]
- O’Brien, F.E.; Dinan, T.G.; Griffin, B.T.; Cryan, J.F. Interactions between antidepressants and P-glycoprotein at the blood-brain barrier: Clinical significance of in vitro and in vivo findings. Br. J. Pharmacol. 2012, 165, 289–312. [Google Scholar] [CrossRef]
- Martínez-Busi, M.; Arredondo, F.; González, D.; Echeverry, C.; Vega-Teijido, M.A.; Carvalho, D.; Rodríguez-Haralambides, A.; Rivera, F.; Dajas, F.; Abin-Carriquiry, J.A. Purification, structural elucidation, antioxidant capacity and neuroprotective potential of the main polyphenolic compounds contained in Achyrocline satureioides (Lam) D.C. (Compositae). Bioorganic Med. Chem. 2019, 27, 2579–2591. [Google Scholar] [CrossRef]
- Burley, S.K.; Berman, H.M.; Kleywegt, G.J.; Markley, J.L.; Nakamura, H.; Velankar, S. Protein Data Bank (PDB): The single global macromolecular structure archive. In Methods in Molecular Biology; Humana Press: New York, NY, USA, 2017; Volume 1607, pp. 627–641. [Google Scholar]
- Irwin, J.J.; Sterling, T.; Mysinger, M.M.; Bolstad, E.S.; Coleman, R.G. ZINC: A free tool to discover chemistry for biology. J. Chem. Inf. Model. 2012, 52, 1757–1768. [Google Scholar] [CrossRef]
- Irwin, J.J.; Tang, K.G.; Young, J.; Dandarchuluun, C.; Wong, B.R.; Khurelbaatar, M.; Moroz, Y.S.; Mayfield, J.; Sayle, R.A. ZINC20—A Free Ultralarge-Scale Chemical Database for Ligand Discovery. J. Chem. Inf. Model. 2020, 60, 6065–6073. [Google Scholar] [CrossRef]
- Thomsen, R.; Christensen, M.H. MolDock: A new technique for high-accuracy molecular docking. J. Med. Chem. 2006, 49, 3315–3321. [Google Scholar] [CrossRef]
- Cruciani, G.; Pastor, M.; Guba, W. VolSurf: A new tool for the pharmacokinetic optimization of lead compounds. Eur. J. Pharm. Sci. 2000, 11, S29–S39. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Wang, P.H.; Tu, Y.S.; Tseng, Y.J. PgpRules: A decision tree based prediction server for P-glycoprotein substrates and inhibitors. Bioinformatics 2019, 35, 4193–4195. [Google Scholar] [CrossRef]


| No. | Compound | MolDock Score 1 [kcal/mol] | LOGP nOct 2 | LOGP cHex 2 | P-gp Substrate 3 | P-gp Inhibitor 3 | LgBB 2 | BBB Permeant 4 |
|---|---|---|---|---|---|---|---|---|
| 1 | ZINC03869685 | −82.233 | 2.078 | −5.609 | No | No | −2.955 | No |
| 2 | ZINC03874317 | −82.088 | 1.815 | −7.256 | No | No | −3.224 | No |
| 3 | ZINC05784821 | −80.472 | 1.942 | −5.924 | No | No | −3.200 | No |
| 4 | ZINC04098600 | −80.627 | 2.255 | −5.273 | No | No | −2.865 | No |
| 5 | ZINC06520226 | −78.471 | 2.669 | −4.283 | No | No | −2.822 | No |
| 6 | ZINC14436449 | −81.632 | 1.783 | −6.806 | No | No | −3.311 | No |
| 7 | ZINC06484604 | −86.261 | 2.275 | −4.212 | No | No | −2.707 | No |
| 8 | ZINC00039111 | −86.966 | 2.518 | −3.626 | No | No | −2.552 | No |
| 9 | ZINC06525297 | −82.619 | 1.873 | −5.635 | No | No | −3.163 | No |
| 10 | ZINC03869768 | −81.960 | 2.238 | −5.530 | No | No | −2.745 | No |
| 11 | ZINC00517261 | −86.485 | 2.275 | −4.212 | No | No | −2.847 | No |
| 12 | ZINC03875620 | −80.734 | 2.570 | −2.354 | No | No | −2.615 | No |
| 13 | ZINC00057845 | −87.179 | 2.692 | −3.660 | No | No | −2.074 | No |
| 14 | ZINC04731234 | −76.973 | 2.600 | −3.994 | No | No | −2.837 | No |
| 15 | ZINC01645590 | −83.643 | 2.767 | −0.957 | No | No | −2.235 | No |
| 16 | ZINC06018683 | −88.909 | 2.472 | −2.815 | No | No | −2.522 | No |
| 17 | ZINC00120273 | −78.290 | 2.670 | −2.890 | No | No | −1.924 | No |
| 18 | ZINC05998785 | −75.133 | 2.275 | −4.212 | No | No | −2.822 | No |
| 19 | ZINC06483609 | −88.006 | 2.209 | −4.462 | No | No | −3.034 | No |
| 20 | ZINC06483700 | −90.616 | 2.337 | −2.776 | No | No | −2.667 | No |
| 21 | ZINC06403375 | −85.664 | 2.767 | −0.957 | No | No | −2.505 | No |
| 22 | ZINC00039321 | −74.655 | 2.936 | −0.873 | No | No | −1.773 | No |
| 23 | ZINC03881558 | −85.134 | 1.919 | −6.491 | No | No | −3.099 | No |
| 24 | ZINC06411540 | −85.910 | 2.730 | −2.275 | No | No | −2.333 | No |
| 25 | ZINC00057752 | −72.415 | 2.642 | −2.664 | No | No | −1.973 | No |
| 26 | ZINC06536276 | −82.025 | 2.033 | −5.556 | No | No | −2.964 | No |
| 27 | ZINC05998596 | −82.584 | 2.249 | −5.092 | Yes | No | −1.525 | No |
| 28 | ZINC00008662 | −82.625 | 2.518 | −3.626 | No | No | −2.400 | No |
| 29 | ZINC02146994 | −89.938 | 2.777 | −0.793 | No | No | −2.267 | No |
| 30 | ZINC05732763 | −91.827 | 2.715 | −2.229 | No | No | −2.513 | No |
| 31 | ZINC48057104 | −81.994 | 2.199 | −4.587 | No | No | −2.683 | No |
| 32 | ZINC05733650 | −75.462 | 2.464 | −1.444 | No | No | −2.376 | No |
| 33 | ZINC05640267 | −87.012 | 2.446 | −3.695 | Yes | No | −1.263 | No |
| 34 | ZINC14644239 | −84.261 | 2.003 | −5.232 | No | No | −2.699 | No |
| 35 | ZINC06095498 | −82.714 | 2.337 | −3.738 | No | No | −2.375 | No |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pavlović, N.; Milošević Sopta, N.; Mitrović, D.; Zaklan, D.; Tomas Petrović, A.; Stilinović, N.; Vukmirović, S. Principal Component Analysis (PCA) of Molecular Descriptors for Improving Permeation through the Blood–Brain Barrier of Quercetin Analogues. Int. J. Mol. Sci. 2024, 25, 192. https://doi.org/10.3390/ijms25010192
Pavlović N, Milošević Sopta N, Mitrović D, Zaklan D, Tomas Petrović A, Stilinović N, Vukmirović S. Principal Component Analysis (PCA) of Molecular Descriptors for Improving Permeation through the Blood–Brain Barrier of Quercetin Analogues. International Journal of Molecular Sciences. 2024; 25(1):192. https://doi.org/10.3390/ijms25010192
Chicago/Turabian StylePavlović, Nebojša, Nastasija Milošević Sopta, Darko Mitrović, Dragana Zaklan, Ana Tomas Petrović, Nebojša Stilinović, and Saša Vukmirović. 2024. "Principal Component Analysis (PCA) of Molecular Descriptors for Improving Permeation through the Blood–Brain Barrier of Quercetin Analogues" International Journal of Molecular Sciences 25, no. 1: 192. https://doi.org/10.3390/ijms25010192
APA StylePavlović, N., Milošević Sopta, N., Mitrović, D., Zaklan, D., Tomas Petrović, A., Stilinović, N., & Vukmirović, S. (2024). Principal Component Analysis (PCA) of Molecular Descriptors for Improving Permeation through the Blood–Brain Barrier of Quercetin Analogues. International Journal of Molecular Sciences, 25(1), 192. https://doi.org/10.3390/ijms25010192







