Chimeric Human Papillomavirus-16 Virus-like Particles Presenting HIV-1 P18I10 Peptide: Expression, Purification, Bio-Physical Properties and Immunogenicity in BALB/c Mice
Abstract
1. Introduction
2. Results
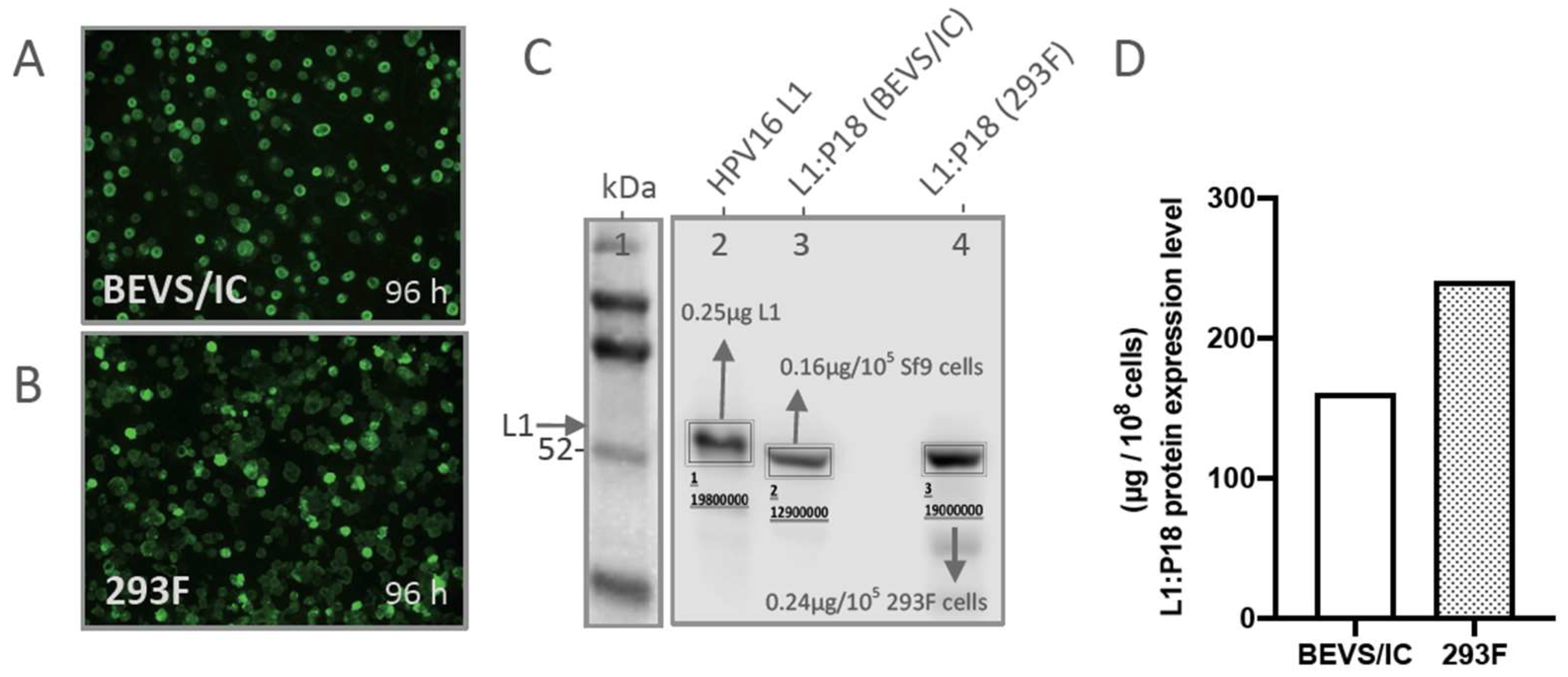
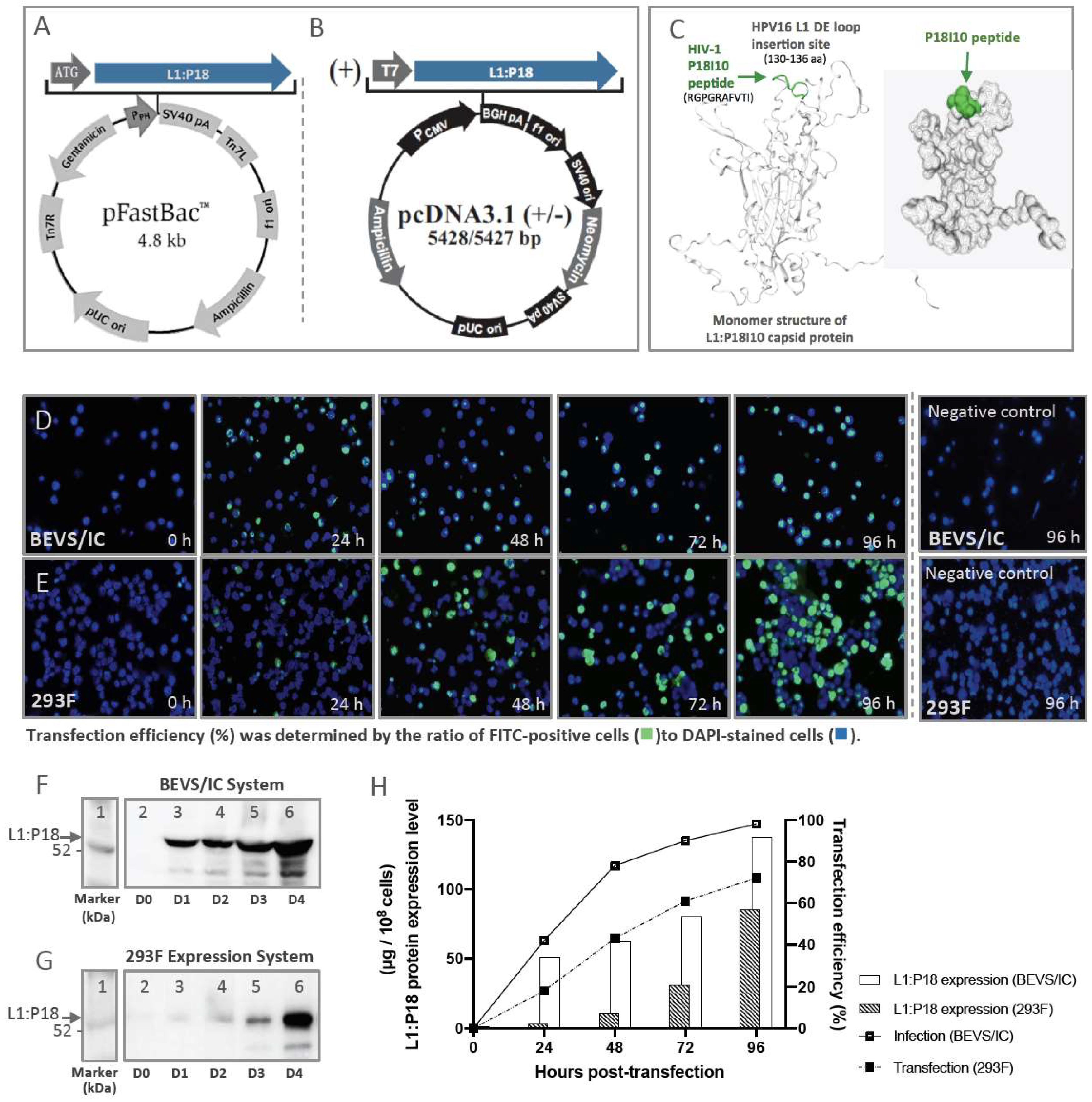
2.1. Comparison of L1:P18I10 Proteins Production in BEVS/IC and 293F Expression Systems
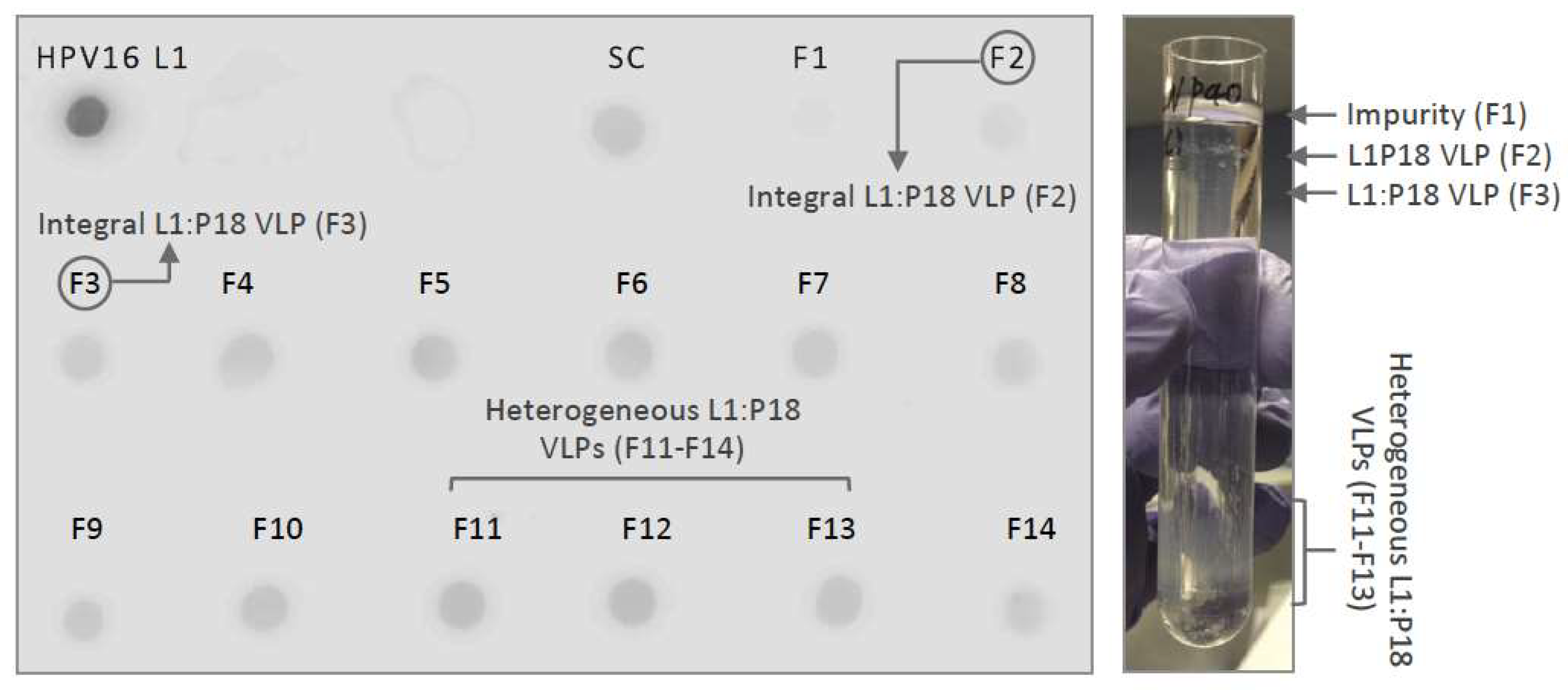
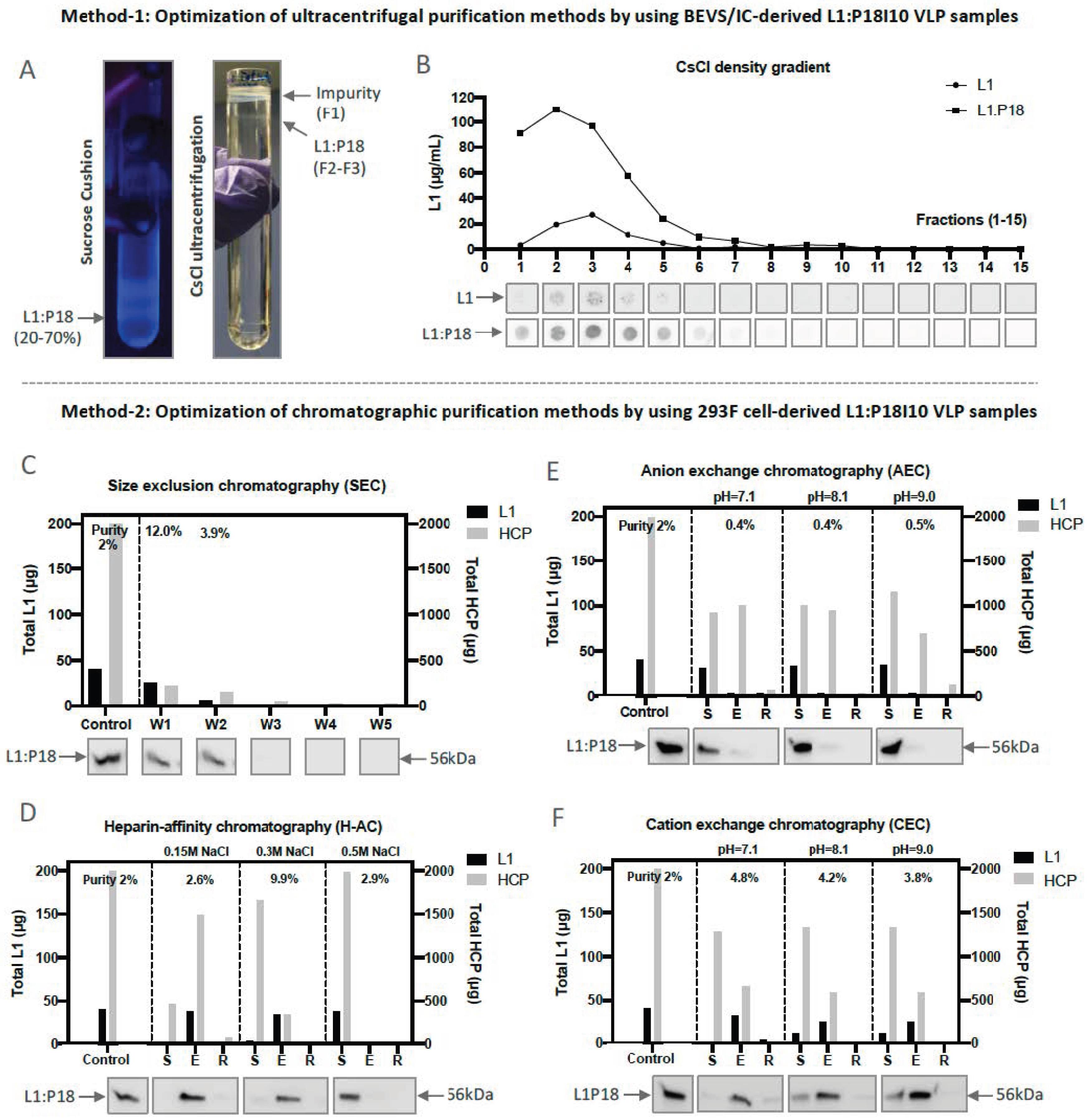
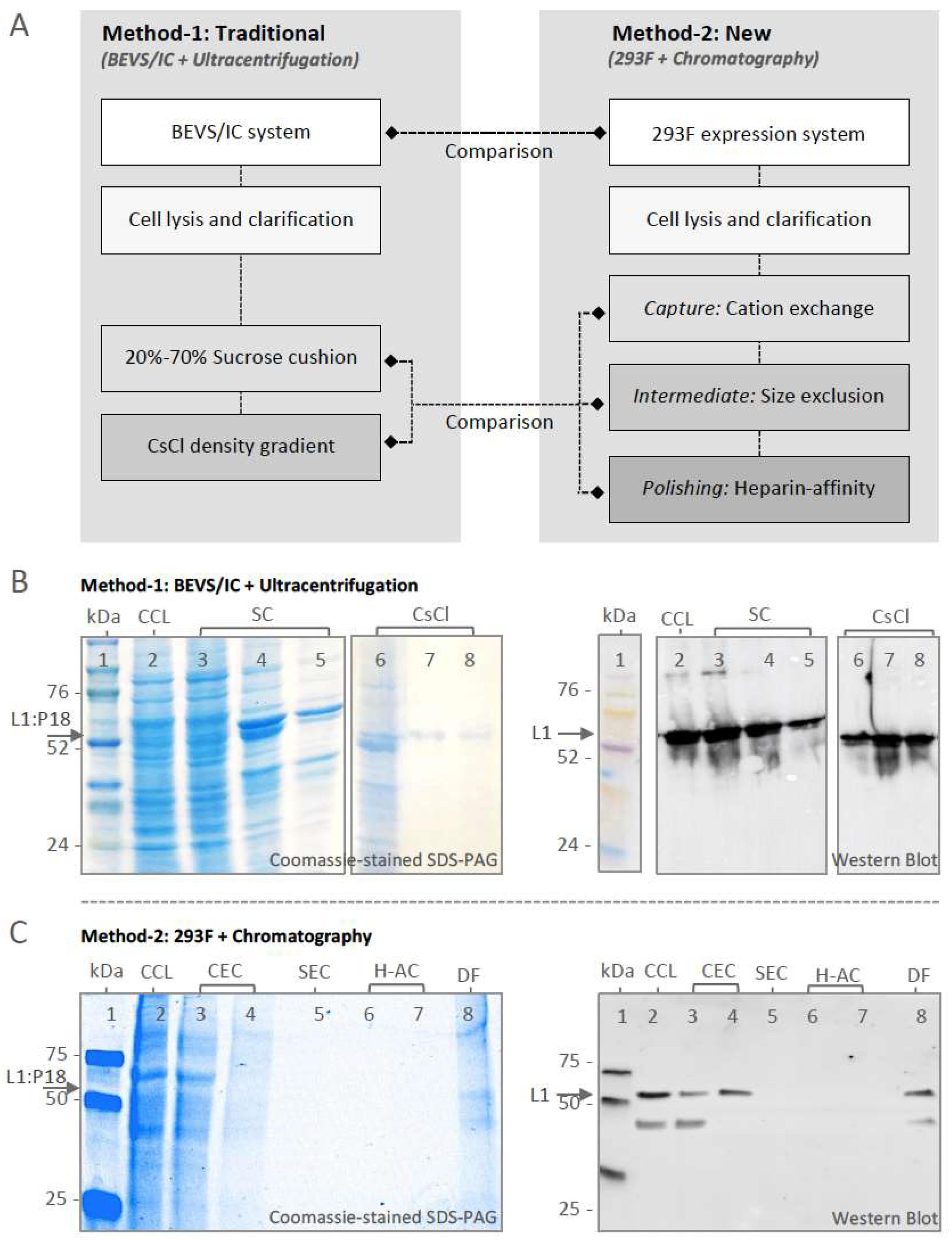
2.2. Optimization of L1:P18I10 VLP Purification Using Ultracentrifugal or Chromatographic Methods
2.2.1. Optimization of Ultracentrifugal Purification Methods by Using BEVS/IC-Derived L1:P18I10 VLP
2.2.2. Optimization of Chromatographic Purification Methods by Using 293F-Derived L1:P18I10 VLP
2.3. Comparison of L1:P18I10 VLP Purification Using Ultracentrifugation or Chromatography
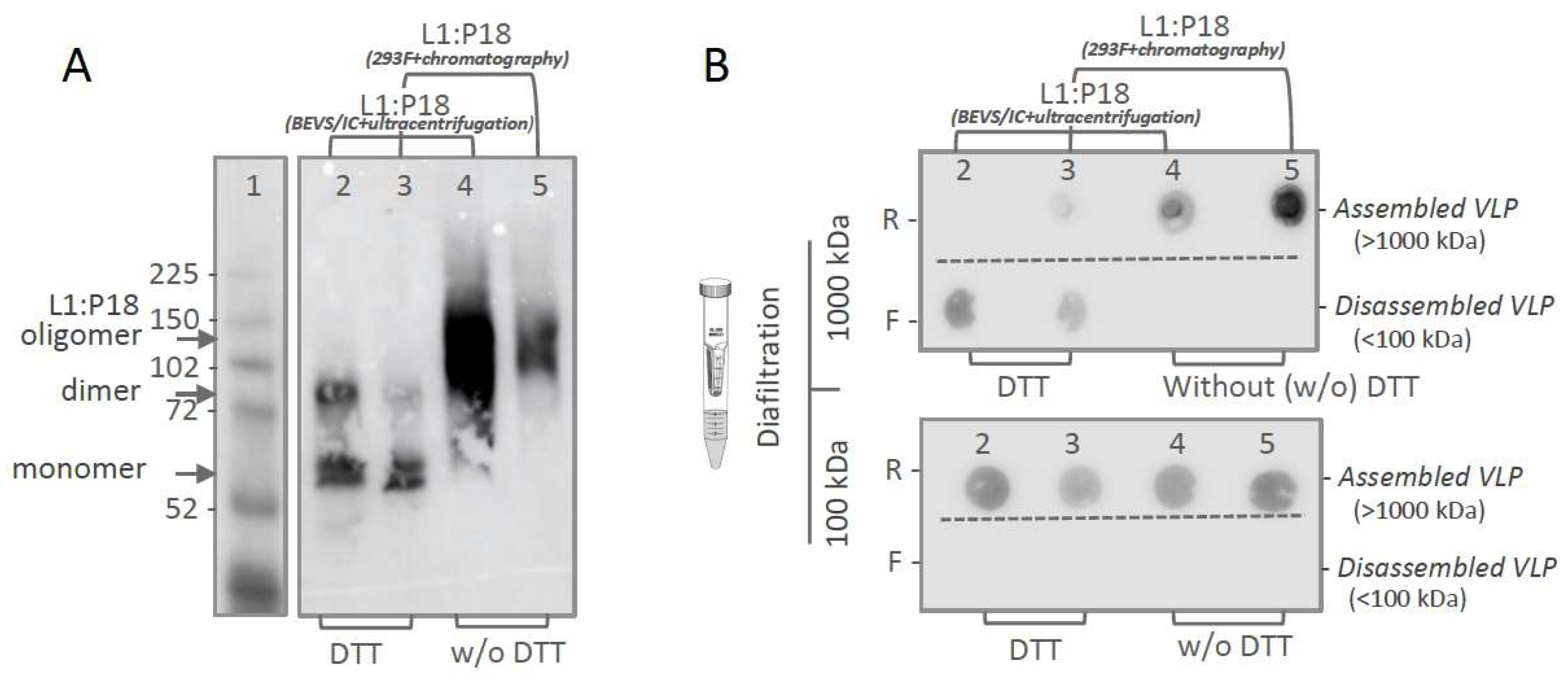
2.4. In Vitro Stability and Self-Assembly of Ultracentrifugation- and Chromatography-Purified L1:P18I10 VLPs
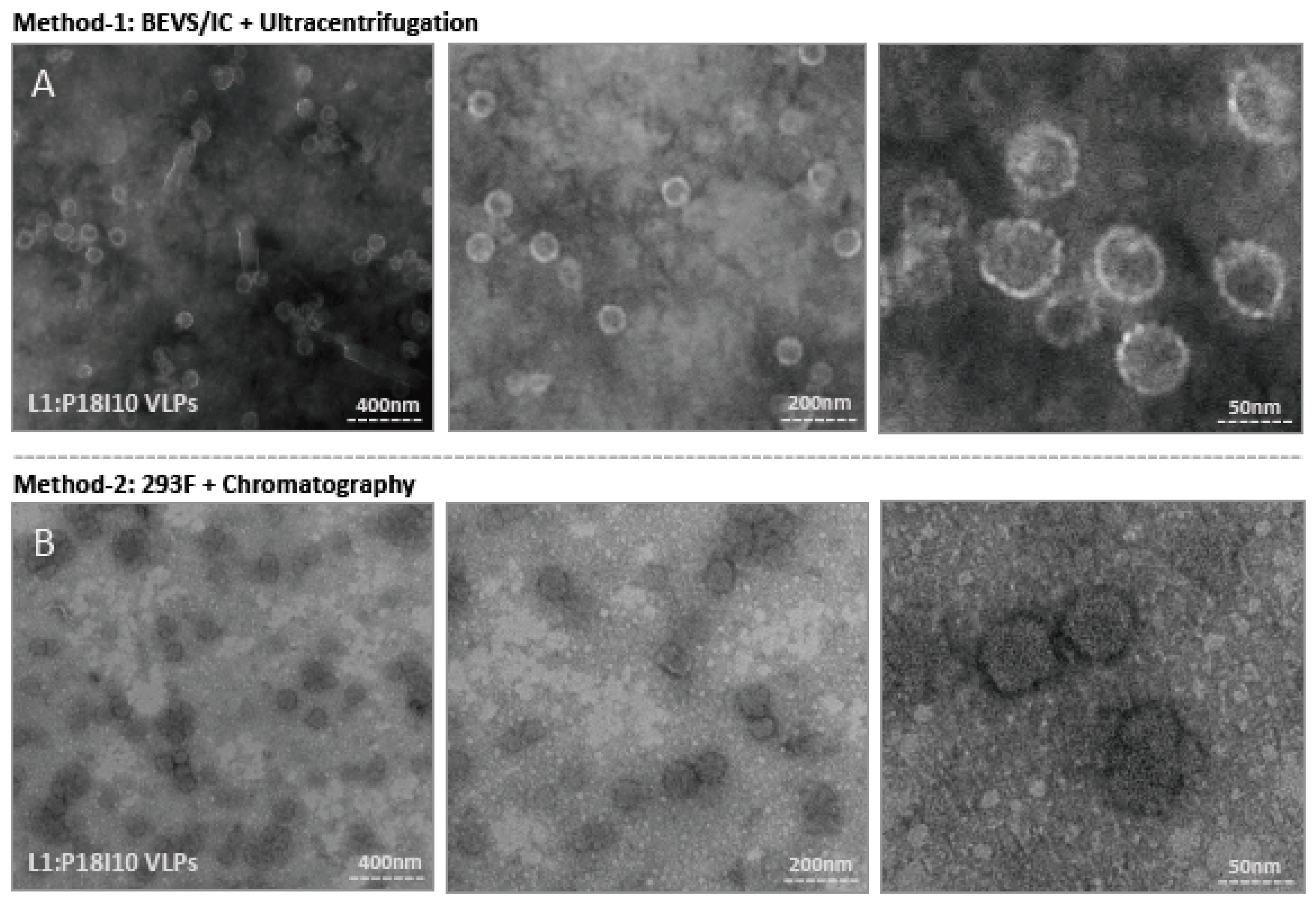
2.5. Morphological Characterization of L1:P18I10 VLPs Purified by Using Ultracentrifugal or Chromatographic Methods
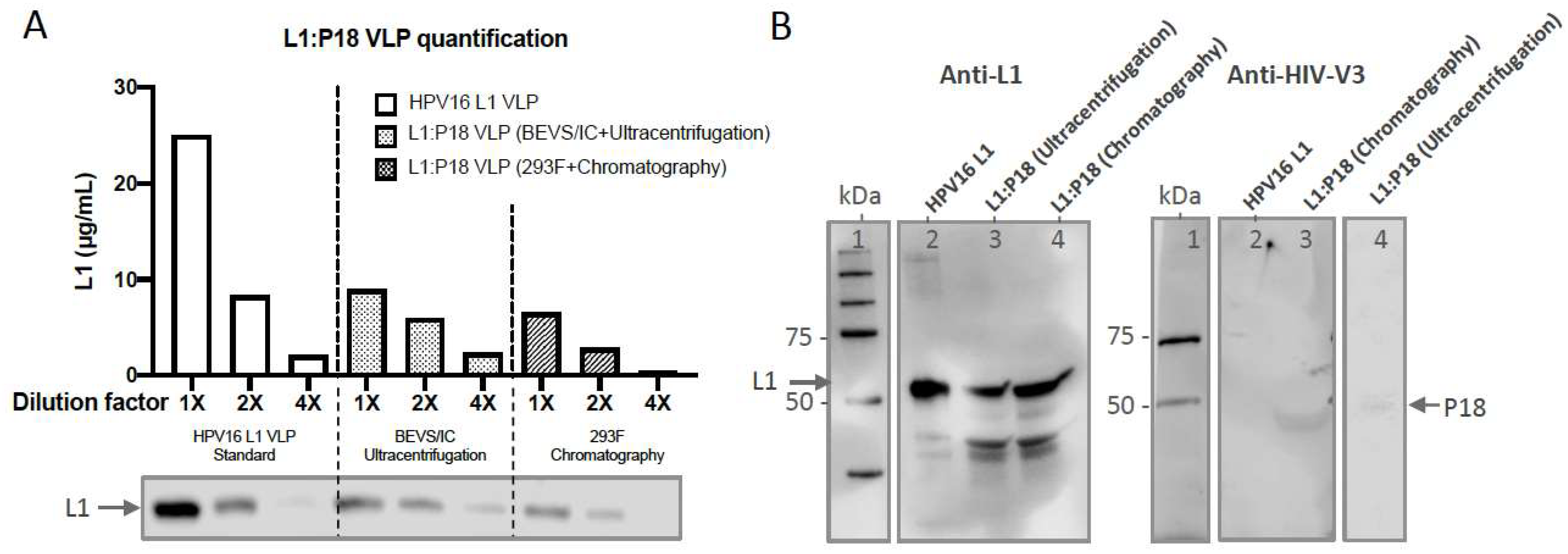
2.6. Quantification and Epitope-Characterization of Ultracentrifugation- and Chromatography- Purified L1:P18I10 VLPs
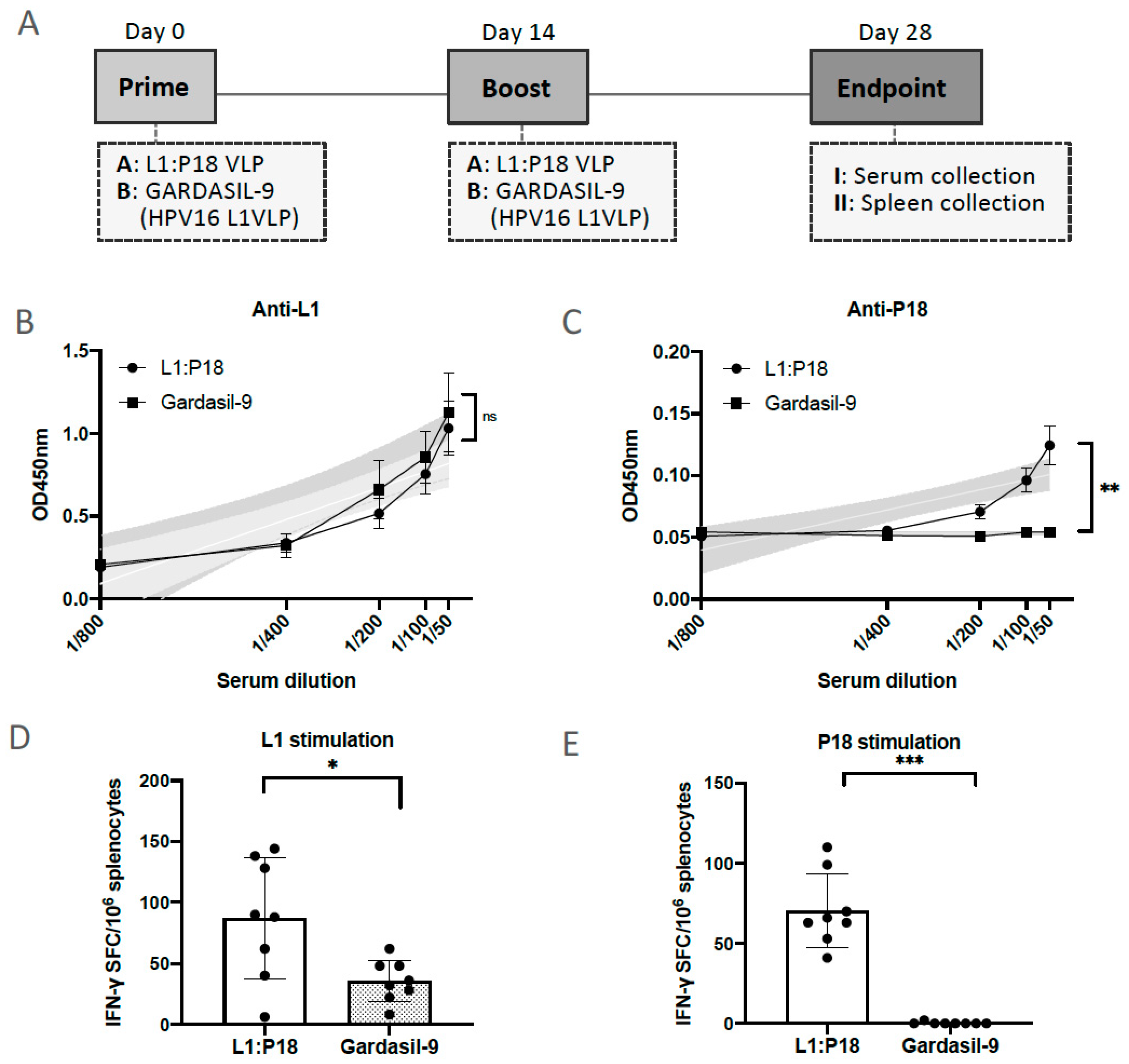
2.7. Evaluation of HPV16- and HIV-1-Specific Humoral and Cellular Immune Responses Induced by 293F Cell-Derived and Chromatographic-Purified L1:P18I10 VLPs
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Cell Culture
4.2. Production of L1:P18I10 Proteins by Using BEVS/IC System
4.3. Production of L1:P18I10 Proteins by Using 293F Expression System
4.4. Immunofluorescence Staining
4.5. Cell Lysis and Clarification
4.6. Sucrose Cushion (SC)
4.7. Cesium Chloride (CsCl) Density Gradient
4.8. Cation Exchange Chromatography (CEC)
4.9. Size Exclusion Chromatography (SEC)
4.10. Heparin-Affinity Chromatography (H-AC)
4.11. Non-Reducing SDS-PAGE
4.12. Molecular Mass Analysis
4.13. Negative Staining and Transmission Electron Microscope
4.14. Quantification of L1:P18I10 VLPs and Host Cellular Proteins
4.15. Western Blotting Analysis
4.16. Immunization of Mice and Sample Collection
4.17. Enzyme-Linked Immunosorbent Assay (ELISA)
4.18. Mouse IFN-γ Enzyme-Linked Immunosorbent Spot Assay (ELISpot)
4.19. Statistical Analysis
4.20. Mice and Ethics Statements
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- UNAIDS Global HIV & AIDS Statistics—2018 Fact Sheet. 2019. Available online: http://www.unaids.org/en/resources/fact-sheet (accessed on 15 June 2020).
- Wang, Q.; Zhang, L. Broadly neutralizing antibodies and vaccine design against HIV-1 infection. Front. Med. 2020, 14, 30–42. [Google Scholar] [CrossRef]
- Streeck, H.; D’souza, M.P.; Littman, D.R.; Crotty, S. Harnessing CD4 + T cell responses in HIV vaccine development. Nat. Med. 2013, 19, 143–149. [Google Scholar] [CrossRef]
- Collins, D.R.; Gaiha, G.D.; Walker, B.D. CD8+ T cells in HIV control, cure and prevention. Nat. Rev. Immunol. 2020, 20, 471–482. [Google Scholar] [CrossRef]
- Rerks-Ngarm, S.; Pitisuttithum, P.; Nitayaphan, S.; Kaewkungwal, J.; Chiu, J.; Paris, R.; Premsri, N.; Namwat, C.; De Souza, M.; Adams, E.; et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 2009, 361, 2209–2220. [Google Scholar] [CrossRef]
- Ng’uni, T.; Chasara, C.; Ndhlovu, Z.M. Major scientific hurdles in HIV vaccine development: Historical perspective and future directions. Front. Immunol. 2020, 11, 590780. [Google Scholar] [CrossRef] [PubMed]
- Robinson, H.L. HIV/AIDS vaccines: 2018. Clin. Pharmacol. Ther. 2018, 104, 1062–1073. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, H.G.; Bensumaidea, S.H.; Alshammari, F.D.; Alenazi, F.S.H.; Almutlaq, B.A.; Alturkstani, M.Z.; Aladani, I.A. Prevalence of human papillomavirus subtypes 16 and 18 among Yemeni patients with cervical cancer. Asian Pacific J. Cancer Prev. 2017, 18, 1543–1548. [Google Scholar] [CrossRef]
- Olcese, V.A.; Chen, Y.; Schlegel, R.; Yuan, H. Characterization of HPV16 L1 loop domains in the formation of a type-specific, conformational epitope. BMC Microbiol. 2004, 4, 29. [Google Scholar] [CrossRef]
- Dupuy, C.; Buzoni-Gate, D.; Touze, A.; Le Cann, P.; Bout, D.; Coursaget, P. Cell mediated immunity induced in mice by HPV 16 L1 virus-like particles. Microb. Pathog. 1997, 22, 219–225. [Google Scholar] [CrossRef]
- Schiller, J.; Lowy, D. Explanations for the high potency of HPV prophylactic vaccines. Vaccine 2018, 36, 4768–4773. [Google Scholar] [CrossRef]
- Markowitz, L.E.; Schiller, J.T. Human Papillomavirus Vaccines. J. Infect. Dis. 2021, 224, S367–S378. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.W.; Saubi, N.; Joseph-Munné, J. Design concepts of virus-like particle-based HIV-1 vaccines. Front. Immunol. 2020, 11, 573157. [Google Scholar] [CrossRef] [PubMed]
- Eto, Y.; Saubi, N.; Ferrer, P.; Joseph, J. Designing chimeric virus-like particle-based vaccines for human papillomavirus and HIV: Lessons learned. AIDS Rev. 2019, 21, 218–232. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.J.; Liu, X.S.; Zhao, K.N.; Leggatt, G.R.; Frazer, I.H. Papillomavirus virus-like particles for the delivery of multiple cytotoxic T cell epitopes. Virology 2000, 273, 374–382. [Google Scholar] [CrossRef]
- Liu, X.S.; Abdul-Jabbar, I.; Qi, Y.M.; Frazer, I.H.; Zhou, J. Mucosal immunisation with papillomavirus virus-like particles elicits systemic and mucosal immunity in mice. Virology 1998, 252, 39–45. [Google Scholar] [CrossRef]
- Peng, S.; Frazer, I.H.; Fernando, G.J.; Zhou, J. Papillomavirus virus-like particles can deliver defined CTL epitopes to the MHC class I pathway. Virology 1998, 240, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Xiao, S.L.; Wen, J.L.; Kong, N.Z.; Yue, H.L.; Leggatt, G.; Frazer, I.H. Route of administration of chimeric BPV1 VLP determines the character of the induced immune responses. Immunol. Cell Biol. 2002, 80, 21–29. [Google Scholar] [CrossRef]
- Zhai, Y.; Zhong, Z.; Zariffard, M.; Spear, G.T.; Qiao, L. Bovine papillomavirus-like particles presenting conserved epitopes from membrane-proximal external region of HIV-1 gp41 induced mucosal and systemic antibodies. Vaccine 2013, 31, 5422–5429. [Google Scholar] [CrossRef]
- Zhang, H.; Huang, Y.; Fayad, R.; Spear, G.T.; Qiao, L. Induction of mucosal and systemic neutralizing antibodies against human immunodeficiency virus type 1 (HIV-1) by oral immunization with bovine papillomavirus-HIV-1 gp41 chimeric virus-like particles. J. Virol. 2004, 78, 8342–8348. [Google Scholar] [CrossRef]
- Chen, C.-W.; Saubi, N.; Kilpeläinen, A.; Joseph-Munné, J. Chimeric Human Papillomavirus-16 Virus-like Particles Presenting P18I10 and T20 Peptides from HIV-1 Envelope Induce HPV16 and HIV-1-Specific Humoral and T Cell-Mediated Immunity in BALB/c Mice. Vaccines 2022, 11, 15. [Google Scholar] [CrossRef]
- Kirnbauer, R.; Taub, J.; Greenstone, H.; Roden, R.; Dürst, M.; Gissmann, L.; Lowy, D.R.; Schiller, J.T. Efficient Self-Assembly of Human Papillomavirus Type 16 LI and L1-L2 into Virus-like Particles. J. Virol. 1993, 67, 6929–6936. [Google Scholar] [CrossRef]
- Zhao, Q.; Potter, C.S.; Carragher, B.; Lander, G.; Sworen, J.; Towne, V.; Abraham, D.; Duncan, P.; Washabaugh, M.W.; Sitrin, R.D. Characterization of virus-like particles in GARDASIL® by cryo transmission electron microscopy. Hum. Vaccines Immunother. 2014, 10, 734–739. [Google Scholar] [CrossRef] [PubMed]
- Le Cann, P.; Coursaget, P.; Iochmann, S.; Touze, A. Self-assembly of human papillomavirus type 16 capsids by expression of the L1 protein in insect cells. FEMS Microbiol. Lett. 1994, 117, 269–274. [Google Scholar] [CrossRef] [PubMed]
- Sasagawa, T.; Pushko, P.; Steers, G.; Gschmeissner, S.E.; Nasser Hajibagheri, M.A.; Finch, J.; Crawford, L.; Tommasino, M. Synthesis and assembly of virus-like particles of human papillomaviruses type 6and Type 16 in fission yeast Schizosaccharomyces pombe. Virology 1995, 206, 126–135. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.N.; Jeong, H.S.; Park, S.N.; Kim, H.J. Purification and immunogenicity study of human papillomavirus type 16 L1 protein in Saccharomyces cerevisiae. J. Virol. Methods 2007, 139, 24–30. [Google Scholar] [CrossRef]
- Kim, H.J.; Kim, S.Y.; Lim, S.J.; Kim, J.Y.; Lee, S.J.; Kim, H.J. One-step chromatographic purification of human papillomavirus type 16 L1 protein from Saccharomyces cerevisiae. Protein Expr. Purif. 2010, 70, 68–74. [Google Scholar] [CrossRef]
- Park, M.A.; Kim, H.J.; Kim, H.J. Optimum conditions for production and purification of human papillomavirus type 16 L1 protein from Saccharomyces cerevisiae. Protein Expr. Purif. 2008, 59, 175–181. [Google Scholar] [CrossRef]
- Zhang, W.; Carmichael, J.; Ferguson, J.; Inglis, S.; Ashrafian, H.; Stanley, M. Expression of human papillomavirus type 16 L1 protein in Escherichia coli: Denaturation, renaturation, and self-assembly of virus-like particles in vitro. Virology 1998, 243, 423–431. [Google Scholar] [CrossRef]
- Schädlich, L.; Senger, T.; Kirschning, C.J.; Müller, M.; Gissmann, L. Refining HPV 16 L1 purification from E. coli: Reducing endotoxin contaminations and their impact on immunogenicity. Vaccine 2009, 27, 1511–1522. [Google Scholar] [CrossRef]
- Chen, X.S.; Casini, G.; Harrison, S.C.; Garcea, R.L. Papillomavirus capsid protein expression in Escherichia coli: Purification and assembly of HPV11 and HPV16 L1. J. Mol. Biol. 2001, 307, 173–182. [Google Scholar] [CrossRef]
- Aires, K.A.; Cianciarullo, A.M.; Carneiro, S.M.; Villa, L.L.; Boccardo, E.; Pérez-Martinez, C.; Perez-Arellano, I.; Oliveira, M.L.S.; Ho, P.L. Production of human papillomavirus type 16 L1 virus-like particles by recombinant Lactobacillus casei cells. Appl. Environ. Microbiol. 2006, 72, 745–752. [Google Scholar] [CrossRef]
- Biemelt, S.; Sonnewald, U.; Galmbacher, P.; Willmitzer, L.; Müller, M. Production of Human Papillomavirus Type 16 Virus-Like Particles in Transgenic Plants. J. Virol. 2003, 77, 9211–9220. [Google Scholar] [CrossRef]
- Zahin, M.; Joh, J.; Khanal, S.; Husk, A.; Mason, H.; Warzecha, H.; Ghim, S.J.; Miller, D.M.; Matoba, N.; Jenson, A.B. Scalable production of HPV16 L1 protein and VLPs from tobacco leaves. PLoS ONE 2016, 11, e0160995. [Google Scholar] [CrossRef]
- Fuenmayor, J.; Gòdia, F.; Cervera, L. Production of virus-like particles for vaccines. N. Biotechnol. 2017, 39, 174–180. [Google Scholar] [CrossRef]
- Vicente, T.; Roldão, A.; Peixoto, C.; Carrondo, M.J.T.; Alves, P.M. Large-scale production and purification of VLP-based vaccines. J. Invertebr. Pathol. 2011, 107, 42–48. [Google Scholar] [CrossRef]
- Monie, A.; Hung, C.F.; Roden, R.; Wu, T.C. CervarixTM:A vaccine for the prevention of HPV 16, 18-associated cervical cancer. Biol. Targets Ther. 2008, 2, 107–113. [Google Scholar] [CrossRef]
- Monograph, P. CERVARIX–Product monograph. Toxicology 2010, 18, 1–55. [Google Scholar]
- Nooraei, S.; Bahrulolum, H.; Hoseini, Z.S.; Katalani, C.; Hajizade, A.; Easton, A.J.; Ahmadian, G. Virus-like particles: Preparation, immunogenicity and their roles as nanovaccines and drug nanocarriers. J. Nanobiotechnology 2021, 19, 59. [Google Scholar] [CrossRef] [PubMed]
- Millipore Sigma. Generic Process of Virus-like Particle (VLP) Based Vaccine Manufacturing; Millipore Sigma: Burlington, MA, USA, 2016; pp. 1–12. [Google Scholar]
- Vlps, A.; Middelberg, A.P.J.; Lua, L.H.L. Virus-like particle bioprocessing: Challenges and opportunities. Pharm. Bioprocess. 2013, 1, 407–409. [Google Scholar]
- Achour, A.; Lemhammedi, S.; Picard, O.; M’bika, J.P.; Zagury, J.F.; Moukrim, Z.; Willer, A.; Beix, F.; Burny, A.; Zagury, D. Cytotoxic T Lymphocytes Specific for HIV-1 gp160 Antigen and Synthetic P18IIIB Peptide in an HLA-A11-Immunized Individual. AIDS Res. Hum. Retrovir. 1994, 10, 19–25. [Google Scholar] [CrossRef]
- Nakagawa, Y.; Kikuchi, H.; Takahashi, H. Molecular analysis of TCR and peptide/MHC interaction using P18-I10-derived peptides with a single D-amino acid substitution. Biophys. J. 2007, 92, 2570–2582. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.S.; Garcea, R.L.; Goldberg, I.; Casini, G.; Harrison, S.C. Structure of Small Virus-like Particles Assembled from the L1 Protein of Human Papillomavirus 16. Mol. Cell 2000, 5, 557–567. [Google Scholar] [CrossRef]
- McLean, C.S.; Churcher, M.J.; Meinke, J.; Smith, G.L.; Higgins, G.; Stanley, M.; Minson, A.C. Production and characterisation of a monoclonal antibody to human papillomavirus type 16 using recombinant vaccinia virus. J. Clin. Pathol. 1990, 43, 488–492. [Google Scholar] [CrossRef] [PubMed]
- Senger, T.; Schädlich, L.; Gissmann, L.; Müller, M. Enhanced papillomavirus-like particle production in insect cells. Virology 2009, 388, 344–353. [Google Scholar] [CrossRef] [PubMed]
- Peyret, H. A protocol for the gentle purification of virus-like particles produced in plants. J. Virol. Methods 2015, 225, 59–63. [Google Scholar] [CrossRef] [PubMed]
- Yazdani, R.; Shams-Bakhsh, M.; Hassani-Mehraban, A.; Arab, S.S.; Thelen, N.; Thiry, M.; Crommen, J.; Fillet, M.; Jacobs, N.; Brans, A.; et al. Production and characterization of virus-like particles of grapevine fanleaf virus presenting L2 epitope of human papillomavirus minor capsid protein. BMC Biotechnol. 2019, 19, 81. [Google Scholar] [CrossRef] [PubMed]
- Park, J.Y.; Pyo, H.M.; Yoon, S.W.; Baek, S.Y.; Park, S.N.; Kim, C.J.; Poo, H. Production and prophylactic efficacy study of human papillomavirus-like particle expressing HPV16 L1 capsid protein. J. Microbiol. 2002, 40, 313–318. [Google Scholar]
- Volpers, C.; Schirmacher, P.; Streeck, R.E.; Sapp, M. Assembly of the Major and the Minor Capsid Protein of Human Papillomavirus Type 33 into Virus-like Particles and Tubular Structures in Insect Cells. Virology 1994, 200, 504–512. [Google Scholar] [CrossRef]
- The use of CaptoTM Core 700 and Capto Q ImpRes in the purification of human papilloma virus like particles. J. Asia’s Pharm. Biopharm. Ind. 2014, 1–4.
- Mukherjee, S.; Thorsteinsson, M.V.; Johnston, L.B.; DePhillips, P.A.; Zlotnick, A. A Quantitative Description of In Vitro Assembly of Human Papillomavirus 16 Virus-Like Particles. J. Mol. Biol. 2008, 381, 229–237. [Google Scholar] [CrossRef]
- McCarthy, M.P.; White, W.I.; Palmer-Hill, F.; Koenig, S.; Suzich, J.A. Quantitative Disassembly and Reassembly of Human Papillomavirus Type 11 Viruslike Particles In Vitro. J. Virol. 1998, 72, 32–41. [Google Scholar] [CrossRef]
- Mistry, N.; Wibom, C.; Evander, M. Cutaneous and mucosal human papillomaviruses differ in net surface charge, potential impact on tropism. Virol. J. 2008, 5, 118. [Google Scholar] [CrossRef]
- Shank-Retzlaff, M.L.; Zhao, Q.; Anderson, C.; Hamm, M.; High, K.; Nguyen, M.; Wang, F.; Wang, N.; Wang, B.; Wang, Y.; et al. Evaluation of the thermal stability of Gardasil®. Hum. Vaccin. 2006, 2, 147–154. [Google Scholar] [CrossRef]
- Eto, Y.; Saubi, N.; Ferrer, P.; Joseph-Munné, J. Expression of chimeric HPV-HIV protein L1P18 in pichia pastoris; purification and characterization of the virus-like particles. Pharmaceutics 2021, 13, 1967. [Google Scholar] [CrossRef]
- Shi, L.; Sanyal, G.; Ni, A.; Luo, Z.; Doshna, S.; Wang, B.; Graham, T.L.; Wang, N.; Volkin, D.B. Stabilization of human papillomavirus virus-like particles by non-ionic surfactants. J. Pharm. Sci. 2005, 94, 1538–1551. [Google Scholar] [CrossRef]
- Carter, J.J.; Wipf, G.C.; Benki, S.F.; Christensen, N.D.; Galloway, D.A.; Al, C.E.T.; Irol, J.V. Identification of a Human Papillomavirus Type 16-Specific Epitope on the C-Terminal Arm of the Major Capsid Protein L1. J. Virol. 2003, 77, 11625–11632. [Google Scholar] [CrossRef]
- Von Brunn, A.; Brand, M.; Reichhuber, C.; Morys-wortmann, C.; Deinhardt, F.; Schdelt, F. Principal neutralizing domain of HIV-I is highly immunogenic when expressed on the surface of hepatitis B core particles. Vaccine 1993, 11, 817–824. [Google Scholar] [CrossRef]
- Shank-Retzlaff, M.; Wang, F.; Morley, T.; Anderson, C.; Hamm, M.; Brown, M.; Rowland, K.; Pancari, G.; Zorman, J.; Lowe, R.; et al. Correlation between mouse potency and in vitro relative potency for human papillomavirus Type 16 virus-like particles and Gardasil vaccine samples. Hum. Vaccin. 2005, 1, 191–197. [Google Scholar] [CrossRef]
- Yang, O.O.; Kalams, S.A.; Trocha, A.; Cao, H.; Luster, A.; Johnson, R.P.; Walker, B.D. Suppression of human immunodeficiency virus type 1 replication by CD8+ cells: Evidence for HLA class I-restricted triggering of cytolytic and noncytolytic mechanisms. J. Virol. 1997, 71, 3120–3128. [Google Scholar] [CrossRef]
- Goulder, P.J.R.; Watkins, D.I. Impact of MHC class I diversity on immune control of immunodeficiency virus replication. Nat. Rev. Immunol. 2008, 8, 619–630. [Google Scholar] [CrossRef]
- Joseph, J.; Saubi, N.; Im, E.J.; Fernández-Lloris, R.; Gil, O.; Cardona, P.J.; Gatell, J.M.; Hanke, T. Newborn mice vaccination with BCG.HIVA222 + MVA.HIVA enhances HIV-1-specific immune responses: Influence of age and immunization routes. Clin. Dev. Immunol. 2011, 2011, 11. [Google Scholar] [CrossRef]
- Saubi, N.; Gea-Mallorquí, E.; Ferrer, P.; Hurtado, C.; Sánchez-Úbeda, S.; Eto, Y.; Gatell, J.M.; Hanke, T.; Joseph, J. Engineering new mycobacterial vaccine design for HIV-TB pediatric vaccine vectored by lysine auxotroph of BCG. Mol. Ther. Methods Clin. Dev. 2014, 1, 14017. [Google Scholar] [CrossRef]
- Mahant, A.; Saubi, N.; Eto, Y.; Guitart, N.; Gatell, J.M.; Hanke, T.; Joseph, J. Preclinical development of BCG.HIVA2auxo.int, harboring an integrative expression vector, for a HIV-TB Pediatric vaccine. Enhancement of stability and specific HIV-1 T-cell immunity. Hum. Vaccines Immunother. 2017, 13, 1798–1810. [Google Scholar] [CrossRef]
- Hanke, T.; McMichael, A.J. Design and construction of an experimental HIV-1 vaccine for a year-2000 clinical trial in Kenya. Nat. Med. 2000, 6, 951–955. [Google Scholar] [CrossRef]
- Clendinen, C.; Zhang, Y.; Warburton, R.N.; Light, D.W. Manufacturing costs of HPV vaccines for developing countries. Vaccine 2016, 34, 5984–5989. [Google Scholar] [CrossRef]
- Deng, F. Advances and challenges in enveloped virus-like particle (VLP)-based vaccines. J. Immunol. Sci. 2018, 2, 36–41. [Google Scholar] [CrossRef]
- Hervas-Stubbs, S.; Rueda, P.; Lopez, L.; Leclerc, C. Insect Baculoviruses Strongly Potentiate Adaptive Immune Responses by Inducing Type I IFN. J. Immunol. 2007, 178, 2361–2369. [Google Scholar] [CrossRef]
- Rueda, P.; Fominaya, J.; Langeveld, J.P.M.; Bruschke, C.; Vela, C.; Casal, J.I. Effect of different baculovirus inactivation procedures on the integrity and immunogenicity of porcine parvovirus-like particles. Vaccine 2000, 19, 726–734. [Google Scholar] [CrossRef]
- Longo, P.A.; Kavran, J.M.; Kim, M.-S.; Leahy, D.J. Transient Mammalian Cell Transfection with Polyethylenimine (PEI). In Methods Enzymol; Academic Press: Cambridge, MA, USA, 2013; Volume 529, pp. 227–240. ISBN 6176321972. [Google Scholar]
- Durocher, Y.; Perret, S.; Kamen, A. High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res. 2002, 30, E9. [Google Scholar] [CrossRef]
- Fang, X.T.; Sehlin, D.; Lannfelt, L.; Syvänen, S.; Hultqvist, G. Efficient and inexpensive transient expression of multispecific multivalent antibodies in Expi293 cells. Biol. Proced. Online 2017, 19, 11. [Google Scholar] [CrossRef]
- Deng, R.; Yue, Y.; Jin, F.; Chen, Y.; Kung, H.F.; Lin, M.C.M.; Wu, C. Revisit the complexation of PEI and DNA—How to make low cytotoxic and highly efficient PEI gene transfection non-viral vectors with a controllable chain length and structure? J. Control. Release 2009, 140, 40–46. [Google Scholar] [CrossRef]
- Oh, Y.K.; Suh, D.; Kim, J.M.; Choi, H.G.; Shin, K.; Ko, J.J. Polyethylenimine-mediated cellular uptake, nucleus trafficking and expression of cytokine plasmid DNA. Gene Ther. 2002, 9, 1627–1632. [Google Scholar] [CrossRef]
- Han, X.; Fang, Q.; Yao, F.; Wang, X.; Wang, J.; Yang, S.; Shen, B.Q. The heterogeneous nature of polyethylenimine-DNA complex formation affects transient gene expression. Cytotechnology 2009, 60, 63–75. [Google Scholar] [CrossRef]
- Zhou, J.; Doorbar, J.; Sun, X.Y.; Crawford, L.V.; McLean, C.S.; Frazer, I.H. Identification of the nuclear localization signal of human papillomavirus type 16 L1 protein. Virology 1991, 185, 625–632. [Google Scholar] [CrossRef]
- Day, P.M.; Weisberg, A.S.; Thompson, C.D.; Hughes, M.M.; Pang, Y.Y.; Lowy, D.R.; Schiller, J.T. Human Papillomavirus 16 Capsids Mediate Nuclear Entry during Infection. J. Virol. 2019, 93, e00454-19. [Google Scholar] [CrossRef]
- Lipin, D.I.; Chuan, Y.P.; Lua, L.H.L.; Middelberg, A.P.J. Encapsulation of DNA and non-viral protein changes the structure of murine polyomavirus virus-like particles. Arch. Virol. 2008, 153, 2027–2039. [Google Scholar] [CrossRef]
- Huhti, L.; Blazevic, V.; Nurminen, K.; Koho, T.; Hytönen, V.P.; Vesikari, T. A comparison of methods for purification and concentration of norovirus GII-4 capsid virus-like particles. Arch. Virol. 2010, 155, 1855–1858. [Google Scholar] [CrossRef]
- Sarubbi, E. Protein batches from downstream processing studies can be cost-effective tools for fast and specific protein quantification assays. Anal. Biochem. 2005, 345, 167–169. [Google Scholar] [CrossRef]
- Cook, J.C.; Joyce, J.G.; George, H.A.; Schultz, L.D.; Hurni, W.M.; Jansen, K.U.; Hepler, R.W.; Ip, C.; Lowe, R.S.; Keller, P.M.; et al. Purification of virus-like particles of recombinant human papillomavirus type 11 major capsid protein L1 from Saccharomyces cerevisiae. Protein Expr. Purif. 1999, 17, 477–484. [Google Scholar] [CrossRef]
- Rommel, O.; Dillner, J.; Fligge, C.; Bergsdorf, C.; Wang, X.; Seiinka, H.C.; Sapp, M. Heparan sulfate proteoglycans interact exclusively with conformationally intact HPV L1 assemblies: Basis for a virus-like particle ELISA. J. Med. Virol. 2005, 75, 114–121. [Google Scholar] [CrossRef]
- Merck Canada Inc. Product Monograph Gardasil® Product Monograph Monopril * Product Monograph; Merck Canada Inc.: Kirkland, QC, Canada, 2015; pp. 1–71. [Google Scholar]
| Method | Purification Stages | Total HCP (μg) a | HCP Removal (%) | Total L1 (μg) b | Recovery (%) | Purity (%) d |
|---|---|---|---|---|---|---|
| 1 Traditional | BEVS/IC CCL | 2637.4 | - | 109.4 | 100 | 4 |
| Ultracentrifugation | ||||||
| Step-1: SC | 264.5 | 90 | 16.6 | 15 | 6 | |
| Step-2: CsCl | 12.0 | 99 | 11.9 | 11 | 99 | |
| 2 New | 293F cell expression CCL | 1848.5 | - | 33.3 | 100 | 2 |
| Chromatography | ||||||
| Step-1: CEC | 649.2 | 65 | 21.5 | 65 | 3 | |
| Step-2: SEC | 193.9 | 90 | 19.4 c | 58 | 10 | |
| Step-3: H-AC | 24.7 | 98 | 18.8 c | 56 | 76 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, C.-W.; Saubi, N.; Joseph-Munné, J. Chimeric Human Papillomavirus-16 Virus-like Particles Presenting HIV-1 P18I10 Peptide: Expression, Purification, Bio-Physical Properties and Immunogenicity in BALB/c Mice. Int. J. Mol. Sci. 2023, 24, 8060. https://doi.org/10.3390/ijms24098060
Chen C-W, Saubi N, Joseph-Munné J. Chimeric Human Papillomavirus-16 Virus-like Particles Presenting HIV-1 P18I10 Peptide: Expression, Purification, Bio-Physical Properties and Immunogenicity in BALB/c Mice. International Journal of Molecular Sciences. 2023; 24(9):8060. https://doi.org/10.3390/ijms24098060
Chicago/Turabian StyleChen, Chun-Wei, Narcís Saubi, and Joan Joseph-Munné. 2023. "Chimeric Human Papillomavirus-16 Virus-like Particles Presenting HIV-1 P18I10 Peptide: Expression, Purification, Bio-Physical Properties and Immunogenicity in BALB/c Mice" International Journal of Molecular Sciences 24, no. 9: 8060. https://doi.org/10.3390/ijms24098060
APA StyleChen, C.-W., Saubi, N., & Joseph-Munné, J. (2023). Chimeric Human Papillomavirus-16 Virus-like Particles Presenting HIV-1 P18I10 Peptide: Expression, Purification, Bio-Physical Properties and Immunogenicity in BALB/c Mice. International Journal of Molecular Sciences, 24(9), 8060. https://doi.org/10.3390/ijms24098060





