Study on the Flower Induction Mechanism of Hydrangea macrophylla
Abstract
1. Introduction
2. Results
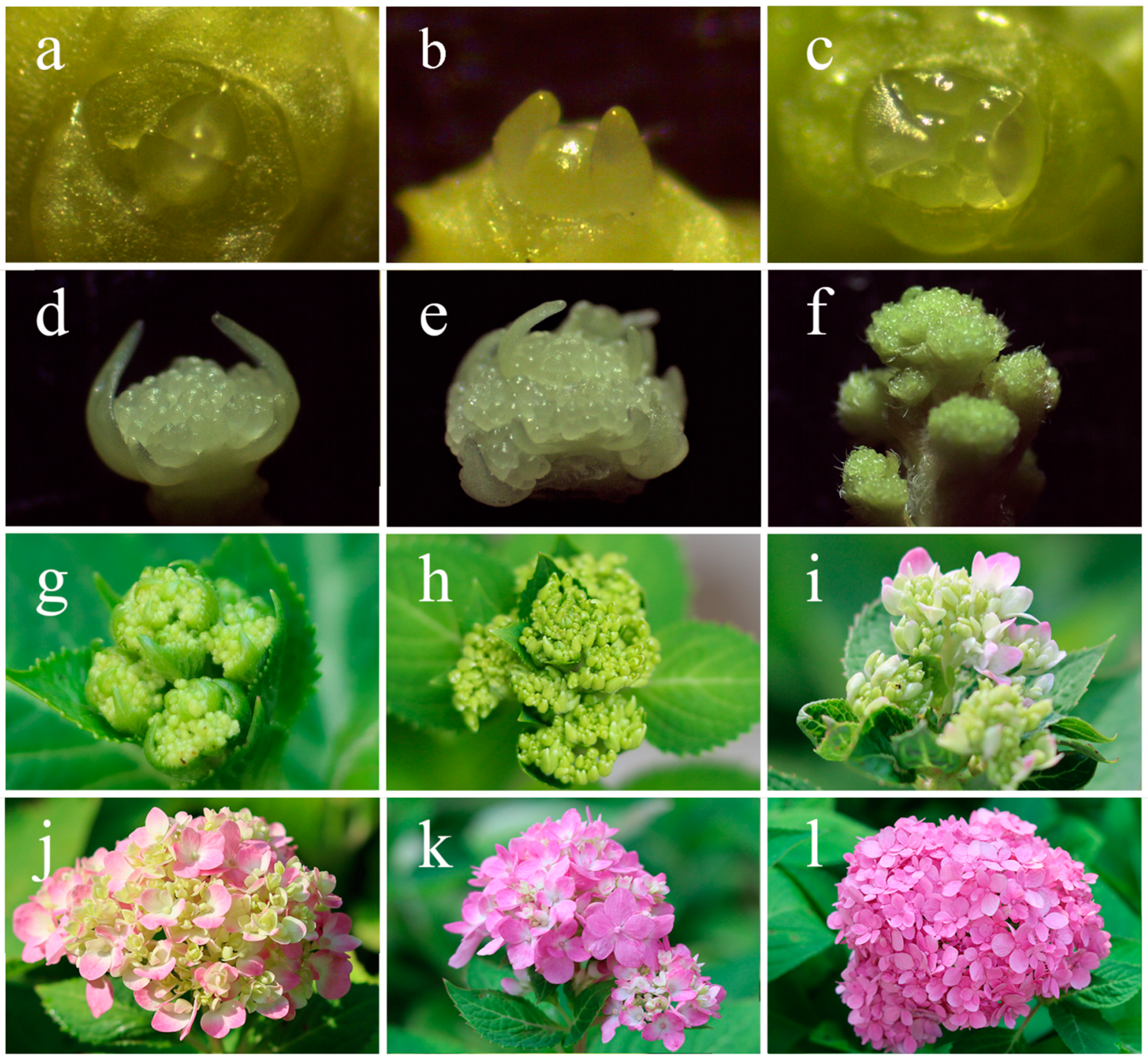
2.1. Phenotypic Observations during Flowering
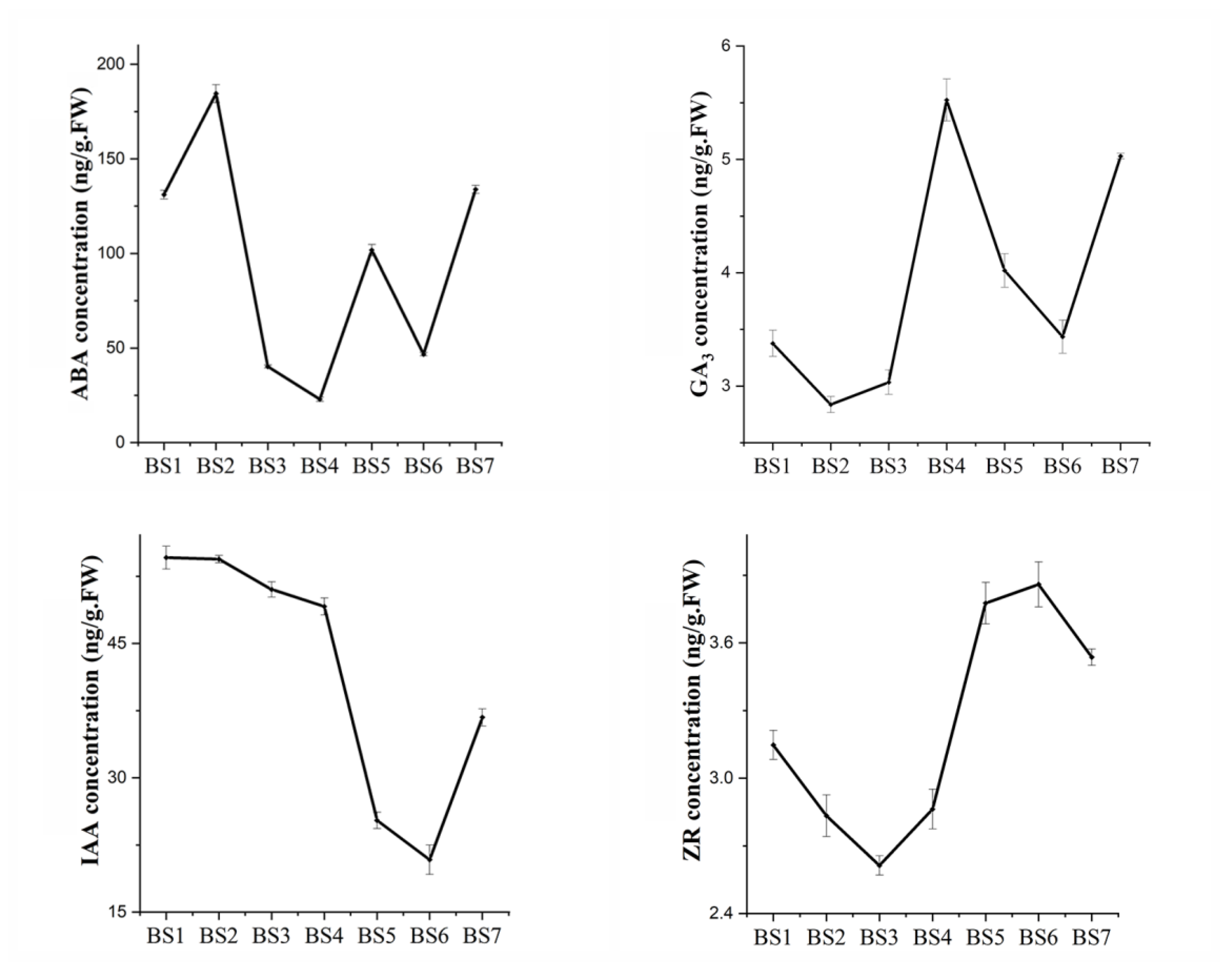
2.2. Endogenous Hormone Response during Flowering
2.3. Quality Assessment of Transcriptome Sequencing
2.4. Analysis of KEGG Metabolic Pathways and GO Functional Classifications
2.5. Expression Profiles of Sugar-Related Genes during Flower Development
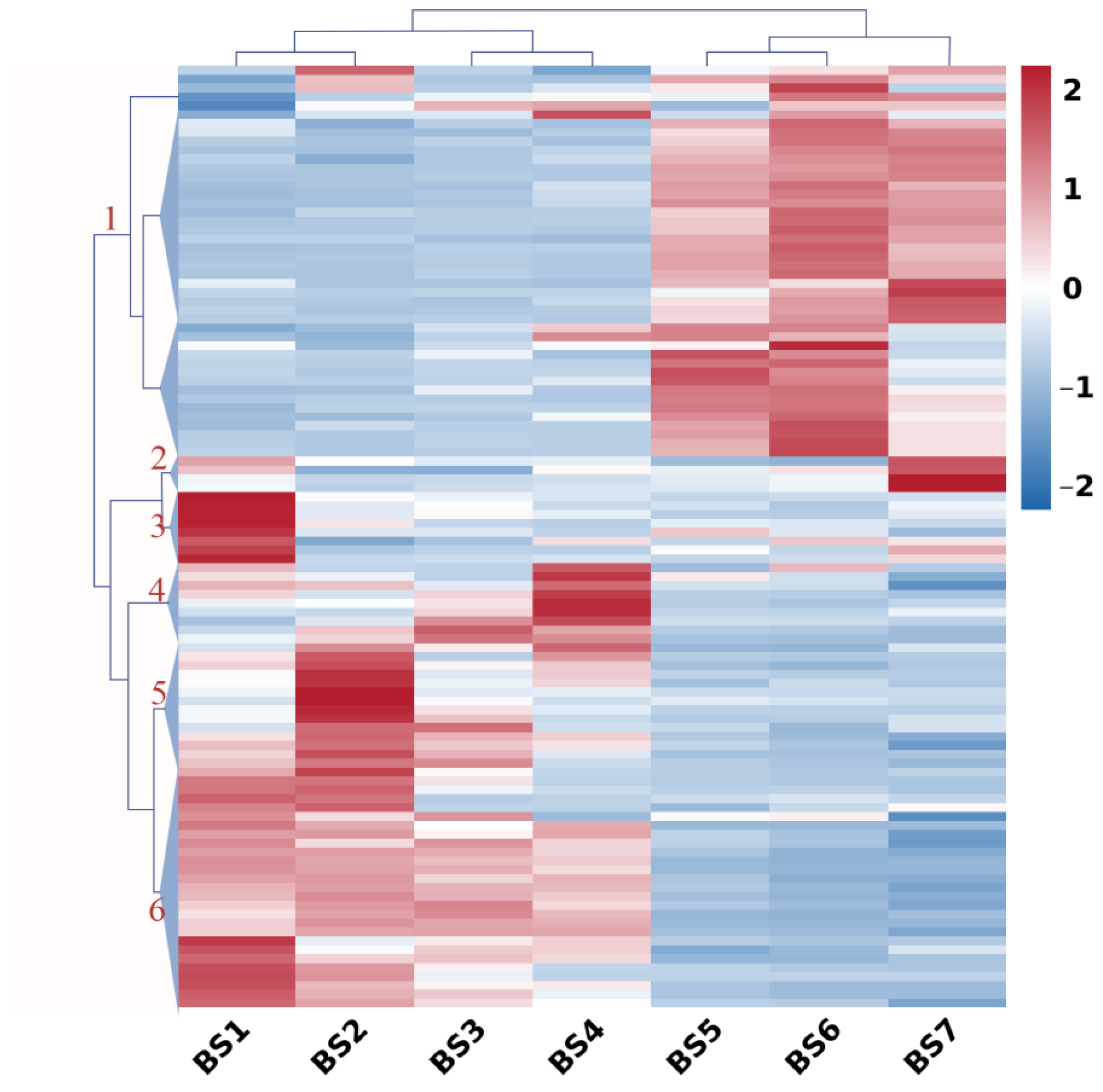
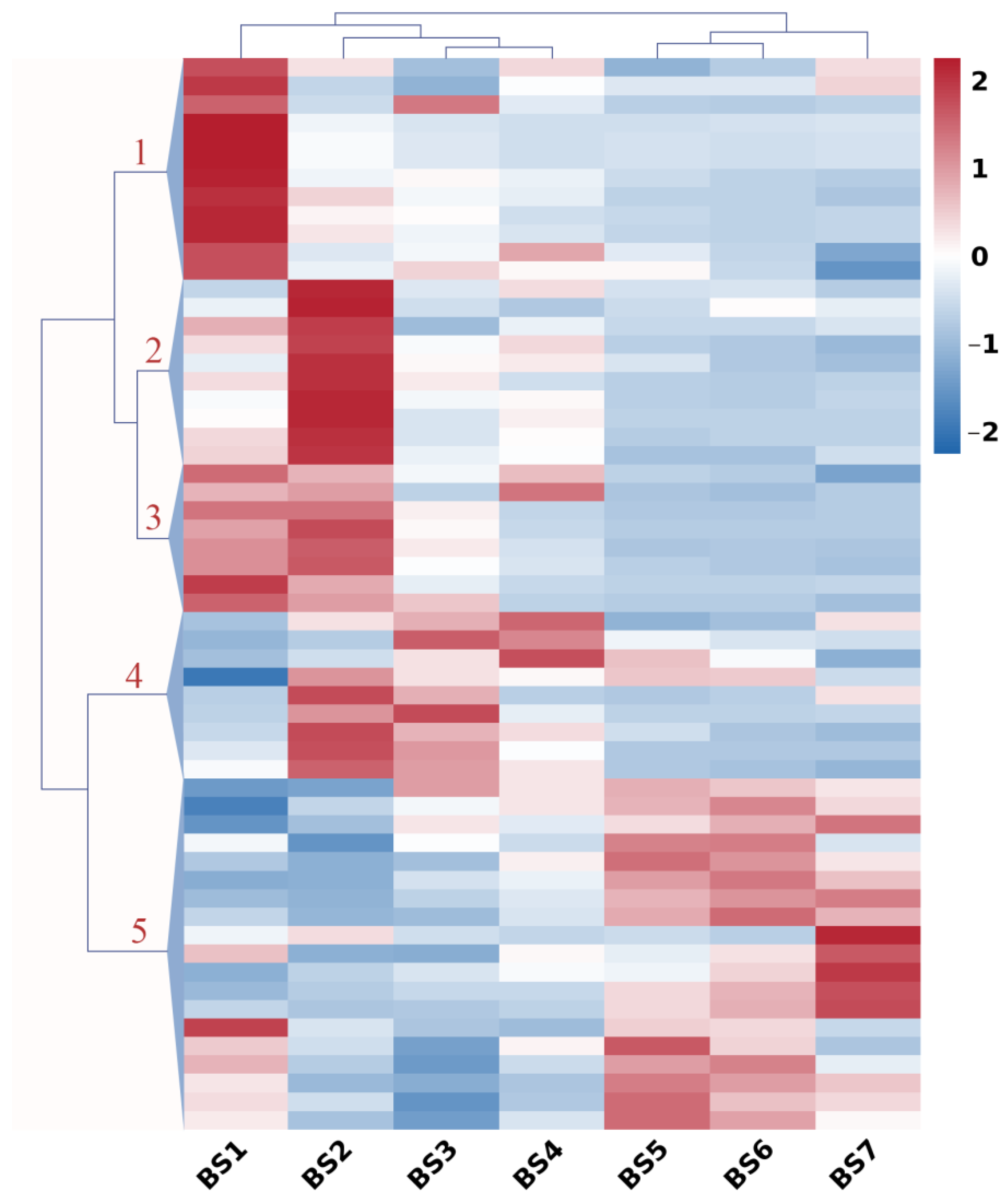
2.6. Expression Profiles of Hormone-Regulatory Genes during Flower Development
2.7. Expression Profiles of Flowering Genes during Flower Development
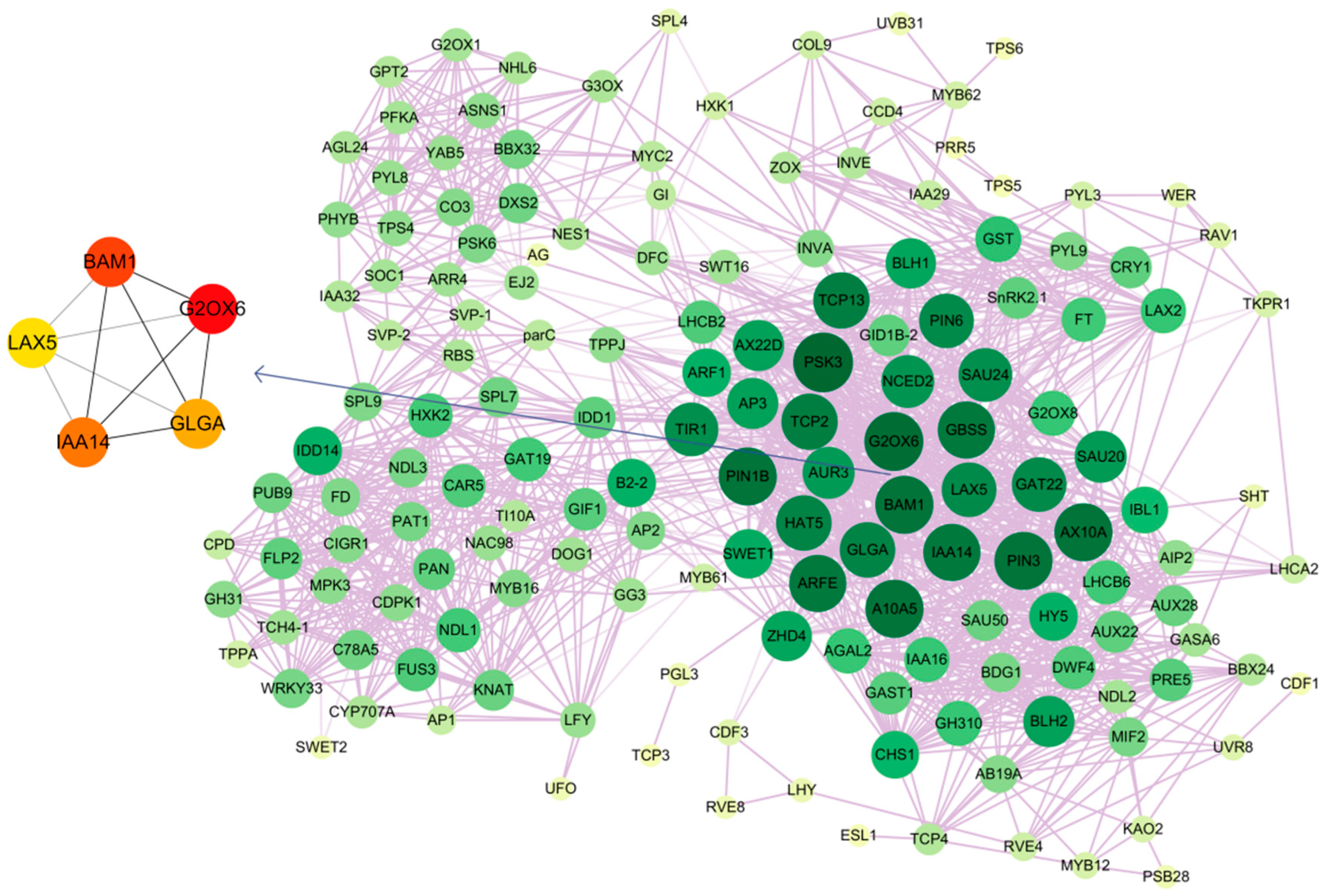
2.8. Co-Expression Pattern Analysis
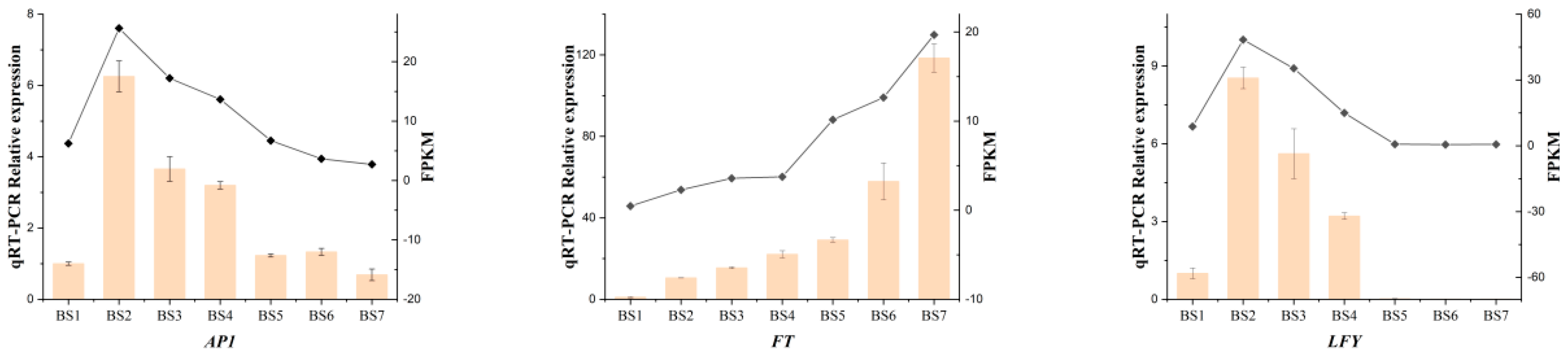
2.9. Real-Time Quantitative PCR Verification of Differentially Expressed Genes
3. Discussion
3.1. Sugar Signaling Mediates Flower Induction in H. macrophylla “Endless Summer”
3.2. Hormone Signal-Mediated Flower Induction in H. macrophylla “Endless Summer”
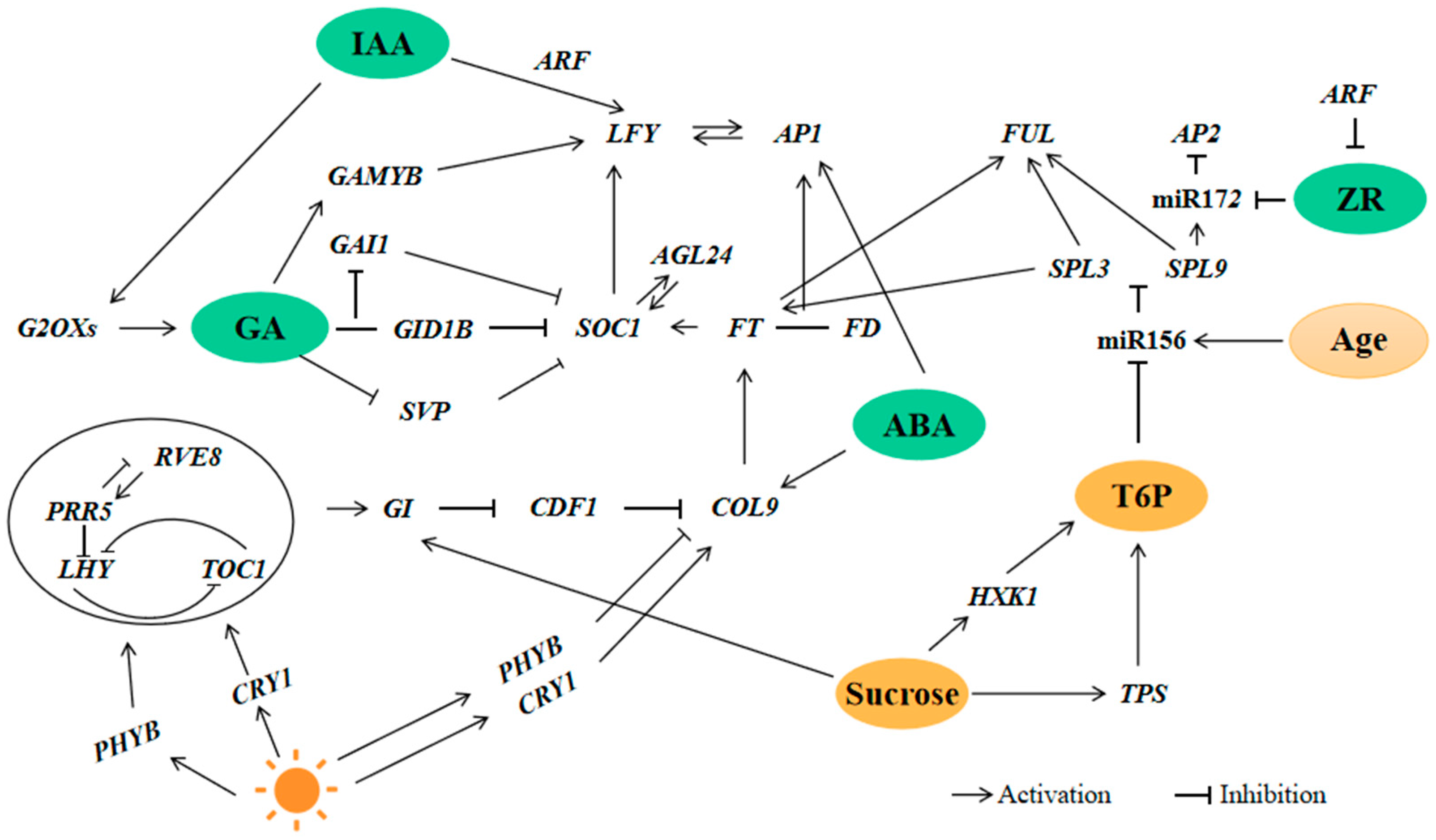
3.3. Flower Induction Pathway in H. macrophylla “Endless Summer”
4. Materials and Methods
4.1. Plant Materials
4.2. Measurement of Hormone Content during Flower Development
4.3. Transcriptome Sequencing
4.4. Functional Annotation of Unigenes and Screening for Differentially Expressed Genes
4.5. Real-Time Quantitative Polymerase Chain Reaction
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhou, Q.; Zhang, S.; Bao, M.; Liu, G. Advances on molecular mechanism of floral initiation in higher plants. Mol. Plant Breed. 2018, 16, 3681–3692. [Google Scholar]
- Shu, H.; Hao, Y.; Cai, Q.; Wang, Z.; Zhu, G.; Chen, S.; Zhou, Y.; Wang, Z. Recent research progress on the molecular regulation of flowering time in Arabidopsis thaliana. Plant Sci. J. 2017, 35, 603–608. [Google Scholar]
- Xing, L.B.; Zhang, D.; Li, Y.M.; Shen, Y.W.; Zhao, C.P.; Ma, J.J.; An, N.; Han, M.Y. Transcription Profiles Reveal Sugar and Hormone Signaling Pathways Mediating Flower Induction in Apple (Malus domestica Borkh.). Plant Cell Physiol. 2015, 56, 2052–2068. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Sun, X.D.; Ni, M. Timing of photoperiodic flowering: Light perception and circadian clock. J. Integr. Plant Biol. 2007, 49, 28–34. [Google Scholar] [CrossRef]
- Rawat, R.; Takahashi, N.; Hsu, P.Y.; Jones, M.A.; Schwartz, J.; Salemi, M.R.; Phinney, B.S.; Harmer, S.L. REVEILLE8 and PSEUDO-REPONSE REGULATOR5 Form a Negative Feedback Loop within the Arabidopsis Circadian Clock. PloS Genet. 2011, 7, e1001350. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Li, Y.H.; Li, Y.; Lu, H.; Hong, H.; Tian, Y.; Li, H.; Zhao, T.; Zhou, X.; Liu, J.; et al. A Domestication-Associated Gene GmPRR3b Regulates the Circadian Clock and Flowering Time in Soybean. Mol. Plants Engl. Version 2020, 13, 15. [Google Scholar] [CrossRef]
- Ren, Y.; Gao, Y.; Gao, S.; Yuan, L.; Wang, X.; Zhang, Q. Genetic characteristics of circadian flowering rhythm in Hemerocallis. Sci. Hortic. 2019, 250, 19–26. [Google Scholar] [CrossRef]
- Yamane, H.; Kashiwa, Y.; Ooka, T.; Tao, R.; Yonemori, K. Suppression Subtractive Hybridization and Differential Screening Reveals Endodormancy-associated Expression of an SVP/AGL24-type MADS-box Gene in Lateral Vegetative Buds of Japanese Apricot. J. Am. Soc. Hortic. Sci. 2008, 133, 708–716. [Google Scholar] [CrossRef]
- Fukazawa, J.; Ohashi, Y.; Takahashi, R.; Nakai, K.; Takahashi, Y. DELLA degradation by gibberellin promotes flowering via GAF1-TPR-dependent repression of floral repressors in Arabidopsis. Plant Cell 2021, 7, 2258–2272. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Park, M.Y.; Conway, S.R.; Wang, J.W.; Weigel, D.; Poethig, R.S. The Sequential Action of miR156 and miR172 Regulates Developmental Timing in Arabidopsis. Cell 2009, 138, 750–759. [Google Scholar] [CrossRef]
- Gou, J.; Tang, C.; Chen, N.; Wang, H.; Debnath, S.; Sun, L.; Flanagan, A.; Tang, Y.; Jiang, Q.; Allen, R.D.; et al. SPL7 and SPL8 represent a novel flowering regulation mechanism in switchgrass. New Phytol. 2019, 222, 1610–1623. [Google Scholar] [CrossRef]
- Jiang, Y.; Peng, J.; Wang, M.; Su, W.; Gan, X.; Jing, Y.; Yang, X.; Lin, S.; Gao, Y. The Role of EjSPL3, EjSPL4, EjSPL5, and EjSPL9 in Regulating Flowering in Loquat (Eriobotrya japonica Lindl.). Int. J. Mol. Sci. 2019, 21, 248. [Google Scholar] [CrossRef]
- Tripathi, R.K.; Bregitzer, P.; Singh, J. Genome-wide analysis of the SPL/miR156 module and its interaction with the AP2/miR172 unit in barley. Sci. Rep. 2018, 8, 7085. [Google Scholar] [CrossRef]
- Moon, J.; Lee, H.; Kim, M.; Lee, I. Analysis of flowering pathway integrators in Arabidopsis. Plant Cell Physiol. 2005, 46, 292–299. [Google Scholar] [CrossRef]
- Wei, Q.; Ma, C.; Xu, Y.; Wang, T.; Chen, Y.; Lü, J.; Zhang, L.; Jiang, C.-Z.; Hong, B.; Gao, J. Control of chrysanthemum flowering through integration with an aging pathway. Nat. Commun. 2017, 8, 829. [Google Scholar] [CrossRef]
- Zou, L.; Pan, C.; Wang, M.X.; Cui, L.; Han, B.Y. Progress on the mechanism of hormones regulating plant flower formation. Yi Chuan = Hered. 2020, 42, 739–751. [Google Scholar]
- Lu, J.C. Comparative transcription profiles reveal that carbohydrates and hormone signalling pathways mediate flower induction in Juglans sigillata after girdling. Ind. Crops Prod. 2020, 153, 112556. [Google Scholar] [CrossRef]
- Matsoukas, I.G.; Massiah, A.J.; Thomas, B. Starch metabolism and antiflorigenic signals modulate the juvenile-to-adult phase transition in Arabidopsis. Plant Cell Environ. 2013, 36, 1802–1811. [Google Scholar] [CrossRef]
- Liu, Y.; Bai, Y.; Li, N.; Li, M.; Liu, W.; Yun, D.J.; Liu, B.; Xu, Z.Y. HEXOKINASE1 forms a nuclear complex with the PRC2 subunits CURLY LEAF and SWINGER to regulate glucose signaling. J. Bot. Engl. Ed. 2022, 64, 13. [Google Scholar] [CrossRef] [PubMed]
- Seo, P.J.; Ryu, J.; Kang, S.K.; Park, C.M. Modulation of sugar metabolism by an INDETERMINATE DOMAIN transcription factor contributes to photoperiodic flowering in Arabidopsis. Plant J. Cell Mol. Biol. 2011, 65, 418–429. [Google Scholar] [CrossRef] [PubMed]
- Wahl, V.; Ponnu, J.; Schlereth, A.; Arrivault, S.; Langenecker, T.; Franke, A.; Feil, R.; Lunn, J.E.; Lunn, M.; Schmid, M. Regulation of Flowering by Trehalose-6-Phosphate Signaling in Arabidopsis thaliana. Science 2013, 339, 704–707. [Google Scholar] [CrossRef]
- Paul, M.; Paul, M.J. Trehalose 6-phosphate: A signal of sucrose status. Biochem. J. 2008, 412, e1. [Google Scholar] [CrossRef]
- Wingler, A.; Henriques, R. Sugars and the speed of life—Metabolic signals that determine plant growth, development and death. Physiol Plant 2022, 174, e13656. [Google Scholar]
- Zhou, Q.; Shi, J.; Li, Z.; Zhang, S.; Zhang, S.; Zhang, J.; Bao, M.; Liu, G. miR156/157 targets SPLs to regulate flowering transition, plant architecture and flower organ size in petunia. Plant Cell Physiol. 2021, 62, 839–857. [Google Scholar] [CrossRef]
- Pallardy, S.G. Plant Hormones and Other Signaling Molecules—ScienceDirect. In Physiology of Woody Plants, 3rd ed.; Elsevier: Amsterdam, The Netherlands, 2008; pp. 367–377. [Google Scholar]
- Zheng, C.; Yang, Y.; Luo, X.; Liu, W.; Yang, W.; Shu, K. Current understanding of the roles of phytohormone abscisic acid in the regulation of plant root growth. Plant Sci. J. 2019, 37, 690–698. [Google Scholar]
- Riboni, M.; Robustelli Test, A.; Galbiati, M.; Tonelli, C.; Conti, L. ABA-dependent control of GIGANTEA signalling enables drought escape via up-regulation of FLOWERING LOCUS T in Arabidopsis thaliana. J. Exp. Bot. 2016, 67, 6309–6322. [Google Scholar] [CrossRef]
- Cui, Z.; Zhou, B.; Zhang, Z.; Hu, Z. Abscisic acid promotes flowering and enhances LcAP1 expression in Litchi chinensis Sonn. S. Afr. J. Bot. 2013, 88, 76–79. [Google Scholar] [CrossRef]
- Chickarmane, V.S.; Gordon, S.P.; Tarr, P.T.; Heisler, M.G.; Meyerowitz, E.M. Cytokinin signaling as a positional cue for patterning the apical–basal axis of the growing Arabidopsis shoot meristem. Proc. Natl. Acad. Sci. USA 2012, 109, 4002–4007. [Google Scholar] [CrossRef]
- Aukerman, M.J.; Sakai, H. Regulation of Flowering Time and Floral Organ Identity by a MicroRNA and Its APETALA2-Like Target Genes. Plant Cell 2003, 15, 2730–2741. [Google Scholar] [CrossRef]
- Zhang, K.; Wang, R.; Zi, H.; Li, Y.; Cao, X.; Li, D.; Guo, L.; Tong, J.; Pan, Y.; Jiao, Y.; et al. AUXIN RESPONSE FACTOR3 Regulates Floral Meristem Determinacy by Repressing Cytokinin Biosynthesis and Signaling. Plant Cell 2018, 30, 324–346. [Google Scholar] [CrossRef]
- Sheng, J.; Li, X.; Zhang, D. Gibberellins, brassinolide, and ethylene signaling were involved in flower differentiation and development in Nelumbo nucifera. Hortic. Plant J. 2022, 8, 243–250. [Google Scholar] [CrossRef]
- Sumitomo, K.; Li, T.; Hisamatsu, T. Gibberellin promotes flowering of chrysanthemum by upregulating CmFL, a chrysanthemum FLORICAULA/LEAFY homologous gene. Plant Sci. 2009, 176, 643–649. [Google Scholar] [CrossRef]
- Krizek, B.A.; Eaddy, M. AINTEGUMENTA-LIKE6 regulates cellular differentiation in flowers. Plant Mol. Biol. 2012, 78, 199–209. [Google Scholar] [CrossRef]
- Okada, K.; Ueda, J.; Komaki, M.K.; Bell, C.J.; Shimura, Y. requirement of the auxin polar transport system in early stages of arabídopsis floral bud formation. Plant CelL 2018, 3, 677–684. [Google Scholar] [CrossRef]
- Yamaguchi, N.; Wu, M.F.; Winter, C.M.; Berns, M.C.; Nole-Wilson, S.; Yamaguchi, A.; Coupland, G.; Krizek, B.A.; Wagner, D. A Molecular Framework for Auxin-Mediated Initiation of Flower Primordia. Dev. Cell 2013, 24, 271–282. [Google Scholar] [CrossRef]
- Frigerio, M.; Alabadí, D.; Pérez-Gómez, J.; García-Cárcel, L.; Phillips, A.L.; Hedden, P.; Blázquez, M.A. Transcriptional regulation of gibberellin metabolism genes by auxin signaling in Arabidopsis. Plant Physiol 2006, 142, 553–563. [Google Scholar] [CrossRef]
- Ó’Maoiléidigh, D.S.; Graciet, E.; Wellmer, F. Gene networks controlling Arabidopsis thaliana flower development. N. Phytol. 2013, 201, 16–30. [Google Scholar] [CrossRef]
- Das, A.; Geetha, G.A.; Ravishankar, K.V.; Shivashankara, K.S.; Roy, T.K.; Dinesh, M.R. Interrelations of growth regulators, carbohydrates and expression of flowering genes (FT, LFY, AP1) in leaf and shoot apex of regular and alternate bearing mango (Mangifera indica L.) cultivars during flowering—ScienceDirect. Sci. Hortic. 2019, 253, 263–269. [Google Scholar] [CrossRef]
- Balcerowicz, M.; Mahjoub, M.; Nguyen, D.; Lan, H.; Stoeckle, D.; Conde, S.; Jaeger, K.E.; Wigge, P.A.; Ezer, D. An early-morning gene network controlled by phytochromes and cryptochromes regulates photomorphogenesis pathways in Arabidopsis. Mol. Plants Engl. Version 2021, 14, 983–996. [Google Scholar] [CrossRef]
- Harmon, F.G.; Chen, J.; Xin, Z. GIGANTEA promotes sorghum flowering by stimulating floral activator gene expression. bioRxiv 2018, 427–492. [Google Scholar] [CrossRef]
- LLi, D.; Zhang, H.; Mou, M.; Chen, Y.; Xiang, S.; Chen, L.; Yu, D. Arabidopsis Class II TCP Transcription Factors Integrate with the FT–FD Module to Control Flowering 1. Plant Physiol. 2019, 181, 97–111. [Google Scholar] [CrossRef] [PubMed]
- Galvão, V.C. Regulation of flowering time by DELLA proteins in Arabidopsis thaliana; Eberhard-Karls-Universitaet Tuebingen: Baden-Württemberg, Germany, 2013. [Google Scholar]
- Woodger, F.J.; Millar, A.; Murray, F.; Jacobsen, J.V.; Gubler, F. The Role of GAMYB Transcription Factors in GA-Regulated Gene Expression. J. Plant Growth Regul. 2003, 22, 176–184. [Google Scholar] [CrossRef]
- Li, D.; Liu, C.; Shen, L.; Wu, Y.; Chen, H.; Robertson, M.; Helliwell, C.A.; Ito, T.; Meyerowitz, E.; Yu, H. A repressor complex governs the integration of flowering signals in Arabidopsis. Dev. Cell 2008, 15, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Roussin-Léveillée, C.; Silva-Martins, G.; Moffett, P. ARGONAUTE5 Represses Age-Dependent Induction of Flowering through Physical and Functional Interaction with miR156 in Arabidopsis. Plant Cell Physiol. 2020, 61, 957–966. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, S.; Oda, A.; Kamada, H.; Coupland, G.; Mizoguchi, T. Circadian clock components in Arabidopsis II. LHY/CCA1 regulate the floral integrator gene SOC1 in both GI-dependent and -independent pathways. Plant Biotechnol. 2006, 22, 319–325. [Google Scholar] [CrossRef]
- He, Z. (Ed.) Guidance to experiment on chemical control in crop plants. In Guidance to Experiment on Chemical Control in Crop Plants; Beijing Agricultural University Publishers: Beijing, China, 1993; pp. 60–68. [Google Scholar]
- Yang, J. Hormonal Changes in the Grains of Rice Subjected to Water Stress during Grain Filling. Plant Physiol. 2001, 127, 315–323. [Google Scholar] [CrossRef]
- Weiler, E.W.; Jourdan, P.S.; Conrad, W. Levels of indole-3-acetic acid in intact and decapitated coleoptiles as determined by a specific and highly sensitive solid-phase enzyme immunoassay. Planta 1981, 153, 561–571. [Google Scholar] [CrossRef]
- Tränkner, C.; Krüger, J.; Wanke, S.; Naumann, J.; Wenke, T.; Engel, F. Rapid identification of inflorescence type markers by genotyping-by-sequencing of diploid and triploid F1 plants of Hydrangea macrophylla. BMC Genet. 2019, 20, 60. [Google Scholar] [CrossRef]
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 2013, 41, e121. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Roberts, A. Ambiguous Fragment Assignment for High-Throughput Sequencing Experiments. Diploma Thesis, University of California, Berkeley, CA, USA, 2013. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Franz, M.; Lopes, C.T.; Huck, G.; Dong, Y.; Sumer, O.; Bader, G.D. Cytoscape.js: A graph theory library for visualisation and analysis. Bioinformatics 2015, 32, 309–311. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G. Cloning and Expression Analysis of the pH-Regulating Gene NHX1 in Sepals of Hydrangea Macrophylla Vacuole; Chinese Academy of Agricultural Sciences: Beijing, China, 2021. [Google Scholar]


| Sample | Raw Reads | Raw Bases | Clean Reads | Clean Bases | Valid Bases | Q30 | GC |
|---|---|---|---|---|---|---|---|
| BS1 | 51.67 M | 7.75 G | 50.52 M | 7.36 G | 94.96% | 88.66% | 45.55% |
| BS2 | 47.87 M | 7.18 G | 46.82 M | 6.84 G | 95.22% | 91.14% | 45.52% |
| BS3 | 48.92 M | 7.34 G | 47.94 M | 6.95 G | 94.68% | 91.03% | 45.38% |
| BS4 | 50.73 M | 7.61 G | 49.60 M | 7.25 G | 95.34% | 90.85% | 45.37% |
| BS5 | 49.45 M | 7.42 G | 48.24 M | 7.06 G | 95.14% | 91.86% | 45.60% |
| BS6 | 48.66 M | 7.30 G | 47.60 M | 6.95 G | 95.22% | 90.84% | 45.57% |
| BS7 | 47.49 M | 7.12 G | 46.46 M | 6.79 G | 95.31% | 91.69% | 45.38% |
| Term | All | N50 | Total Length | Max Length | Min Length | Average Length |
|---|---|---|---|---|---|---|
| Unigene | 47,793 | 1785 | 56,267,846 | 15,827 | 301 | 1177.32 |
| Pathway Definition | Pathway | Gene Number |
|---|---|---|
| Glycolysis/gluconeogenesis | ko00010 | 165 |
| Starch and sucrose metabolism | ko00500 | 159 |
| Amino sugar and nucleotide sugar metabolism | ko00520 | 133 |
| Pentose and glucuronate interconversions | ko00040 | 86 |
| Citrate cycle (TCA cycle) | ko00020 | 69 |
| Pentose phosphate pathway | ko00030 | 66 |
| Plant hormone signal transduction | ko04075 | 274 |
| GO ID | Term | Gene Number |
|---|---|---|
| GO:0009908 | Flower development | 234 |
| GO:0030154 | Cell differentiation | 183 |
| GO:0040008 | Regulation of growth | 172 |
| GO:0009555 | Pollen development | 146 |
| GO:0007623 | Circadian rhythm | 104 |
| GO:0009909 | Regulation of flower development | 104 |
| GO:0009860 | Pollen tube growth | 86 |
| GO:0009846 | Pollen germination | 76 |
| GO:0048573 | Photoperiodism, flowering | 52 |
| GO:0009911 | Positive regulation of flower development | 41 |
| GO:0042752 | Regulation of circadian rhythm | 41 |
| GO:0080092 | Regulation of pollen tube growth | 33 |
| GO:0010073 | Meristem maintenance | 32 |
| GO:0010584 | Pollen exine formation | 32 |
| GO:0048653 | Anther development | 28 |
| GO:0048443 | Stamen development | 25 |
| GO:0010229 | Inflorescence development | 21 |
| GO:0010228 | Vegetative to reproductive phase transition of meristem | 87 |
| GO:0009910 | Negative regulation of flower development | 38 |
| GO:2000028 | Regulation of photoperiodism, flowering | 37 |
| Gene Name | Forward Primer Sequence (5′–3′) | Reverse Primer Sequence (5′–3′) |
|---|---|---|
| RPL34 | ACCCCCGGTGGAAAGCTAGT | ACGGTTCACAGTCCTCCGGT |
| AP1 | GGCAATCCAAGACCAGAAT | GGCAGCAGGAATGAAGATG |
| FT | CCTAGTGACCCGAACCTTA | GACAGTCTGACGACCCAAC |
| LFY | TGAGCAGTGCCGTGATTT | GCCTTCTTGGCATACCTG |
| COL9 | CTATAAACTCGGTGGCTGAC | TGACACGACCACATCTCACT |
| G2OX6 | GAACCCTACACTGAGTCTTACC | GGAGGTCGATTACAGGAAG |
| SPL9 | TCTATTGTCATCTCGCTACGG | TGCTCTGCCAAGGATGTGG |
| The Reaction System | The Reaction Process | ||||
|---|---|---|---|---|---|
| TB Green Premix Ex Taq II | 10 μL | Pre-denaturation | 95 °C | 30 s | 1 cycle |
| Forward Primer | 4 μL | PCR reaction | 95 °C | 5 s | 40 cycles |
| Reverse Primer | 4 μL | 60 °C | 30 s | ||
| cDNA | 2 μL | 72 °C | 30 s | ||
| Total volume | 20 μL | Melting curve | 60–95 °C | 15 s | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Lyu, T.; Lyu, Y. Study on the Flower Induction Mechanism of Hydrangea macrophylla. Int. J. Mol. Sci. 2023, 24, 7691. https://doi.org/10.3390/ijms24097691
Liu Y, Lyu T, Lyu Y. Study on the Flower Induction Mechanism of Hydrangea macrophylla. International Journal of Molecular Sciences. 2023; 24(9):7691. https://doi.org/10.3390/ijms24097691
Chicago/Turabian StyleLiu, Yun, Tong Lyu, and Yingmin Lyu. 2023. "Study on the Flower Induction Mechanism of Hydrangea macrophylla" International Journal of Molecular Sciences 24, no. 9: 7691. https://doi.org/10.3390/ijms24097691
APA StyleLiu, Y., Lyu, T., & Lyu, Y. (2023). Study on the Flower Induction Mechanism of Hydrangea macrophylla. International Journal of Molecular Sciences, 24(9), 7691. https://doi.org/10.3390/ijms24097691





