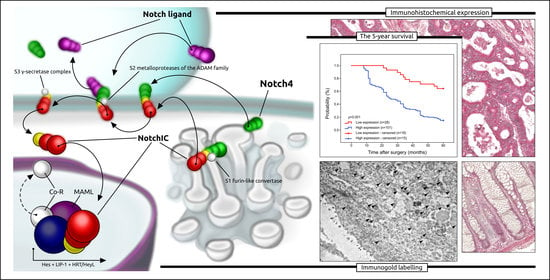
The Clinical Application of Immunohistochemical Expression of Notch4 Protein in Patients with Colon Adenocarcinoma
Abstract
1. Introduction
2. Results
2.1. Patients’ Characteristics
2.2. Correlations between Notch4 Immunohistochemical Expression and Clinicopathological Parameters
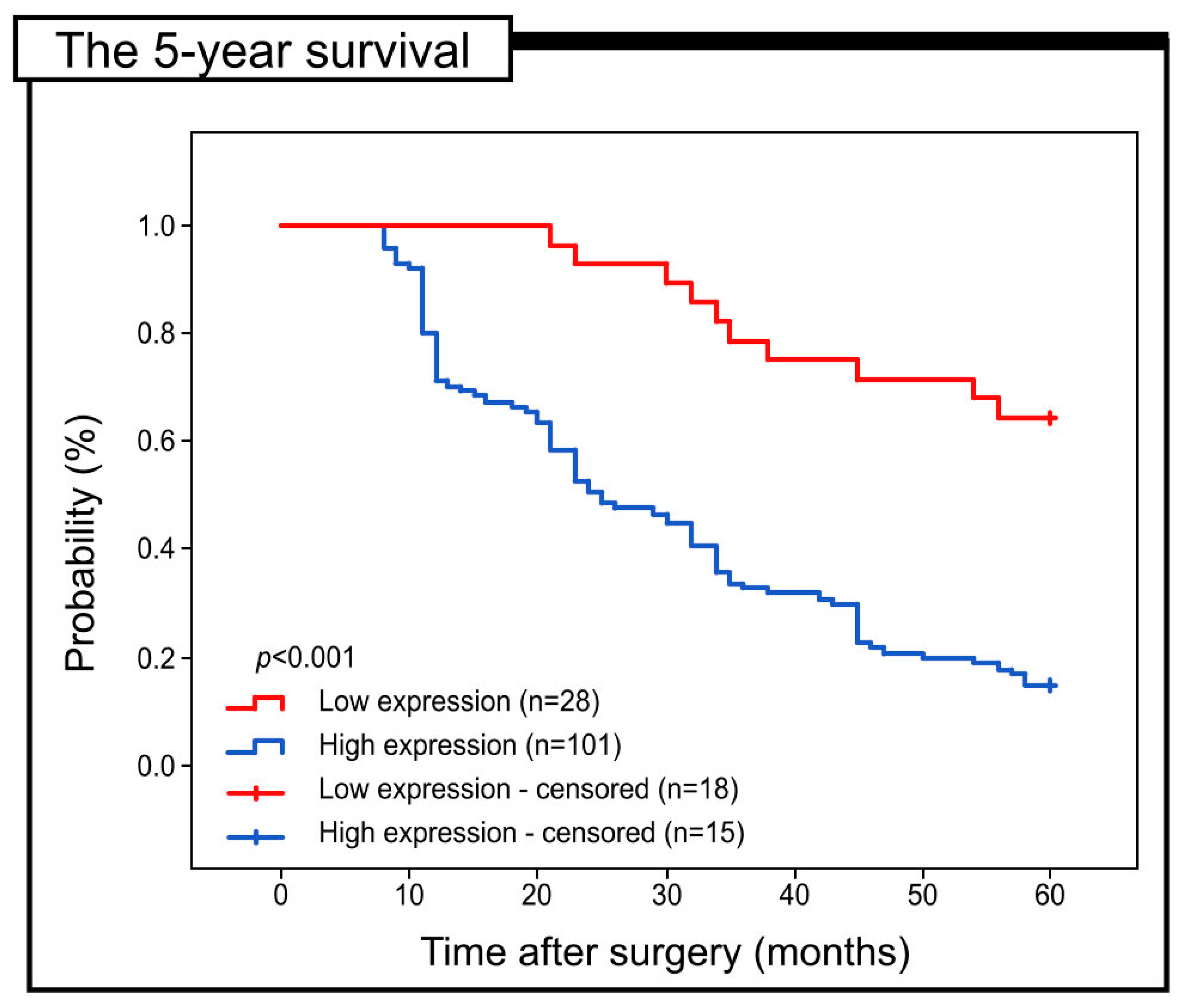
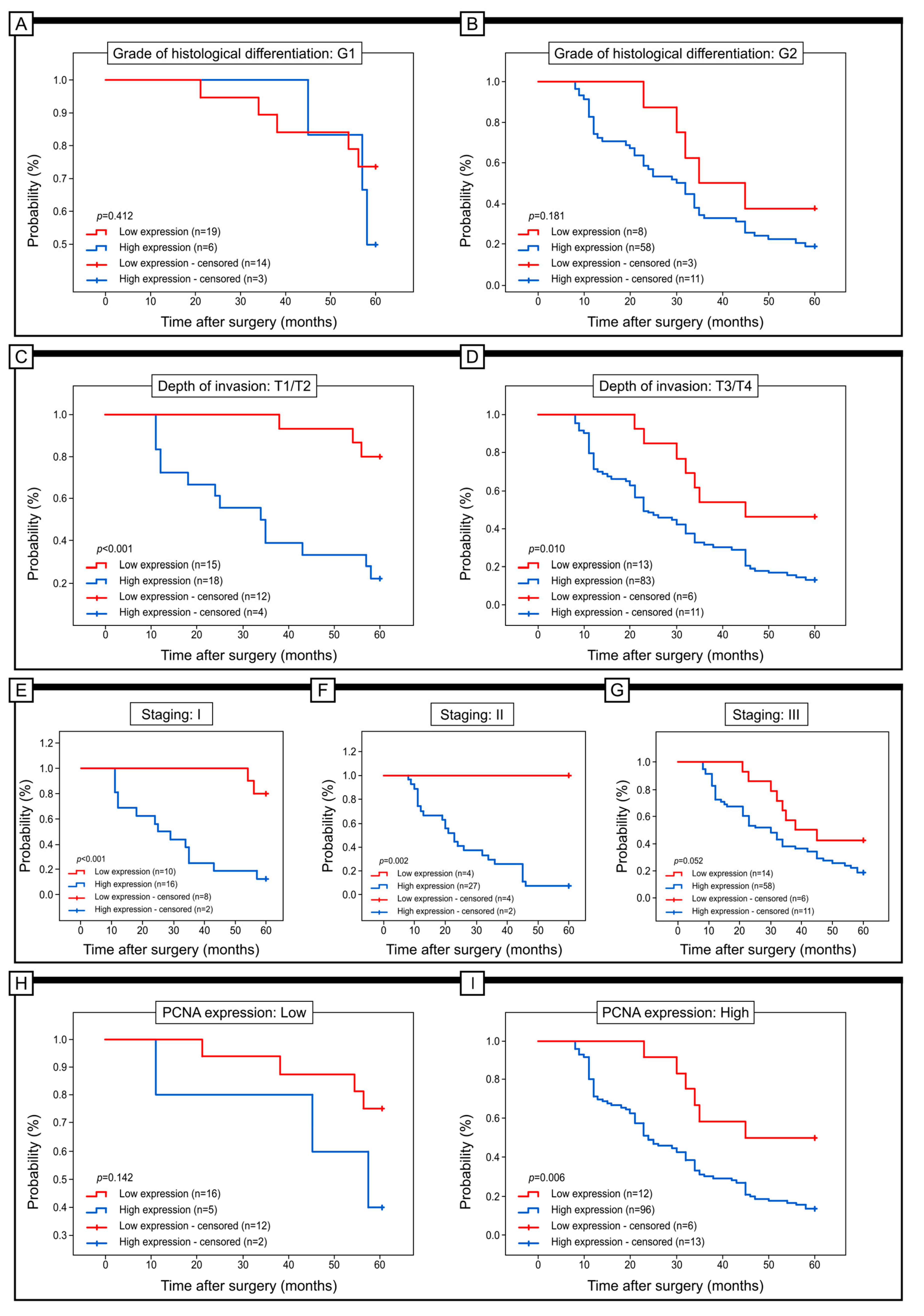
2.3. Prognostic Role of Notch4 Expression in Colon Adenocarcinoma
2.4. Immunofluorescence Staining
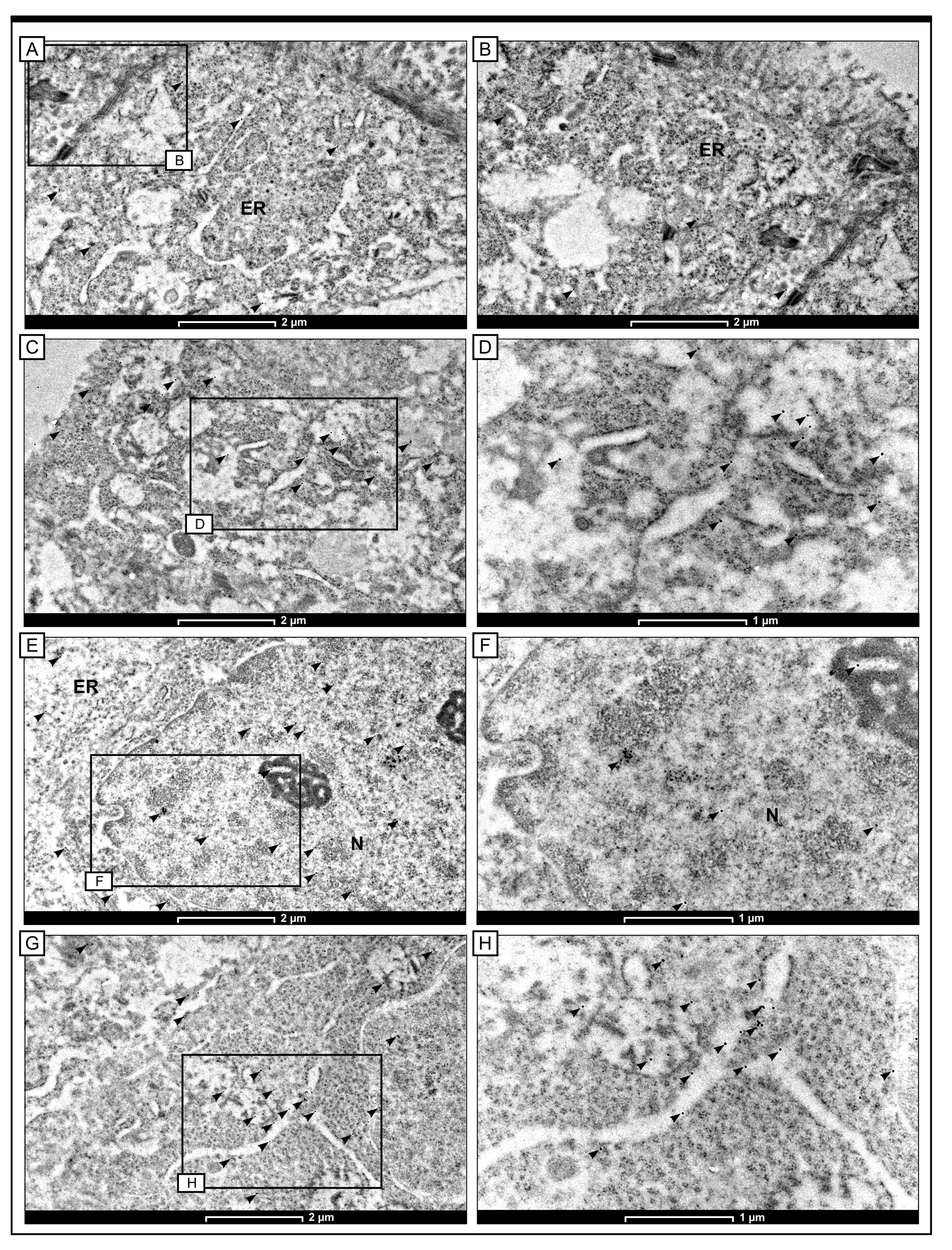
2.5. Intracellular Localisation of Notch4 by the Method of Immunogold Labelling with the Use of Transmission Electron Microscopy (TEM)
3. Discussion
4. Materials and Methods
4.1. Patients and Tumour Samples
4.2. Immunohistochemical and Immunofluorescence Staining
4.3. Immunogold Electron Microscopy
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Morgan, E.; Arnold, M.; Gini, A.; Lorenzoni, V.; Cabasag, C.J.; Laversanne, M.; Vignat, J.; Ferlay, J.; Murphy, N.; Bray, F. Global burden of colorectal cancer in 2020 and 2040: Incidence and mortality estimates from GLOBOCAN. Gut 2023, 72, 338–344. [Google Scholar] [CrossRef] [PubMed]
- Pericleous, S.; Bhogal, R.H.; Mavroeidis, V.K. The Role of Circulating Biomarkers in the Early Detection of Recurrent Colorectal Cancer Following Resection of Liver Metastases. Front. Biosci. Landmark Ed. 2022, 27, 189. [Google Scholar] [CrossRef]
- Daniel, C.L.; Gilreath, K.; Keyes, D. Colorectal cancer disparities beyond biology: Screening, treatment, access. Front. Biosci. Landmark Ed. 2017, 22, 465–478. [Google Scholar] [CrossRef]
- Harbiyeli, I.F.C.; Burtea, D.E.; Ivan, E.T.; Streață, I.; Nicoli, E.R.; Uscatu, D.; Șerbănescu, M.S.; Ioana, M.; Vilmann, P.; Săftoiu, A. Assessing Putative Markers of Colorectal Cancer Stem Cells: From Colonoscopy to Gene Expression Profiling. Diagnostics 2022, 12, 2280. [Google Scholar] [CrossRef] [PubMed]
- Latos, W.; Aebisher, D.; Latos, M.; Krupka-Olek, M.; Dynarowicz, K.; Chodurek, E.; Cieślar, G.; Kawczyk-Krupka, A. Colonoscopy: Preparation and Potential Complications. Diagnostics 2022, 12, 747. [Google Scholar] [CrossRef] [PubMed]
- Sawicki, T.; Ruszkowska, M.; Danielewicz, A.; Niedźwiedzka, E.; Arłukowicz, T.; Przybyłowicz, K.E. A Review of Colorectal Cancer in Terms of Epidemiology, Risk Factors, Development, Symptoms and Diagnosis. Cancers 2021, 13, 2025. [Google Scholar] [CrossRef]
- Xi, Y.; Xu, P. Global colorectal cancer burden in 2020 and projections to 2040. Transl. Oncol. 2021, 14, 101174. [Google Scholar] [CrossRef] [PubMed]
- Keum, N.; Giovannucci, E. Global burden of colorectal cancer: Emerging trends, risk factors and prevention strategies. Nature reviews. Gastroenterol. Hepatol. 2019, 16, 713–732. [Google Scholar] [CrossRef]
- Brzozowa-Zasada, M.; Piecuch, A.; Michalski, M.; Segiet, O.; Kurek, J.; Harabin-Słowińska, M.; Wojnicz, R. Notch and its oncogenic activity in human malignancies. Eur. Surg. ACA Acta Chir. Austriaca 2017, 49, 199–209. [Google Scholar] [CrossRef]
- Brzozowa-Zasada, M.; Piecuch, A.; Dittfeld, A.; Mielańczyk, Ł.; Michalski, M.; Wyrobiec, G.; Harabin-Słowińska, M.; Kurek, J.; Wojnicz, R. Notch signalling pathway as an oncogenic factor involved in cancer development. Contemp. Oncol. Pozn. Pol. 2016, 20, 267–272. [Google Scholar] [CrossRef]
- Misiorek, J.O.; Przybyszewska-Podstawka, A.; Kałafut, J.; Paziewska, B.; Rolle, K.; Rivero-Müller, A.; Nees, M. Context Matters: NOTCH Signatures and Pathway in Cancer Progression and Metastasis. Cells 2021, 10, 94. [Google Scholar] [CrossRef] [PubMed]
- Moore, G.; Annett, S.; McClements, L.; Robson, T. Top Notch Targeting Strategies in Cancer: A Detailed Overview of Recent Insights and Current Perspectives. Cells 2020, 9, 1503. [Google Scholar] [CrossRef]
- Ahn, S.; Hyeon, J.; Park, C.K. Notch1 and Notch4 are markers for poor prognosis of hepatocellular carcinoma. Hepatobiliary Pancreat. Dis. Int. HBPD INT 2013, 12, 286–294. [Google Scholar] [CrossRef]
- Bao, S.; Jin, S.; Wang, C.; Tu, P.; Hu, K.; Lu, J. Androgen receptor suppresses vasculogenic mimicry in hepatocellular carcinoma via circRNA7/miRNA7-5p/VE-cadherin/Notch4 signalling. J. Cell. Mol. Med. 2020, 24, 14110–14120. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Han, C.; Zhu, G.; Liao, X.; Qin, W.; Yang, C.; Liu, Z.; Su, H.; Liu, X.; Yu, L.; et al. Prognostic value of Notch receptors in postsurgical patients with hepatitis B virus-related hepatocellular carcinoma. Cancer Med. 2017, 6, 1587–1600. [Google Scholar] [CrossRef]
- Wu, W.R.; Shi, X.D.; Zhang, R.; Zhu, M.S.; Xu, L.B.; Yu, X.H.; Zeng, H.; Wang, J.; Liu, C. Clinicopathological significance of aberrant Notch receptors in intrahepatic cholangiocarcinoma. Int. J. Clin. Exp. Pathol. 2014, 7, 3272–3279. [Google Scholar]
- Lin, X.; Sun, B.; Zhu, D.; Zhao, X.; Sun, R.; Zhang, Y.; Zhang, D.; Dong, X.; Gu, Q.; Li, Y.; et al. Notch4+ cancer stem-like cells promote the metastatic and invasive ability of melanoma. Cancer Sci. 2016, 107, 1079–1091. [Google Scholar] [CrossRef]
- Bonyadi Rad, E.; Hammerlindl, H.; Wels, C.; Popper, U.; Ravindran Menon, D.; Breiteneder, H.; Kitzwoegerer, M.; Hafner, C.; Herlyn, M.; Bergler, H.; et al. Notch4 Signaling Induces a Mesenchymal-Epithelial-like Transition in Melanoma Cells to Suppress Malignant Behaviors. Cancer Res. 2016, 76, 1690–1697. [Google Scholar] [CrossRef]
- Li, H.; Zhang, Q.; Duan, Q.; Tan, Y.; Sun, T.; Qi, C. NOTCH4 mutation as predictive biomarker for immunotherapy benefits in NRAS wildtype melanoma. Front. Immunol. 2022, 13, 894110. [Google Scholar] [CrossRef]
- Patel, K.; Bhat, F.A.; Patil, S.; Routray, S.; Mohanty, N.; Nair, B.; Sidransky, D.; Ganesh, M.S.; Ray, J.G.; Gowda, H.; et al. Whole-Exome Sequencing Analysis of Oral Squamous Cell Carcinoma Delineated by Tobacco Usage Habits. Front. Oncol. 2021, 11, 660696. [Google Scholar] [CrossRef]
- Mk, H.; Prince, S.; Mohan, A.M.; Krishnan, K.V.; Devi, A. Association of Notch4 with metastasis in human oral squamous cell carcinoma. Life Sci. 2016, 156, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Hofman, P. Expression of NOTCH1, NOTCH4, HLA-DMA and HLA-DRA is synergistically associated with T cell exclusion, immune checkpoint blockade efficacy and recurrence risk in ER-negative breast cancer. Cell. Oncol. 2022, 45, 463–477. [Google Scholar] [CrossRef]
- Xu, J.; Song, F.; Jin, T.; Qin, J.; Wu, J.; Wang, M.; Wang, Y.; Liu, J. Prognostic values of Notch receptors in breast cancer. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2016, 37, 1871–1877. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.W.; Wei, X.L.; Dou, X.W.; Huang, W.H.; Du, C.W.; Zhang, G.J. The association between Notch4 expression, and clinicopathological characteristics and clinical outcomes in patients with breast cancer. Oncol. Lett. 2018, 15, 8749–8755. [Google Scholar] [CrossRef]
- Nagamatsu, I.; Onishi, H.; Matsushita, S.; Kubo, M.; Kai, M.; Imaizumi, A.; Nakano, K.; Hattori, M.; Oda, Y.; Tanaka, M.; et al. NOTCH4 is a potential therapeutic target for triple-negative breast cancer. Anticancer. Res. 2014, 34, 69–80. [Google Scholar] [PubMed]
- Hu, J.; Yu, J.; Gan, J.; Song, N.; Shi, L.; Liu, J.; Zhang, Z.; Du, J. Notch1/2/3/4 are prognostic biomarker and correlated with immune infiltrates in gastric cancer. Aging 2020, 12, 2595–2609. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, R.; Wang, X.; Ci, H.; Zhou, L.; Zhu, B.; Wu, S.; Wang, D. Evaluation of the correlation of vasculogenic mimicry, Notch4, DLL4, and KAI1/CD82 in the prediction of metastasis and prognosis in non-small cell lung cancer. Medicine 2018, 97, e13817. [Google Scholar] [CrossRef]
- Li, X.; Cao, X.; Zhao, H.; Guo, M.; Fang, X.; Li, K.; Qin, L.; He, Y.; Liu, X. Hypoxia Activates Notch4 via ERK/JNK/P38 MAPK Signaling Pathways to Promote Lung Adenocarcinoma Progression and Metastasis. Front. Cell Dev. Biol. 2021, 9, 780121. [Google Scholar] [CrossRef]
- Chen, C.Y.; Chen, Y.Y.; Hsieh, M.S.; Ho, C.C.; Chen, K.Y.; Shih, J.Y.; Yu, C.J. Expression of Notch Gene and Its Impact on Survival of Patients with Resectable Non-small Cell Lung Cancer. J. Cancer 2017, 8, 1292–1300. [Google Scholar] [CrossRef]
- Láinez-González, D.; Serrano-López, J.; Alonso-Dominguez, J.M. Understanding the Notch Signaling Pathway in Acute Myeloid Leukemia Stem Cells: From Hematopoiesis to Neoplasia. Cancers 2022, 14, 1459. [Google Scholar] [CrossRef]
- Tsaouli, G.; Ferretti, E.; Bellavia, D.; Vacca, A.; Felli, M.P. Notch/CXCR4 Partnership in Acute Lymphoblastic Leukemia Progression. J. Immunol. Res. 2019, 2019, 5601396. [Google Scholar] [CrossRef] [PubMed]
- Shaik, J.P.; Alanazi, I.O.; Pathan, A.A.K.; Parine, N.R.; Almadi, M.A.; Azzam, N.A.; Aljebreen, A.M.; Alharbi, O.; Alanazi, M.S.; Khan, Z. Frequent Activation of Notch Signaling Pathway in Colorectal Cancers and Its Implication in Patient Survival Outcome. J. Oncol. 2020, 2020, 6768942. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Chen, Z.; Li, J.; Ye, F.; Chen, G.; Fan, Q.; Dong, H.; Yuan, S.; Zhu, X. NOTCH4 Is a Novel Prognostic Marker that Correlates with Colorectal Cancer Progression and Prognosis. J. Cancer 2018, 9, 2374–2379. [Google Scholar] [CrossRef] [PubMed]
- Reedijk, M.; Odorcic, S.; Zhang, H.; Chetty, R.; Tennert, C.; Dickson, B.C.; Lockwood, G.; Gallinger, S.; Egan, S.E. Activation of Notch signaling in human colon adenocarcinoma. Int. J. Oncol. 2008, 33, 1223–1229. [Google Scholar] [CrossRef]
- Saini, N.; Sarin, A. Nucleolar localization of the Notch4 intracellular domain underpins its regulation of the cellular response to genotoxic stressors. Cell Death Discov. 2020, 6, 7. [Google Scholar] [CrossRef]
- Frithiof, H.; Welinder, C.; Larsson, A.M.; Rydén, L.; Aaltonen, K. A novel method for downstream characterization of breast cancer circulating tumor cells following CellSearch isolation. J. Transl. Med. 2015, 13, 126. [Google Scholar] [CrossRef]
- Toribio, M.L.; González-García, S. Notch Partners in the Long Journey of T-ALL Pathogenesis. Int. J. Mol. Sci. 2023, 24, 1383. [Google Scholar] [CrossRef]
- Zhou, X.; Teng, L.; Wang, M. Distinct prognostic values of four-Notch-receptor mRNA expression in ovarian cancer. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2016, 37, 6979–6985. [Google Scholar] [CrossRef]
- Jung, S.G.; Kwon, Y.D.; Song, J.A.; Back, M.J.; Lee, S.Y.; Lee, C.; Hwang, Y.Y.; An, H.J. Prognostic significance of Notch 3 gene expression in ovarian serous carcinoma. Cancer Sci. 2010, 101, 1977–1983. [Google Scholar] [CrossRef]
- Capodanno, Y.; Chen, Y.; Schrader, J.; Tomosugi, M.; Sumi, S.; Yokoyama, A.; Hiraoka, N.; Ohki, R. Cross-talk among MEN1, p53 and Notch regulates the proliferation of pancreatic neuroendocrine tumor cells by modulating INSM1 expression and subcellular localization. Neoplasia 2021, 23, 979–992. [Google Scholar] [CrossRef]
- Kelman, Z. PCNA: Structure, functions and interactions. Oncogene 1997, 14, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Qian, C.; Liu, F.; Ye, B.; Zhang, X.; Liang, Y.; Yao, J. Notch4 promotes gastric cancer growth through activation of Wnt1/β-catenin signaling. Mol. Cell. Biochem. 2015, 401, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Kuang, Y.; Wang, Y.; Xu, Q.; Ren, Q. Notch-4 silencing inhibits prostate cancer growth and EMT via the NF-κB pathway. Apoptosis Int. J. Program. Cell Death 2017, 22, 877–884. [Google Scholar] [CrossRef]
- Fukusumi, T.; Guo, T.W.; Sakai, A.; Ando, M.; Ren, S.; Haft, S.; Liu, C.; Amornphimoltham, P.; Gutkind, J.S.; Califano, J.A. The NOTCH4-HEY1 Pathway Induces Epithelial-Mesenchymal Transition in Head and Neck Squamous Cell Carcinoma. Clin. Cancer Res. 2018, 24, 619–633. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Qiao, L.; Liang, N.; Xie, J.; Luo, H.; Deng, G.; Zhang, J. Vasculogenic mimicry and tumor metastasis. J. BUON 2016, 21, 533–541. [Google Scholar]
- Remmele, W.; Stegner, H.E. Vorschlag zur einheitlichen Definition eines Immunreaktiven Score (IRS) für den immunhistochemischen Ostrogenrezeptor-Nachweis (ER-ICA) im Mammakarzinomgewebe [Recommendation for uniform definition of an immunoreactive score (IRS) for immunohistochemical oestrogen receptor detection (ER-ICA) in breast cancer tissue]. Der Pathol. 1987, 8, 138–140. [Google Scholar]
- Alnuaimi, A.R.; Bottner, J.; Nair, V.A.; Ali, N.; Alnakhli, R.; Dreyer, E.; Talaat, I.M.; Busch, H.; Perner, S.; Kirfel, J.; et al. Immunohistochemical Expression Analysis of Caldesmon Isoforms in Colorectal Carcinoma Reveals Interesting Correlations with Tumor Characteristics. Int. J. Mol. Sci. 2023, 24, 2275. [Google Scholar] [CrossRef]


| N (Number of Cases) | % | ||
|---|---|---|---|
| Gender | Females | 62 | 48.06 |
| Males | 67 | 51.94 | |
| Age [years] | ≤60 years | 44 | 34.11 |
| 61–75 years | 47 | 36.43 | |
| >75 years | 38 | 29.46 | |
| Grade of histological differentiation | G1 | 25 | 19.38 |
| G2 | 66 | 51.16 | |
| G3 | 38 | 29.46 | |
| Depth of invasion | T1 | 15 | 11.63 |
| T2 | 18 | 13.95 | |
| T3 | 73 | 56.59 | |
| T4 | 23 | 17.83 | |
| Regional Lymph Node involvement | N0 | 57 | 44.18 |
| N1 | 38 | 29.46 | |
| N2 | 34 | 26.36 | |
| Location of tumour | Right-sided tumours | 66 | 51.16 |
| Left-sided tumours | 63 | 48.84 | |
| Notch4 expression in healthy tissue | Low | 118 | 91.47 |
| High | 11 | 8.53 | |
| Angioinvasion | No | 30 | 23.26 |
| Yes | 99 | 76.74 | |
| PCNA expression | Low | 21 | 16.28 |
| High | 108 | 83.72 | |
| Staging | I | 26 | 20.16 |
| II | 31 | 24.03 | |
| III | 72 | 55.81 | |
| The Immunoexpression Level of Notch4 | p-Value | |||||
|---|---|---|---|---|---|---|
| Low | High | |||||
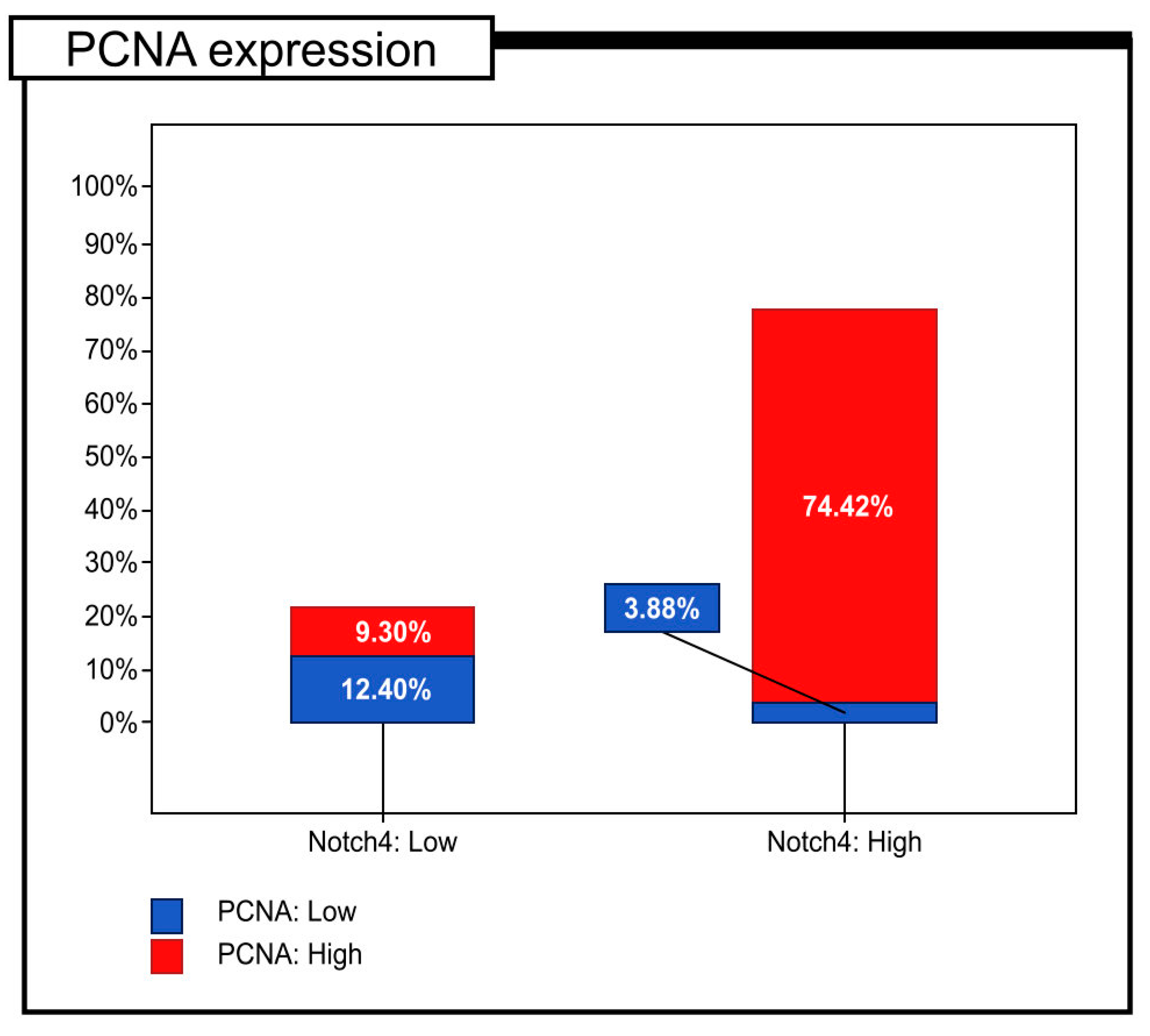
| The immunoexpression level of PCNA | Low | 16 | (12.40%) | 5 | (3.88%) | p < 0.001 |
| High | 12 | (9.30%) | 96 | (74.42%) | p = 0.146 | |
| The Immunoexpression Level of Notch4 | p-Value | |||||
|---|---|---|---|---|---|---|
| Low | High | |||||
| Age [Years] | ≤60 years | 8 | (18.18%) | 36 | (81.82%) | p = 0.670 |
| 61–75 years | 10 | (21.28%) | 37 | (78.72%) | ||
| >75 years | 10 | (26.32%) | 28 | (73.68%) | ||
| Gender | Females | 15 | (24.19%) | 47 | (75.81%) | p = 0.510 |
| Males | 13 | (19.40%) | 54 | (80.60%) | ||
| Grade of histological differentiation | G1 | 19 | (76.00%) | 6 | (24.00%) | p < 0.001 |
| G2 | 8 | (12.12%) | 58 | (87.88%) | ||
| G3 | 1 | (2.63%) | 37 | (97.37%) | ||
| Depth of invasion | T1/T2 | 15 | (45.45%) | 18 | (54.55%) | p < 0.001 |
| T3/T4 | 13 | (13.54%) | 83 | (86.46%) | ||
| Regional Lymph Node involvement | N0 | 14 | (24.56%) | 43 | (75.44%) | p = 0.484 |
| N1/N2 | 14 | (19.44%) | 58 | (80.56%) | ||
| Localisation | Left-sided tumours | 11 | (16.67%) | 55 | (83.33%) | p = 0.155 |
| Right-sided tumours | 17 | (26.98%) | 46 | (73.02%) | ||
| Angioinvasion | Yes | 18 | (60.00%) | 12 | (40.00%) | p < 0.001 |
| No | 10 | (10.10%) | 89 | (89.90%) | ||
| PCNA expression | Low | 16 | (76.19%) | 5 | (23.81%) | p < 0.001 |
| High | 12 | (11.11%) | 96 | (88.89%) | ||
| Staging | I | 10 | (38.46%) | 16 | (61.54%) | p = 0.052 |
| II | 14 | (19.44%) | 58 | (80.56%) | ||
| III | 4 | (12.90%) | 27 | (87.10%) | ||
| Prognostic Parameter | Univariate Analysis | Multivariate Analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | p-Value | HR | 95% CI | p-Value | |
| Gender | 0.883 | 0.591–1.318 | 0.543 | – | – | – |
| Age | 0.999 | 0.985–1.014 | 0.929 | – | – | – |
| Staging | 1.152 | 0.896–1.481 | 0.269 | – | – | – |
| Histological differentiation | 2.415 | 1.785–3.267 | <0.001 | 1.907 | 1.251–2.907 | 0.003 |
| Depth of invasion | 1.602 | 1.251–2.052 | <0.001 | 0.985 | 0.715–1.358 | 0.928 |
| Regional Lymph Node involvement | 1.108 | 0.869–1.412 | 0.409 | – | – | – |
| Localisation | 1.091 | 0.731–1.629 | 0.668 | – | – | – |
| Immunohistochemical expression of Notch4 in cancer tissue | 4.389 | 2.269–8.487 | <0.001 | 2.414 | 1.134–5.141 | 0.022 |
| Angioinvasion | 2.865 | 1.618–5.074 | <0.001 | 0.896 | 0.427–1.880 | 0.772 |
| PCNA expression | 4.527 | 2.087–9.820 | <0.001 | 1.454 | 0.486–4.355 | 0.503 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brzozowa-Zasada, M.; Piecuch, A.; Michalski, M.; Matysiak, N.; Kucharzewski, M.; Łos, M.J. The Clinical Application of Immunohistochemical Expression of Notch4 Protein in Patients with Colon Adenocarcinoma. Int. J. Mol. Sci. 2023, 24, 7502. https://doi.org/10.3390/ijms24087502
Brzozowa-Zasada M, Piecuch A, Michalski M, Matysiak N, Kucharzewski M, Łos MJ. The Clinical Application of Immunohistochemical Expression of Notch4 Protein in Patients with Colon Adenocarcinoma. International Journal of Molecular Sciences. 2023; 24(8):7502. https://doi.org/10.3390/ijms24087502
Chicago/Turabian StyleBrzozowa-Zasada, Marlena, Adam Piecuch, Marek Michalski, Natalia Matysiak, Marek Kucharzewski, and Marek J. Łos. 2023. "The Clinical Application of Immunohistochemical Expression of Notch4 Protein in Patients with Colon Adenocarcinoma" International Journal of Molecular Sciences 24, no. 8: 7502. https://doi.org/10.3390/ijms24087502
APA StyleBrzozowa-Zasada, M., Piecuch, A., Michalski, M., Matysiak, N., Kucharzewski, M., & Łos, M. J. (2023). The Clinical Application of Immunohistochemical Expression of Notch4 Protein in Patients with Colon Adenocarcinoma. International Journal of Molecular Sciences, 24(8), 7502. https://doi.org/10.3390/ijms24087502