Clinical-Grade Patches as a Medium for Enrichment of Sweat-Extracellular Vesicles and Facilitating Their Metabolic Analysis
Abstract
1. Introduction
2. Results
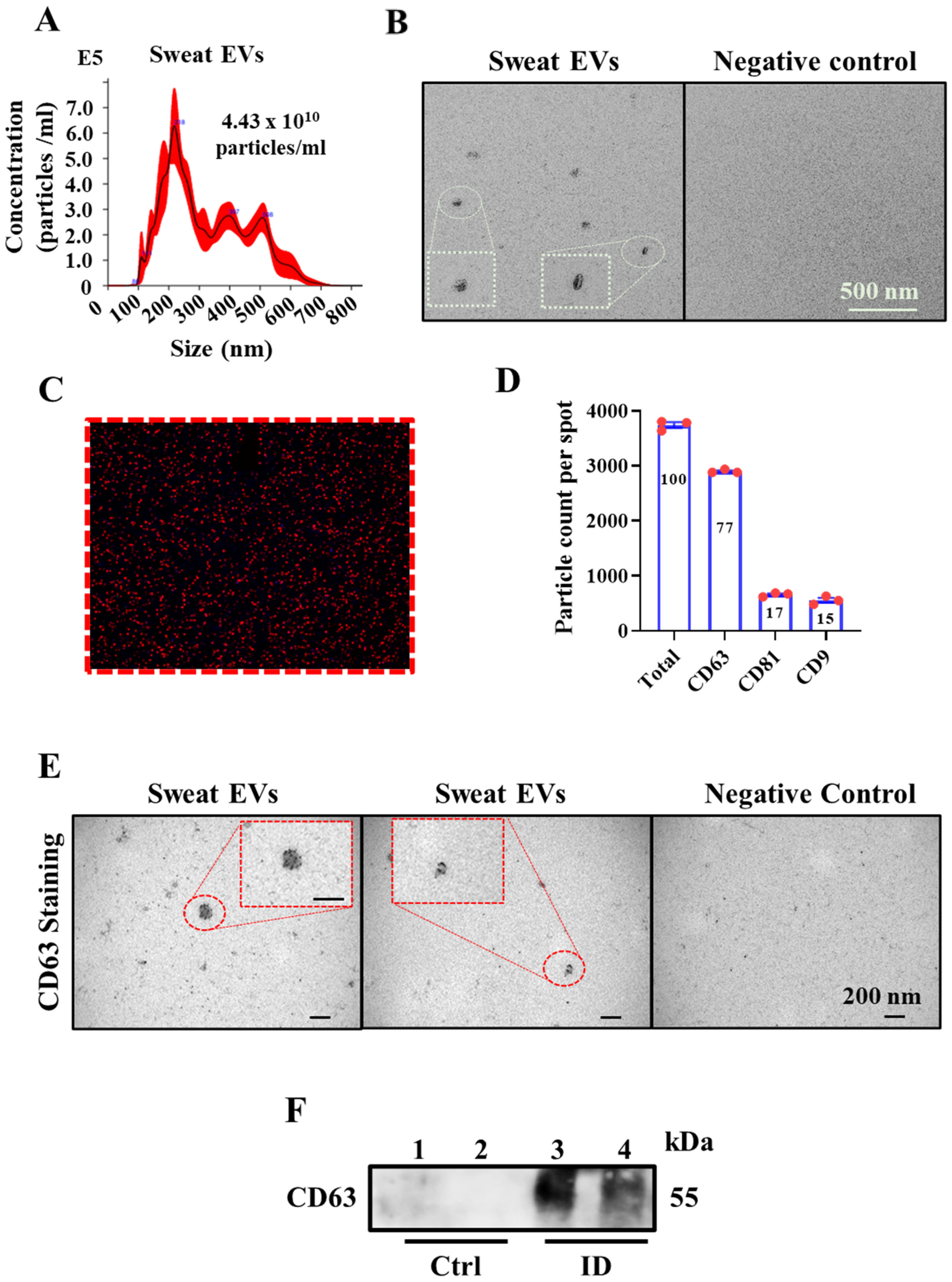
2.1. Sweat Extracellular Vesicles (EVs) Can Be Enriched and Isolated via Clinical Grade Patches
2.2. Sweat Extracted EVs Express Cargo Metabolites
2.3. Metabolite Levels in EVs Extracted from Sweat May Provide a Means to Study their Association with Diseases
2.4. Association of the Metabolite Levels in Sweat EVs with Blood Glucose Level
2.5. Changes in the Metabolites Contained in Sweat EVs in Relation to BMI
3. Discussion
4. Material and Methods
4.1. Study Design
4.2. Heat Exposure Trial in the Laboratory
4.3. Isolation of Sweat EVs
4.4. Targeted LC-MS Metabolomics Analysis
4.5. Metabolomics Data Analysis
4.6. Western Blot
4.7. NanoSight Nanoparticle Tracking Analysis
4.8. ExoView Analysis
4.9. Electron Microscopy_Negative Staining
4.10. Immunoelectron Microscopy
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baker, L.B. Physiology of sweat gland function: The roles of sweating and sweat composition in human health. Temperature 2019, 6, 211–259. [Google Scholar] [CrossRef]
- Murota, H.; Matsui, S.; Ono, E.; Kijima, A.; Kikuta, J.; Ishii, M.; Katayama, I. Sweat, the driving force behind normal skin: An emerging perspective on functional biology and regulatory mechanisms. J. Dermatol. Sci. 2015, 77, 3–10. [Google Scholar] [CrossRef]
- Xuan, X.; Pérez-Ràfols, C.; Chen, C.; Cuartero, M.; Crespo, G.A. Lactate biosensing for reliable on-body sweat analysis. ACS Sens. 2021, 6, 2763–2771. [Google Scholar] [CrossRef]
- Cuartero, M.; Parrilla, M.; Crespo, G. Wearable potentiometric sensors for medical applications. Sensors 2019, 19, 363. [Google Scholar] [CrossRef]
- Raiszadeh, M.M.; Ross, M.M.; Russo, P.S.; Schaepper, M.A.; Zhou, W.; Deng, J.; Ng, D.; Dickson, A.; Dickson, C.; Strom, M.; et al. Proteomic analysis of eccrine sweat: Implications for the discovery of schizophrenia biomarker proteins. J. Proteome Res. 2012, 11, 2127–2139. [Google Scholar] [CrossRef]
- Adewole, O.O.; Erhabor, G.E.; Adewole, T.O.; Ojo, A.O.; Oshokoya, H.; Wolfe, L.M.; Prenni, J.E. Proteomic profiling of eccrine sweat reveals its potential as a diagnostic biofluid for active tuberculosis. Proteom.—Clin. Appl. 2016, 10, 547–553. [Google Scholar] [CrossRef]
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef] [PubMed]
- Bart, G.; Fischer, D.; Samoylenko, A.; Zhyvolozhnyi, A.; Stehantsev, P.; Miinalainen, I.; Kaakinen, M.; Nurmi, T.; Singh, P.; Kosamo, S.; et al. Characterization of nucleic acids from extracellular vesicle-enriched human sweat. BMC Genom. 2021, 22, 425. [Google Scholar] [CrossRef] [PubMed]
- Malik, S.; Saltzman, W.M.; Bahal, R. Extracellular vesicles mediated exocytosis of antisense peptide nucleic acids. Mol. Ther. Nucleic Acids 2021, 25, 302–315. [Google Scholar] [CrossRef] [PubMed]
- Prieto-Vila, M.; Yoshioka, Y.; Ochiya, T. Biological functions driven by mRNAs carried by extracellular vesicles in cancer. Front. Cell Dev. Biol. 2021, 9, 620498. [Google Scholar] [CrossRef] [PubMed]
- Piombino, C.; Mastrolia, I.; Omarini, C.; Candini, O.; Dominici, M.; Piacentini, F.; Toss, A. The role of exosomes in breast cancer diagnosis. Biomedicines 2021, 9, 312. [Google Scholar] [CrossRef]
- Zhao, Z.; Yang, S.; Zhou, A.; Li, X.; Fang, R.; Zhang, S.; Zhao, G.; Li, P. Small extracellular vesicles in the development, diagnosis, and possible therapeutic application of esophageal squamous cell carcinoma. Front. Oncol. 2021, 11, 732702. [Google Scholar] [CrossRef] [PubMed]
- Testa, A.; Venturelli, E.; Brizzi, M.F. Extracellular vesicles: New tools for early diagnosis of breast and genitourinary cancers. Int. J. Mol. Sci. 2021, 22, 8430. [Google Scholar] [CrossRef] [PubMed]
- Speedy, D.B.; Noakes, T.D.; Schneider, C. Exercise-associated hyponatremia: A review. Emerg. Med. 2001, 13, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Emaminejad, S.; Nyein, H.Y.Y.; Challa, S.; Chen, K.; Peck, A.; Fahad, H.M.; Ota, H.; Shiraki, H.; Kiriya, D.; et al. Fully integrated wearable sensor arrays for multiplexed in situ perspiration analysis. Nature 2016, 529, 509–514. [Google Scholar] [CrossRef]
- Martínez-Reyes, I.; Chandel, N.S. Mitochondrial TCA cycle metabolites control physiology and disease. Nat. Commun. 2020, 11, 102. [Google Scholar] [CrossRef]
- Hosseini, M.; Dousset, L.; Mahfouf, W.; Serrano-Sanchez, M.; Redonnet-Vernhet, I.; Mesli, S.; Kasraian, Z.; Obre, E.; Bonneu, M.; Claverol, S.; et al. Energy metabolism rewiring precedes UVB-induced primary skin tumor formation. Cell Rep. 2018, 23, 3621–3634. [Google Scholar] [CrossRef]
- Kuehne, A.; Emmert, H.; Soehle, J.; Winnefeld, M.; Fischer, F.; Wenck, H.; Gallinat, S.; Terstegen, L.; Lucius, R.; Hildebrand, J.; et al. Acute Activation of Oxidative Pentose Phosphate Pathway as First-Line Response to Oxidative Stress in Human Skin Cells. Mol. Cell 2015, 59, 359–371. [Google Scholar] [CrossRef]
- James, K.-D. Chapter 19—Animal metabolites: From amphibians, reptiles, aves/birds, and invertebrates. In Pharmacognosy; Badal, S., Delgoda, R., Eds.; Academic Press: Boston, MA, USA, 2017; pp. 401–411. [Google Scholar] [CrossRef]
- Altman, B.J.; Stine, Z.E.; Dang, C.V. From krebs to clinic: Glutamine metabolism to cancer therapy. Nat. Rev. Cancer 2016, 16, 619–634. [Google Scholar] [CrossRef]
- Mamas, M.; Dunn, W.B.; Neyses, L.; Goodacre, R. The role of metabolites and metabolomics in clinically applicable biomarkers of disease. Arch. Toxicol. 2011, 85, 5–17. [Google Scholar] [CrossRef]
- Elia, I.; Haigis, M.C. Metabolites and the tumor microenvironment: From cellular mechanisms to systemic metabolism. Nat. Metab. 2021, 3, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Hier, S.W.; Cornbleet, T.; Bergeim, O. The amino acids of human sweat. J. Biol. Chem. 1946, 166, 327–333. [Google Scholar] [CrossRef] [PubMed]
- Abu-Soud, H.M.; Gachhui, R.; Raushel, F.M.; Stuehr, D.J. The ferrous-dioxy complex of neuronal nitric oxide synthase: Divergent effects of l-arginine and tetrahydrobiopterin on its stability*. J. Biol. Chem. 1997, 272, 17349–17353. [Google Scholar] [CrossRef]
- Cinelli, M.A.; Do, H.T.; Miley, G.P.; Silverman, R.B. Inducible nitric oxide synthase: Regulation, structure, and inhibition. Med. Res. Rev. 2020, 40, 158–189. [Google Scholar] [CrossRef] [PubMed]
- Molina, M.N.; Ferder, L.; Manucha, W. Emerging Role of Nitric Oxide and Heat Shock Proteins in Insulin Resistance. Curr. Hypertens Rep. 2015, 18, 1. [Google Scholar] [CrossRef]
- Fealey, R.D.; Low, P.A.; Thomas, J.E. Thermoregulatory sweating abnormalities in diabetes mellitus. Mayo Clin. Proc. 1989, 64, 617–628. [Google Scholar] [CrossRef]
- Kennedy, W.R.; Sakuta, M.; Sutherland, D.; Goetz, F.C. The sweating deficiency in diabetes mellitus: Methods of quantitation and clinical correlation. Neurology 1984, 34, 758–763. [Google Scholar] [CrossRef]
- Cuschieri, S.; Calleja Agius, J. The interaction between diabetes and climate change—A review on the dual global phenomena. Early Hum. Dev. 2021, 155, 105220. [Google Scholar] [CrossRef]
- Moon, J. The effect of the heatwave on the morbidity and mortality of diabetes patients; a meta-analysis for the era of the climate crisis. Environ. Res. 2021, 195, 110762. [Google Scholar] [CrossRef]
- Galicia-Garcia, U.; Benito-Vicente, A.; Jebari, S.; Larrea-Sebal, A.; Siddiqi, H.; Uribe, K.B.; Ostolaza, H.; Martín, C. Pathophysiology of Type 2 Diabetes Mellitus. Int. J. Mol. Sci. 2020, 21, 6275. [Google Scholar] [CrossRef]
- Zheng, Y.; Ley, S.H.; Hu, F.B. Global aetiology and epidemiology of type 2 diabetes mellitus and its complications. Nat. Rev. Endocrinol. 2018, 14, 88–98. [Google Scholar] [CrossRef] [PubMed]
- Zorena, K.; Jachimowicz-Duda, O.; Ślęzak, D.; Robakowska, M.; Mrugacz, M. Adipokines and Obesity. Potential Link to Metabolic Disorders and Chronic Complications. Int. J. Mol. Sci. 2020, 21, 3570. [Google Scholar] [CrossRef] [PubMed]
- Haczeyni, F.; Bell-Anderson, K.S.; Farrell, G.C. Causes and mechanisms of adipocyte enlargement and adipose expansion. Obes. Rev. 2018, 19, 406–420. [Google Scholar] [CrossRef]
- Vangipurapu, J.; Fernandes Silva, L.; Kuulasmaa, T.; Smith, U.; Laakso, M. Microbiota-related metabolites and the risk of type 2 diabetes. Diabetes Care 2020, 43, 1319–1325. [Google Scholar] [CrossRef]
- Okawara, H.; Sawada, T.; Nakashima, D.; Maeda, Y.; Minoji, S.; Morisue, T.; Katsumata, Y.; Matsumoto, M.; Nakamura, M.; Nagura, T. Kinetic changes in sweat lactate following fatigue during constant workload exercise. Physiol. Rep. 2022, 10, e15169. [Google Scholar] [CrossRef]
- Spurway, N.C. Aerobic exercise, anaerobic exercise and the lactate thsreshold. Br. Med. Bull. 1992, 48, 569–591. [Google Scholar] [CrossRef]
- Garcia, S.O.; Ulyanova, Y.V.; Figueroa-Teran, R.; Bhatt, K.H.; Singhal, S.; Atanassov, P. Wearable sensor system powered by a biofuel cell for detection of lactate levels in sweat. ECS J. Solid State Sci. Technol. 2016, 5, M3075–M3081. [Google Scholar] [CrossRef]
- Currano, L.J.; Sage, F.C.; Hagedon, M.; Hamilton, L.; Patrone, J.; Gerasopoulos, K. Wearable sensor system for detection of lactate in sweat. Sci. Rep. 2018, 8, 15890. [Google Scholar] [CrossRef]
- Luo, X.; Shi, W.; Yu, H.; Xie, Z.; Li, K.; Cui, Y. Wearable carbon nanotube-based biosensors on gloves for lactate. Sensors 2018, 18, 3398. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Park, I.; Pack, S.P.; Lee, G.; Hong, Y. Colorimetric sensing of lactate in human sweat using polyaniline nanoparticles-based sensor platform and colorimeter. Biosensors 2022, 12, 248. [Google Scholar] [CrossRef] [PubMed]
- Krause, M.; Rak-Raszewska, A.; Naillat, F.; Saarela, U.; Schmidt, C.; Ronkainen, V.-P.; Bart, G.; Ylä-Herttuala, S.; Vainio, S.J. Exosomes as secondary inductive signals involved in kidney organogenesis. J. Extracell. Vesicles 2018, 7, 1422675. [Google Scholar] [CrossRef] [PubMed]




| Variable | Healthy Participants |
|---|---|
| Number | 11 |
| Gender | Male |
| Age/years | 62.64 |
| Height/cm | 176.45 ± 6.74 |
| Weight/Kg | 78.87 ± 7.79 |
| Blood glucose/mmol/L | 5.7 ± 0.57 |
| Body mass index BMI/Kg/m2 | 25.43 ± 2.04 |
| Sweat EV’s Concentration (particles/mL) | Blood Glucose Levels (mmol/L) | Body Mass Index (BMI) (Kg·m2) | |
|---|---|---|---|
| Number | |||
| 1 | 4.02 × 1010 | 5.9 | 25 |
| 2 | 2.14 × 1011 | 5 | 28.5 |
| 3 | 1.18 × 1010 | 6 | 26.3 |
| 4 | 2.24 × 1010 | 5.4 | 23.6 |
| 5 | 2.28 × 1010 | 5.4 | 23 |
| 6 | 3.2 × 1010 | 5.4 | 26 |
| 7 | 5.1 × 1010 | 4.5 | 25 |
| 8 | 4.77 × 1010 | 6.2 | 28 |
| 9 | 4.4 × 1010 | 5.2 | 27.3 |
| 10 | 1.42 × 1011 | 4.9 | 22.3 |
| 11 | 6.84 × 1010 | 5.3 | 24.3 |
| Variable | T2D Participants |
|---|---|
| Number | 10 |
| Gender | Male |
| Age/years | 64 |
| Height/cm | 176.2 ± 5.73 |
| Weight/Kg | 93.63 ± 16.28 |
| Blood glucose/mmol/L | 8.04 ± 2.58 |
| HbA1c/mmol/mol | 49 ± 9.68 |
| Body mass index BMI/Kg/m2 | 30.13 ± 4.83 |
| Variable | Correlation with BG (Healthy) | Lower Band H | Upper Band H | p Value |
|---|---|---|---|---|
| Particles/mL | −0.688 | −0.924 | −0.073 | 0.019 * |
| Pyroglutamate | −0.789 | −0.953 | −0.269 | 0.004 ** |
| Glycine | −0.541 | −0.873 | 0.136 | 0.085 ns |
| Alanine | −0.807 | −0.958 | −0.31 | 0.003** |
| Arginine | −0.798 | −0.955 | −0.289 | 0.003 ** |
| Asparagine | −0.807 | −0.958 | −0.31 | 0.003 ** |
| Leucine | −0.615 | −0.9 | 0.039 | 0.044 * |
| Glutamate | −0.615 | −0.9 | 0.039 | 0.044 * |
| Glutamine | −0.56 | −0.881 | 0.112 | 0.073 ns |
| Linoleate | −0.587 | −0.89 | 0.077 | 0.058 ns |
| Lactate | −0.495 | −0.856 | 0.189 | 0.121 |
| Lysine | −0.615 | −0.9 | 0.039 | 0.044 * |
| Methionine | −0.642 | −0.909 | −0.001 | 0.033 * |
| Proline | −0.716 | −0.932 | −0.122 | 0.013 * |
| Serine | −0.688 | −0.924 | −0.073 | 0.019 * |
| Threonine | −0.752 | −0.943 | −0.19 | 0.008 ** |
| Tyrosine | −0.505 | −0.86 | 0.178 | 0.113 |
| Malate | −0.716 | −0.932 | −0.122 | 0.013 * |
| Myristate | −0.358 | −0.797 | 0.328 | 0.28 |
| Valine | −0.697 | −0.927 | −0.089 | 0.017 * |
| Palmitate | −0.321 | −0.779 | 0.361 | 0.336 |
| Succinate | −0.642 | −0.909 | −0.001 | 0.033 * |
| Tryptophan | −0.606 | −0.897 | 0.051 | 0.048 * |
| Aspartate | −0.385 | −0.809 | 0.302 | 0.242 |
| Isoleucine | −0.817 | −0.96 | −0.334 | 0.002 ** |
| Variable | Correlation with BG (T2D) | Lower Band H | Upper Band H | p Value |
|---|---|---|---|---|
| Particles/mL | 0.317 | −0.391 | 0.789 | 0.372 |
| Pyroglutamate | 0.287 | −0.418 | 0.776 | 0.422 |
| Glycine | −0.165 | −0.72 | 0.519 | 0.649 |
| Alanine | 0.287 | −0.418 | 0.776 | 0.422 |
| Arginine | 0.043 | −0.603 | 0.655 | 0.907 |
| Asparagine | 0.079 | −0.579 | 0.675 | 0.828 |
| Leucine | 0.348 | −0.361 | 0.802 | 0.325 |
| Glutamate | 0.244 | −0.456 | 0.757 | 0.497 |
| Glutamine | 0.079 | −0.579 | 0.675 | 0.828 |
| Linoleate | 0.049 | −0.599 | 0.658 | 0.894 |
| Lactate | −0.012 | −0.637 | 0.622 | 0.973 |
| Lysine | 0.165 | −0.519 | 0.72 | 0.649 |
| Methionine | 0.628 | −0.003 | 0.901 | 0.052 ns |
| Proline | −0.091 | −0.682 | 0.571 | 0.802 |
| Serine | 0.348 | −0.361 | 0.802 | 0.325 |
| Threonine | 0.287 | −0.418 | 0.776 | 0.422 |
| Tyrosine | 0.323 | −0.385 | 0.792 | 0.362 |
| Malate | −0.012 | −0.637 | 0.622 | 0.973 |
| Myristate | −0.134 | −0.704 | 0.541 | 0.712 |
| Valine | −0.116 | −0.695 | 0.554 | 0.75 |
| Palmitate | −0.433 | −0.835 | 0.27 | 0.211 |
| Succinate | 0.061 | −0.591 | 0.665 | 0.867 |
| Tryptophan | 0.366 | −0.343 | 0.809 | 0.298 |
| Aspartate | −0.03 | −0.647 | 0.611 | 0.933 |
| Isoleucine | 0.238 | −0.461 | −0.755 | 0.508 |
| Variable | Correlation with BMI (Healthy) | Lower Band H | Upper Band H | p Value |
|---|---|---|---|---|
| Particles/mL | 0.127 | −0.514 | 0.677 | 0.714 |
| Pyroglutamate | 0.118 | −0.52 | 0.672 | 0.734 |
| Glycine | 0.036 | −0.576 | 0.623 | 0.924 |
| Alanine | 0.109 | −0.527 | 0.667 | 0.755 |
| Arginine | 0.164 | −0.487 | 0.698 | 0.634 |
| Asparagine | 0.055 | −0.564 | 0.634 | 0.881 |
| Leucine | 0.182 | −0.474 | 0.708 | 0.595 |
| Glutamate | 0.064 | −0.558 | 0.64 | 0.86 |
| Glutamine | −0.127 | −0.677 | 0.514 | 0.714 |
| Linoleate | 0.009 | −0.594 | 0.606 | 0.989 |
| Lactate | 0 | −0.6 | 0.6 | 1 |
| Lysine | −0.155 | −0.693 | 0.494 | 0.654 |
| Methionine | 0.245 | −0.425 | 0.741 | 0.468 |
| Proline | −0.009 | −0.606 | 0.594 | 0.989 |
| Serine | 0.091 | −0.539 | 0.656 | 0.797 |
| Threonine | 0.018 | −0.588 | 0.611 | 0.968 |
| Tyrosine | 0.145 | −0.501 | 0.687 | 0.673 |
| Malate | −0.109 | −0.667 | 0.527 | 0.755 |
| Myristate | −0.164 | −0.698 | 0.487 | 0.634 |
| Valine | −0.282 | −0.67 | 0.394 | 0.402 |
| Palmitate | −0.164 | −0.698 | 0.487 | 0.634 |
| Succinate | −0.136 | −0.682 | 0.507 | 0.694 |
| Tryptophan | 0.291 | −0.387 | 0.765 | 0.386 |
| Aspartate | −0.027 | −0.617 | 0.582 | 0.946 |
| Isoleucine | 0 | −0.6 | 0.6 | 1 |
| Variable | Correlation with BMI (T2D) | Lower Band H | Upper Band H | p Value |
|---|---|---|---|---|
| Particles/mL | 0.77 | 0.175 | 0.953 | 0.014 * |
| Pyroglutamate | 0.527 | −0.202 | 0.88 | 0.123 |
| Glycine | 0.552 | −0.172 | 0.889 | 0.104 |
| Alanine | 0.527 | −0.202 | 0.88 | 0.123 |
| Arginine | 0.345 | −0.382 | 0.808 | 0.331 |
| Asparagine | 0.309 | −0.413 | 0.792 | 0.387 |
| Leucine | 0.442 | −0.293 | 0.849 | 0.204 |
| Glutamate | 0.564 | −0.158 | 0.893 | 0.096 |
| Glutamine | 0.37 | −0.36 | 0.819 | 0.296 |
| Linoleate | 0.127 | −0.548 | 0.702 | 0.733 |
| Lactate | 0.382 | −0.35 | 0.824 | 0.279 |
| Lysine | 0.515 | −0.215 | 0.876 | 0.133 |
| Methionine | 0.309 | −0.413 | 0.792 | 0.387 |
| Proline | 0.394 | −0.339 | 0.829 | 0.263 |
| Serine | 0.442 | −0.293 | 0.849 | 0.204 |
| Threonine | 0.345 | −0.382 | 0.808 | 0.331 |
| Tyrosine | 0.43 | −0.304 | 0.844 | 0.218 |
| Malate | 0.636 | −0.061 | 0.916 | 0.054 ns |
| Myristate | 0.309 | −0.413 | 0.792 | 0.387 |
| Palmitate | 0.297 | −0.423 | 0.787 | 0.407 |
| Succinate | 0.648 | −0.043 | 0.92 | 0.049 * |
| Tryptophan | 0.382 | −0.35 | 0.824 | 0.279 |
| Aspartate | 0.152 | −0.531 | 0.715 | 0.682 |
| Isoleucine | 0.333 | −0.393 | 0.803 | 0.349 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rahat, S.T.; Mäkelä, M.; Nasserinejad, M.; Ikäheimo, T.M.; Hyrkäs-Palmu, H.; Valtonen, R.I.P.; Röning, J.; Sebert, S.; Nieminen, A.I.; Ali, N.; et al. Clinical-Grade Patches as a Medium for Enrichment of Sweat-Extracellular Vesicles and Facilitating Their Metabolic Analysis. Int. J. Mol. Sci. 2023, 24, 7507. https://doi.org/10.3390/ijms24087507
Rahat ST, Mäkelä M, Nasserinejad M, Ikäheimo TM, Hyrkäs-Palmu H, Valtonen RIP, Röning J, Sebert S, Nieminen AI, Ali N, et al. Clinical-Grade Patches as a Medium for Enrichment of Sweat-Extracellular Vesicles and Facilitating Their Metabolic Analysis. International Journal of Molecular Sciences. 2023; 24(8):7507. https://doi.org/10.3390/ijms24087507
Chicago/Turabian StyleRahat, Syeda Tayyiba, Mira Mäkelä, Maryam Nasserinejad, Tiina M. Ikäheimo, Henna Hyrkäs-Palmu, Rasmus I. P. Valtonen, Juha Röning, Sylvain Sebert, Anni I. Nieminen, Nsrein Ali, and et al. 2023. "Clinical-Grade Patches as a Medium for Enrichment of Sweat-Extracellular Vesicles and Facilitating Their Metabolic Analysis" International Journal of Molecular Sciences 24, no. 8: 7507. https://doi.org/10.3390/ijms24087507
APA StyleRahat, S. T., Mäkelä, M., Nasserinejad, M., Ikäheimo, T. M., Hyrkäs-Palmu, H., Valtonen, R. I. P., Röning, J., Sebert, S., Nieminen, A. I., Ali, N., & Vainio, S. (2023). Clinical-Grade Patches as a Medium for Enrichment of Sweat-Extracellular Vesicles and Facilitating Their Metabolic Analysis. International Journal of Molecular Sciences, 24(8), 7507. https://doi.org/10.3390/ijms24087507










