Glycemia-Induced miRNA Changes: A Review
Abstract
1. Introduction
2. The Pathophysiology of Diabetes Mellitus
3. The Pathophysiology of Hypoglycemia
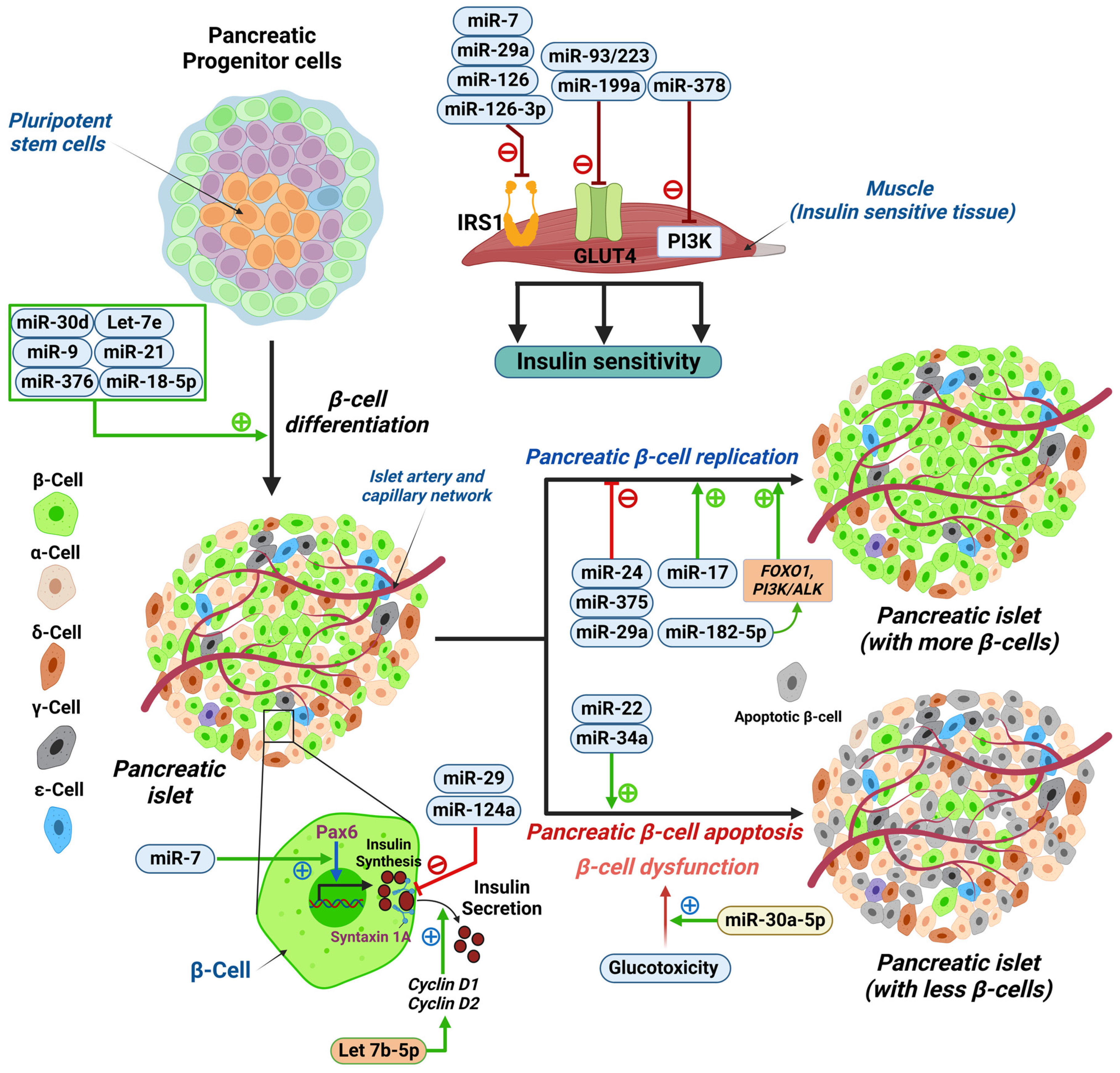
4. miRNA Biology
5. Relationship between Glycemic Changes and miRNAs
5.1. Hypoglycemia-Induced miRNA Changes
5.2. Hyperglycemia-Induced miRNA Changes
5.2.1. Endothelial Dysfunction
5.2.2. Pancreatic Beta-Cell Dysfunction
5.2.3. IR
6. The Implications of miRNAs in Hypoglycemia
7. Future Directions
7.1. miRNAs as Potential Biomarkers for T2DM
7.2. Biomarkers in T2DM Macrovascular Complications
7.3. Biomarkers in T2DM Microvascular Complications
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nutter, C.A.; Kuyumcu-Martinez, M.N. Emerging roles of RNA-binding proteins in diabetes and their therapeutic potential in diabetic complications. Wiley Interdiscip. Rev. RNA. 2018, 9, e1459. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Xing, W.; Xie, L. Regulatory Roles of MicroRNAs in Diabetes. Int. J. Mol. Sci. 2016, 17, 1729. [Google Scholar] [CrossRef] [PubMed]
- Banday, M.Z.; Sameer, A.S.; Nissar, S. Pathophysiology of diabetes: An overview. Avicenna. J. Med. 2020, 10, 174–188. [Google Scholar] [CrossRef] [PubMed]
- Caballero, A.E. The “A to Z” of Managing Type 2 Diabetes in Culturally Diverse Populations. Front. Endocrinol. 2018, 9, 479. [Google Scholar] [CrossRef]
- Kim, M.; Zhang, X. The Profiling and Role of miRNAs in Diabetes Mellitus. J. Diabetes Clin. Res. 2019, 1, 5–23. [Google Scholar]
- Flowers, E.; Allen, I.E.; Kanaya, A.M.; Aouizerat, B.E. Circulating MicroRNAs predict glycemic improvement and response to a behavioral intervention. Biomark. Res. 2021, 9, 65. [Google Scholar] [CrossRef]
- Mononen, N.; Lyytikäinen, L.P.; Seppälä, I.; Mishra, P.P.; Juonala, M.; Waldenberger, M.; Klopp, N.; Illig, T.; Leiviskä, J.; Loo, B.M.; et al. Whole blood microRNA levels associate with glycemic status and correlate with target mRNAs in pathways important to type 2 diabetes. Sci. Rep. 2019, 9, 8887. [Google Scholar] [CrossRef]
- Yang, W.M.; Jeong, H.J.; Park, S.Y.; Lee, W. Saturated fatty acid-induced miR-195 impairs insulin signaling and glycogen metabolism in HepG2 cells. FEBS Lett. 2014, 588, 3939–3946. [Google Scholar] [CrossRef]
- Butler, A.E.; Ramachandran, V.; Cunningham, T.K.; David, R.; Gooderham, N.J.; Benurwar, M.; Dargham, S.R.; Hayat, S.; Sathyapalan, T.; Najafi-Shoushtari, S.H.; et al. Increased MicroRNA Levels in Women With Polycystic Ovarian Syndrome but Without Insulin Resistance: A Pilot Prospective Study. Front. Endocrinol. 2020, 11, 571357. [Google Scholar] [CrossRef]
- Ramanjaneya, M.; Bettahi, I.; Pawar, K.; Halabi, N.M.; Moin, A.S.M.; Sathyapalan, T.; Abou-Samra, A.B.; Atkin, S.L.; Butler, A.E. MicroRNA Changes Up to 24 h following Induced Hypoglycemia in Type 2 Diabetes. Int. J. Mol. Sci. 2022, 23, 14696. [Google Scholar] [CrossRef]
- Agrawal, R.; Durupt, G.; Verma, D.; Montgomery, M.; Vieira-de Abreu, A.; Taylor, C.; Swaminathan, S.; Fisher, S.J. MicroRNA-7a overexpression in VMH restores the sympathoadrenal response to hypoglycemia. JCI Insight 2019, 4, 130521. [Google Scholar] [CrossRef]
- Cryer, P.E. Hypoglycemia-associated autonomic failure in diabetes. Am. J. Physiol. Endocrinol. Metab. 2001, 281, E1115–E1121. [Google Scholar] [CrossRef]
- Kalra, S.; Mukherjee, J.J.; Venkataraman, S.; Bantwal, G.; Shaikh, S.; Saboo, B.; Das, A.K.; Ramachandran, A. Hypoglycemia: The neglected complication. Indian J. Endocrinol. Metab. 2013, 17, 819–834. [Google Scholar] [CrossRef]
- Heller, S.P.; Choudhary, P.; Davies, C.; Emery, C.; Campbell, M.J.; Freeman, J.; Amiel, S.A.; Malik, R.; Frier, B.M.; Allen, K.V.; et al. Risk of hypoglycaemia in types 1 and 2 diabetes: Effects of treatment modalities and their duration. Diabetologia 2007, 50, 1140–1147. [Google Scholar]
- UK Prospective Diabetes Study (UKPDS) Group. Intensive blood-glucose control with sulphonylureas or insulin compared with conventional treatment and risk of complications in patients with type 2 diabetes (UKPDS 33). Lancet 1998, 352, 837–853. [Google Scholar] [CrossRef]
- Nathan, D.M.; Genuth, S.; Lachin, J.; Cleary, P.; Crofford, O.; Davis, M.; Rand, L.; Siebert, C. The effect of intensive treatment of diabetes on the development and progression of long-term complications in insulin-dependent diabetes mellitus. N. Engl. J. Med. 1993, 329, 977–986. [Google Scholar]
- Mirra, P.; Nigro, C.; Prevenzano, I.; Leone, A.; Raciti, G.A.; Formisano, P.; Beguinot, F.; Miele, C. The Destiny of Glucose from a MicroRNA Perspective. Front. Endocrinol. 2018, 9, 46. [Google Scholar] [CrossRef]
- Cryer, P.E. Mechanisms of hypoglycemia-associated autonomic failure and its component syndromes in diabetes. Diabetes 2005, 54, 3592–3601. [Google Scholar] [CrossRef]
- Ranganathan, K.; Sivasankar, V. MicroRNAs-Biology and clinical applications. J. Oral Maxillofac. Pathol. 2014, 18, 229–234. [Google Scholar] [CrossRef]
- Slezak-Prochazka, I.; Durmus, S.; Kroesen, B.J.; van den Berg, A. MicroRNAs, macrocontrol: Regulation of miRNA processing. RNA 2010, 16, 1087–1095. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- Mussa, B.M.; Taneera, J.; Mohammed, A.K.; Srivastava, A.; Mukhopadhyay, D.; Sulaiman, N. Potential role of hypothalamic microRNAs in regulation of FOS and FTO expression in response to hypoglycemia. J. Physiol. Sci. 2019, 69, 981–991. [Google Scholar] [CrossRef] [PubMed]
- Favaro, R.R.; Morales-Prieto, D.M.; Herrmann, J.; Sonnemann, J.; Schleussner, E.; Markert, U.R.; Zorn, T.M.T. Influence of high glucose in the expression of miRNAs and IGF1R signaling pathway in human myometrial explants. Arch. Gynecol. Obstet. 2021, 303, 1513–1522. [Google Scholar] [CrossRef] [PubMed]
- Tam, W.; Ben-Yehuda, D.; Hayward, W.S. bic a novel gene activated by proviral insertions in avian leukosis virus-induced lymphomas is likely to function through its noncoding, R.N.A. Mol. Cell Biol. 1997, 17, 1490–1502. [Google Scholar] [CrossRef]
- Lin, X.; Qin, Y.; Jia, J.; Lin, T.; Lin, X.; Chen, L.; Zeng, H.; Han, Y.; Wu, L.; Huang, S.; et al. MiR-155 Enhances Insulin Sensitivity by Coordinated Regulation of Multiple Genes in Mice. PLoS Genet. 2016, 12, e1006308. [Google Scholar] [CrossRef]
- Zhang, B.-H.; Shen, C.-A.; Zhu, B.-W.; An, H.-Y.; Zheng, B.; Xu, S.-B.; Sun, J.-C.; Sun, P.-C.; Zhang, W.; Wang, J.; et al. Insight into miRNAs related with glucometabolic disorder. Biomed. Pharmacother. 2019, 111, 657–665. [Google Scholar] [CrossRef]
- Ramanjaneya, M.; Priyanka, R.; Bensila, M.; Jerobin, J.; Pawar, K.; Sathyapalan, T.; Abou-Samra, A.B.; Halabi, N.M.; Moin, A.S.M.; Atkin, S.L.; et al. MiRNA and associated inflammatory changes from baseline to hypoglycemia in type 2 diabetes. Front. Endocrinol. 2022, 13, 917041. [Google Scholar] [CrossRef]
- Chen, H.; Lan, H.Y.; Roukos, D.H.; Cho, W.C. Application of microRNAs in diabetes mellitus. J. Endocrinol. 2014, 222, R1–R10. [Google Scholar] [CrossRef]
- Zhang, W.; Yan, L.; Li, Y.; Chen, W.; Hu, N.; Wang, H.; Ou, H. Roles of miRNA-24 in regulating endothelial nitric oxide synthase expression and vascular endothelial cell proliferation. Mol. Cell. Biochem. 2015, 405, 281–289. [Google Scholar] [CrossRef]
- Ait-Aissa, K.; Nguyen, Q.M.; Gabani, M.; Kassan, A.; Kumar, S.; Choi, S.-K.; Gonzalez, A.A.; Khataei, T.; Sahyoun, A.M.; Chen, C.; et al. MicroRNAs and obesity-induced endothelial dysfunction: Key paradigms in molecular therapy. Cardiovasc. Diabetol. 2020, 19, 136. [Google Scholar] [CrossRef]
- Li, H.T.; Wang, J.; Li, S.F.; Cheng, L.; Tang, W.Z.; Feng, Y.G. Upregulation of microRNA-24 causes vasospasm following subarachnoid hemorrhage by suppressing the expression of endothelial nitric oxide synthase. Mol. Med. Rep. 2018, 18, 1181–1187. [Google Scholar] [CrossRef]
- Chawla, A.; Chawla, R.; Jaggi, S. Microvasular and macrovascular complications in diabetes mellitus: Distinct or continuum? Indian J. Endocrinol. Metab. 2016, 20, 546–551. [Google Scholar] [CrossRef]
- Fowler, M.J. Microvascular and Macrovascular Complications of Diabetes. Clin. Diabetes 2008, 26, 77–82. [Google Scholar] [CrossRef]
- Shantikumar, S.; Caporali, A.; Emanueli, C. Role of microRNAs in diabetes and its cardiovascular complications. Cardiovasc. Res. 2012, 93, 583–593. [Google Scholar] [CrossRef]
- Santovito, D.; De Nardis, V.; Marcantonio, P.; Mandolini, C.; Paganelli, C.; Vitale, E.; Buttitta, F.; Bucci, M.; Mezzetti, A.; Consoli, A.; et al. Plasma exosome microRNA profiling unravels a new potential modulator of adiponectin pathway in diabetes: Effect of glycemic control. J. Clin. Endocrinol. Metab. 2014, 99, E1681–E1685. [Google Scholar] [CrossRef]
- Song, Y.; Wu, L.; Li, M.; Xiong, X.; Fang, Z.; Zhou, J.; Yan, G.; Chen, X.; Yang, J.; Li, Y. Down-regulation of MicroRNA-592 in obesity contributes to hyperglycemia and insulin resistance. EBioMedicine 2019, 42, 494–503. [Google Scholar] [CrossRef]
- He, X.; Kuang, G.; Wu, Y.; Ou, C. Emerging roles of exosomal miRNAs in diabetes mellitus. Clin. Transl. Med. 2021, 11, e468. [Google Scholar] [CrossRef]
- Villegas-Valverde, C.C.; Kokuina, E.; Breff-Fonseca, M.C. Strengthening National Health Priorities for Diabetes Prevention and Management. MEDICC Rev. 2018, 20, 5. [Google Scholar]
- Zheng, Y.; Wang, Z.; Tu, Y.; Shen, H.; Dai, Z.; Lin, J.; Zhou, Z. miR-101a and miR-30b contribute to inflammatory cytokine-mediated β-cell dysfunction. Lab Investig. 2015, 95, 1387–1397. [Google Scholar] [CrossRef]
- Belgardt, B.F.; Ahmed, K.; Spranger, M.; Latreille, M.; Denzler, R.; Kondratiuk, N.; von Meyenn, F.; Villena, F.N.; Herrmanns, K.; Bosco, D.; et al. The microRNA-200 family regulates pancreatic beta cell survival in type 2 diabetes. Nat. Med. 2015, 21, 619–627. [Google Scholar] [CrossRef]
- Lin, X.; Guan, H.; Huang, Z.; Liu, J.; Li, H.; Wei, G.; Cao, X.; Li, Y. Downregulation of Bcl-2 expression by miR-34a mediates palmitate-induced Min6 cells apoptosis. J. Diabetes Res. 2014, 2014, 258695. [Google Scholar] [CrossRef] [PubMed]
- Liao, X.; Xue, H.; Wang, Y.C.; Nazor, K.L.; Guo, S.; Trivedi, N.; Peterson, S.E.; Liu, Y.; Loring, J.F.; Laurent, L.C. Matched miRNA and mRNA signatures from an hESC-based in vitro model of pancreatic differentiation reveal novel regulatory interactions. J. Cell. Sci. 2013, 126 Pt 17, 3848–3861. [Google Scholar]
- Gomes, P.R.; Graciano, M.F.; Pantaleão, L.C.; Rennó, A.L.; Rodrigues, S.C.; Velloso, L.A.; Latorraca, M.Q.; Carpinelli, A.R.; Anhê, G.F.; Bordin, S. Long-term disruption of maternal glucose homeostasis induced by prenatal glucocorticoid treatment correlates with miR-29 upregulation. Am. J. Physiol. Endocrinol. Metab. 2014, 306, E109–E120. [Google Scholar] [CrossRef] [PubMed]
- Locke, J.M.; da Silva Xavier, G.; Dawe, H.R.; Rutter, G.A.; Harries, L.W. Increased expression of miR-187 in human islets from individuals with type 2 diabetes is associated with reduced glucose-stimulated insulin secretion. Diabetologia 2014, 57, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.W.; You, Y.H.; Jung, S.; Suh-Kim, H.; Lee, I.K.; Cho, J.H.; Yoon, K.H. miRNA-30a-5p-mediated silencing of Beta2/NeuroD expression is an important initial event of glucotoxicity-induced beta cell dysfunction in rodent models. Diabetologia 2013, 56, 847–855. [Google Scholar] [CrossRef]
- Weale, C.J.; Matshazi, D.M.; Davids, S.F.G.; Raghubeer, S.; Erasmus, R.T.; Kengne, A.P.; Davison, G.M.; Matsha, T.E. Expression Profiles of Circulating microRNAs in South African Type 2 Diabetic Individuals on Treatment. Front. Genet. 2021, 12, 702410. [Google Scholar] [CrossRef]
- Li, Z.Y.; Na, H.M.; Peng, G.; Pu, J.; Liu, P. Alteration of microRNA expression correlates to fatty acid-mediated insulin resistance in mouse myoblasts. Mol. Biosyst. 2011, 7, 871–877. [Google Scholar] [CrossRef]
- Yang, W.M.; Jeong, H.J.; Park, S.Y.; Lee, W. Induction of miR-29a by saturated fatty acids impairs insulin signaling and glucose uptake through translational repression of IRS-1 in myocytes. FEBS Lett. 2014, 588, 2170–2176. [Google Scholar] [CrossRef]
- Fernandez-Twinn, D.S.; Alfaradhi, M.Z.; Martin-Gronert, M.S.; Duque-Guimaraes, D.E.; Piekarz, A.; Ferland-McCollough, D.; Bushell, M.; Ozanne, S.E. Downregulation of IRS-1 in adipose tissue of offspring of obese mice is programmed cell-autonomously through post-transcriptional mechanisms. Mol. Metab. 2014, 3, 325–333. [Google Scholar] [CrossRef]
- Aljaibeji, H.; Elemam, N.M.; Mohammed, A.K.; Hasswan, H.; Thahyabat, M.A.; Alkhayyal, N.; Sulaiman, N.; Taneera, J. Let7b-5p is Upregulated in the Serum of Emirati Patients with Type 2 Diabetes and Regulates Insulin Secretion in INS-1 Cells. Exp. Clin. Endocrinol. Diabetes 2022, 130, 22–29. [Google Scholar] [CrossRef]
- Ji, H.; Fan, L.; Shan, A.; Wang, W.; Ning, G.; Cao, Y.; Jiang, X. Let7b-5p inhibits insulin secretion and decreases pancreatic β-cell mass in mice. Mol. Cell. Endocrinol. 2022, 540, 111506. [Google Scholar] [CrossRef]
- Elemam, N.M.; Hasswan, H.; Aljaibeji, H.; Sulaiman, N. Circulating Soluble ACE2 and Upstream microRNA Expressions in Serum of Type 2 Diabetes Mellitus Patients. Int. J. Mol. Sci. 2021, 22, 5263. [Google Scholar] [CrossRef]
- Liu, W.; Cao, H.; Ye, C.; Chang, C.; Lu, M.; Jing, Y.; Zhang, D.; Yao, X.; Duan, Z.; Xia, H.; et al. Hepatic miR-378 targets p110α and controls glucose and lipid homeostasis by modulating hepatic insulin signalling. Nat. Commun. 2014, 5, 5684. [Google Scholar] [CrossRef]
- Yan, S.T.; Li, C.L.; Tian, H.; Li, J.; Pei, Y.; Liu, Y.; Gong, Y.-P.; Fang, F.-S.; Sun, B.-R. MiR-199a is overexpressed in plasma of type 2 diabetes patients which contributes to type 2 diabetes by targeting GLUT4. Mol. Cell. Biochem. 2014, 397, 45–51. [Google Scholar] [CrossRef]
- Sempere, L.F.; Azmi, A.S.; Moore, A. microRNA-based diagnostic and therapeutic applications in cancer medicine. Wiley Interdiscip. Rev. RNA 2021, 12, e1662. [Google Scholar] [CrossRef]
- Dagogo-Jack, S.E.; Craft, S.; Cryer, P.E. Hypoglycemia-associated autonomic failure in insulin-dependent diabetes mellitus. Recent antecedent hypoglycemia reduces autonomic responses to, symptoms of, and defense against subsequent hypoglycemia. J. Clin. Investig. 1993, 91, 819–828. [Google Scholar] [CrossRef]
- Rickels, M.R. Hypoglycemia-associated autonomic failure, counterregulatory responses, and therapeutic options in type 1 diabetes. Ann. N. Y. Acad. Sci. 2019, 1454, 68–79. [Google Scholar] [CrossRef]
- Krammer, T.L.; Mayr, M.; Hackl, M. microRNAs as promising biomarkers of platelet activity in antiplatelet therapy monitoring. Int. J. Mol. Sci. 2020, 21, 3477. [Google Scholar] [CrossRef]
- Pordzik, J.; Pisarz, K.; De Rosa, S.; Jones, A.D.; Eyileten, C.; Indolfi, C.; Malek, L.; Postula, M. The Potential Role of Platelet-Related microRNAs in the Development of Cardiovascular Events in High-Risk Populations, Including Diabetic Patients: A Review. Front. Endocrinol. 2018, 9, 74. [Google Scholar] [CrossRef]
- Kraczkowska, W.; Stachowiak, L.; Pławski, A.; Jagodziński, P.P. Circulating miRNA as potential biomarkers for diabetes mellitus type 2: Should we focus on searching for sex differences? J. Appl. Genet. 2022, 63, 293–303. [Google Scholar] [CrossRef]
- Pordzik, J.; Jakubik, D.; Jarosz-Popek, J.; Wicik, Z.; Eyileten, C.; De Rosa, S.; Indolfi, C.; Siller-Matula, J.M.; Czajka, P.; Postula, M. Significance of circulating microRNAs in diabetes mellitus type 2 and platelet reactivity: Bioinformatic analysis and review. Cardiovasc. Diabetol. 2019, 18, 113. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Zhu, J.; Han, W.; Jiang, X.; Xu, M.; Zhao, Y.; Dong, Q.; Pang, Z.; Guan, Q.; Gao, L.; et al. Significance of serum microRNAs in pre-diabetes and newly diagnosed type 2 diabetes: A clinical study. Acta Diabetol. 2011, 48, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; Huang, K.H.; Lee, M.J.; Galas, D.J.; Wang, K. The microRNA spectrum in 12 body fluids. Clin. Chem. 2010, 56, 1733–1741. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Guo, F. MicroRNAs and type 2 diabetes. ExRNA 2019, 1, 36. [Google Scholar] [CrossRef]
- Filios, S.R.; Shalev, A. β-Cell MicroRNAs: Small but Powerful. Diabetes 2015, 64, 3631–3644. [Google Scholar] [CrossRef]
- Wen, Z.; Zou, X.; Xie, X.; Zheng, S.; Chen, X.; Zhu, K.; Dong, S.; Liang, J.; Huang, X.; Liu, D.; et al. Association of Polymorphisms in miRNA Processing Genes With Type 2 Diabetes Mellitus and Its Vascular Complications in a Southern Chinese Population. Front. Endocrinol. 2019, 10, 461. [Google Scholar] [CrossRef]
- Chang, Y.C.; Chang, E.Y.; Chuang, L.M. Recent progress in the genetics of diabetic microvascular complications. World J. Diabetes 2015, 6, 715–725. [Google Scholar] [CrossRef]
- Yan, S.; Wang, T.; Huang, S.; Di, Y.; Huang, Y.; Liu, X.; Luo, Z.; Han, W.; An, B. Differential expression of microRNAs in plasma of patients with prediabetes and newly diagnosed type 2 diabetes. Acta Diabetol. 2016, 53, 693–702. [Google Scholar] [CrossRef]
- Zhang, T.; Li, L.; Shang, Q.; Lv, C.; Wang, C.; Su, B. Circulating miR-126 is a potential biomarker to predict the onset of type 2 diabetes mellitus in susceptible individuals. Biochem. Biophys. Res. Commun. 2015, 463, 60–63. [Google Scholar] [CrossRef]
- Chien, H.Y.; Chen, C.Y.; Chiu, Y.H.; Lin, Y.C.; Li, W.C. Differential microRNA Profiles Predict Diabetic Nephropathy Progression in Taiwan. Int. J. Med. Sci. 2016, 13, 457–465. [Google Scholar] [CrossRef]
- Mukhadi, S.; Hull, R.; Mbita, Z.; Dlamini, Z. The Role of MicroRNAs in Kidney Disease. Noncoding RNA 2015, 1, 192–221. [Google Scholar] [CrossRef]
- An, Y.; Zhang, C.; Xu, F.; Li, W.; Zeng, C.; Xie, L.; Liu, Z. Increased urinary miR-196a level predicts the progression of renal injury in patients with diabetic nephropathy. Nephrol. Dial. Transplant. 2020, 35, 1009–1016. [Google Scholar] [CrossRef]
- Ciccacci, C.; Latini, A.; Colantuono, A.; Politi, C.; D’Amato, C.; Greco, C.; Rinaldi, M.E.; Lauro, D.; Novelli, G.; Spallone, V.; et al. Expression study of candidate miRNAs and evaluation of their potential use as biomarkers of diabetic neuropathy. Epigenomics 2020, 12, 575–585. [Google Scholar] [CrossRef]
- Zang, J.; Maxwell, A.P.; Simpson, D.A.; McKay, G.J. Differential Expression of Urinary Exosomal MicroRNAs miR-21-5p and miR-30b-5p in Individuals with Diabetic Kidney Disease. Sci. Rep. 2019, 9, 10900. [Google Scholar] [CrossRef]
- Smit-McBride, Z.; Morse, L.S. MicroRNA and diabetic retinopathy-biomarkers and novel therapeutics. Ann. Transl. Med. 2021, 9, 1280. [Google Scholar] [CrossRef]
- Yuan, Y.; Peng, W.; Liu, Y.; Xu, Z. Circulating miR-130 and its target PPAR-γ may be potential biomarkers in patients of coronary artery disease with type 2 diabetes mellitus. Mol. Genet. Genom. Med. 2019, 7, e909. [Google Scholar] [CrossRef]
- Bielska, A.; Niemira, M.; Kretowski, A. Recent Highlights of Research on miRNAs as Early Potential Biomarkers for Cardiovascular Complications of Type 2 Diabetes Mellitus. Int. J. Mol. Sci. 2021, 22, 3153. [Google Scholar] [CrossRef]
- Kuschnerus, K.; Straessler, E.T.; Müller, M.F.; Lüscher, T.F.; Landmesser, U.; Kränkel, N. Increased Expression of miR-483-3p Impairs the Vascular Response to Injury in Type 2 Diabetes. Diabetes 2019, 68, 349–360. [Google Scholar] [CrossRef]
- Angelescu, M.A.; Andronic, O.; Dima, S.O.; Popescu, I.; Meivar-Levy, I.; Ferber, S.; Lixandru, D. miRNAs as Biomarkers in Diabetes: Moving towards Precision Medicine. Int. J. Mol. Sci. 2022, 23, 12843. [Google Scholar] [CrossRef]
- Assmann, T.S.; Recamonde-Mendoza, M.; de Souza, B.M.; Bauer, A.C.; Crispim, D. MicroRNAs and diabetic kidney disease: Systematic review and bioinformatic analysis. Mol. Cell. Endocrinol. 2018, 477, 90–102. [Google Scholar] [CrossRef]
- Jordan, N.P.; Nicholson, M.L.; Hosgood, S.A. MicroRNA-126-3p is Downregulated in Human Kidneys in a Model of Reperfusion Injury. Kidney Int. Rep. 2020, 5, 2357–2360. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.H.; Huang, S.; Britton, W.R.; Chen, J. MicroRNAs in Vascular Eye Diseases. Int. J. Mol. Sci. 2020, 21, 649. [Google Scholar] [CrossRef] [PubMed]
- Greco, M.; Chiefari, E.; Accattato, F.; Corigliano, D.M.; Arcidiacono, B.; Mirabelli, M.; Liguori, R.; Brunetti, F.S.; Pullano, S.A.; Scorcia, V.; et al. MicroRNA-1281 as a Novel Circulating Biomarker in Patients With Diabetic Retinopathy. Front. Endocrinol. 2020, 11, 528. [Google Scholar] [CrossRef] [PubMed]
- Santovito, D.; Toto, L.; De Nardis, V.; Marcantonio, P.; D’Aloisio, R.; Mastropasqua, A.; De Cesare, D.; Bucci, M.; Paganelli, C.; Natarelli, L.; et al. Plasma microRNA signature associated with retinopathy in patients with type 2 diabetes. Sci. Rep. 2021, 11, 4136. [Google Scholar] [CrossRef]
- Jiang, Q.; Lyu, X.-M.; Yuan, Y.; Wang, L. Plasma miR-21 expression: An indicator for the severity of Type 2 diabetes with diabetic retinopathy. Biosci. Rep. 2017, 37, BSR20160589. [Google Scholar] [CrossRef]
| Biomarker | Sample | Diagnostic Value | Reference |
|---|---|---|---|
| miR-126 | Peripheral whole blood | Differentiates between T2DM with CAD from T2DM | [77] |
| miR-130 | Serum | Differentiates between T2DM with CAD from CAD patients | [76] |
| miR-210 | Plasma | Differentiates between T2DM with CAD from T2DM | [77] |
| miR-483-3p | Peripheral blood mononuclear cell (PBMC) | Differentiates between T2DM with CAD from CAD | [77,78] |
| Biomarker | Sample | Diagnostic Value | Reference |
|---|---|---|---|
| miR-21 | Plasma | Specify severity of diabetic retinopathy in T2DM patients | [85] |
| miR-21-5p | Urinary exosome | Indicator of renal function | [74] |
| miR-30b-5p | Urinary exosome | Indicator of renal function | [74] |
| miR-196a, miR-121 | Urine | Prognostic marker for renal fibrosis patients with DN | [72] |
| miR-320b, miR25-3p and miR-495 | Plasma exosomes | Useful for diagnosing patients with diabetic retinopathy in T2DM | [84] |
| miR-128a, miR-155 and miR-499 | PBMCs | Monitoring progression | [73] |
| miR-1281 | Serum | Early diagnosis of diabetic retinopathy in T2DM patients | [83] |
| miR-320c | Peripheral blood | Monitoring progression of DN | [72] |
| miR-192 | Peripheral blood | Early detection of T2DM DN | [70] |
| miR-29a/b/c | Peripheral blood | Potential biomarker, especially miR-29a | [67] |
| miR-429 | Plasma | Potential marker for proteinuria and kidney function | [79] |
| miR-126, miR 29 | Peripheral blood | Indicator of renal dysfunction | [79] |
| miR-199a-3p | Plasma | Indicator of neuropathy progression | [79] |
| miR-146 | Plasma | Linked to peripheral neuropathy | [79] |
| miR-21, miR-320-a, mir-320-b | Post-mortem vitreous humor | Linked to DR development and pathogenesis | [79] |
| let-7a-5p | Serum | Linked to retinal proliferation | [79] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Mahayni, S.; Ali, M.; Khan, M.; Jamsheer, F.; Moin, A.S.M.; Butler, A.E. Glycemia-Induced miRNA Changes: A Review. Int. J. Mol. Sci. 2023, 24, 7488. https://doi.org/10.3390/ijms24087488
Al-Mahayni S, Ali M, Khan M, Jamsheer F, Moin ASM, Butler AE. Glycemia-Induced miRNA Changes: A Review. International Journal of Molecular Sciences. 2023; 24(8):7488. https://doi.org/10.3390/ijms24087488
Chicago/Turabian StyleAl-Mahayni, Sara, Mohamed Ali, Muhammad Khan, Fatema Jamsheer, Abu Saleh Md Moin, and Alexandra E. Butler. 2023. "Glycemia-Induced miRNA Changes: A Review" International Journal of Molecular Sciences 24, no. 8: 7488. https://doi.org/10.3390/ijms24087488
APA StyleAl-Mahayni, S., Ali, M., Khan, M., Jamsheer, F., Moin, A. S. M., & Butler, A. E. (2023). Glycemia-Induced miRNA Changes: A Review. International Journal of Molecular Sciences, 24(8), 7488. https://doi.org/10.3390/ijms24087488






