Testing the Utility of Polygenic Risk Scores for Type 2 Diabetes and Obesity in Predicting Metabolic Changes in a Prediabetic Population: An Observational Study
Abstract
1. Introduction
2. Results
2.1. Subjects’ Characteristics and Metabolic Parameters at Baseline and Follow-Up
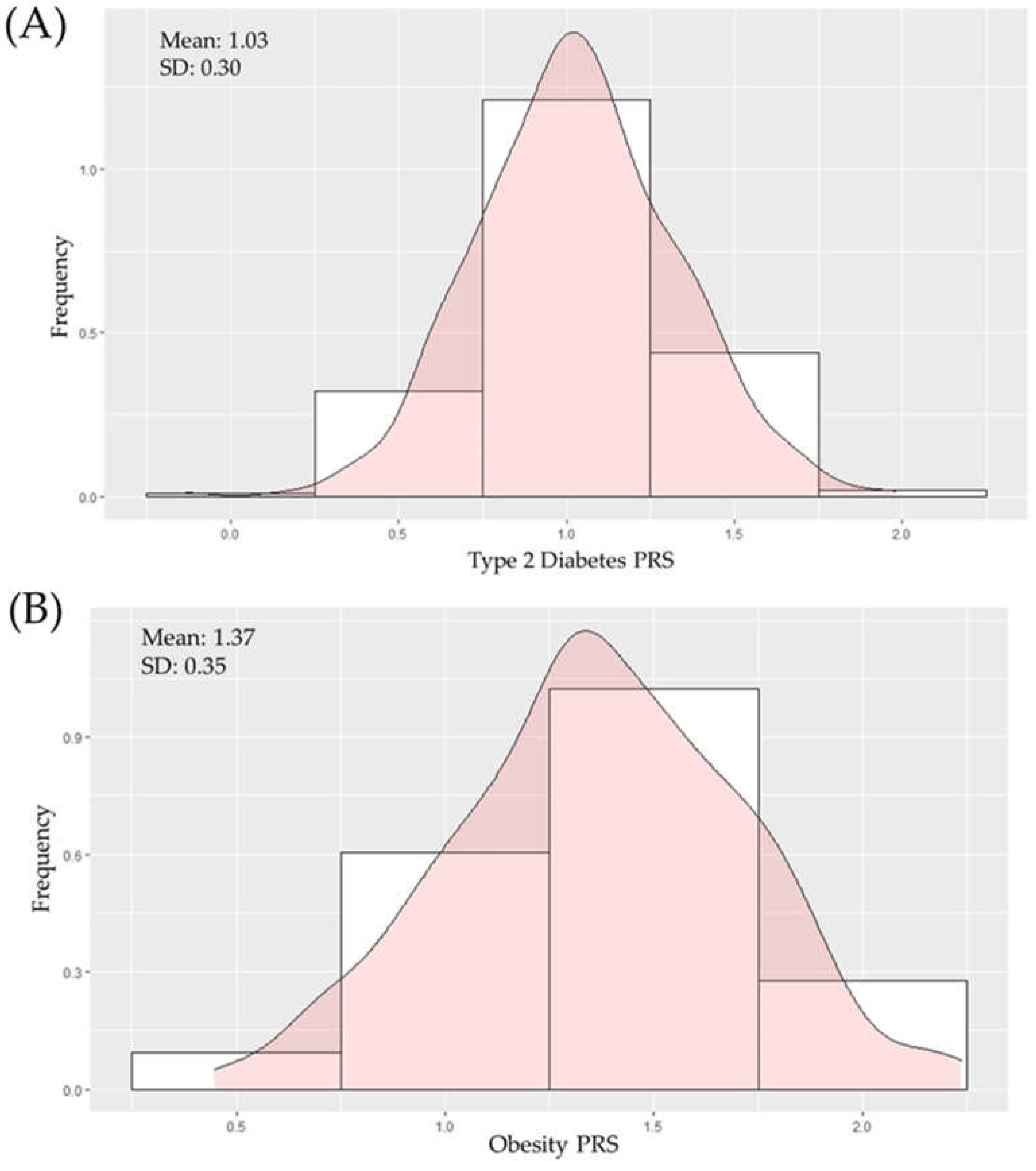
2.2. Construction of Polygenic Scores for T2D
2.3. Association between T2D PRS and Metabolic Parameters
2.4. Association between Obesity PRS and Metabolic Parameters
2.5. Association of Genotypes’ Frequencies with Changes in Metabolic Parameters
3. Materials and Methods
3.1. Study Design and Participants
3.2. Sample Collection and Measurement
3.3. Genotyping
3.4. Risk Score Analysis
3.5. Statistical Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bansal, N. Prediabetes Diagnosis and Treatment: A Review. World J. Diabetes 2015, 6, 296. [Google Scholar] [CrossRef]
- Zand, A. Prediabetes: Why Should We Care? Mech. ACTION 2018, 9, 289. [Google Scholar] [CrossRef] [PubMed]
- Galaviz, K.I.; Narayan, K.M.V.; Lobelo, F.; Weber, M.B. Lifestyle and the Prevention of Type 2 Diabetes: A Status Report. Am. J. Lifestyle Med. 2018, 12, 4–20. [Google Scholar] [CrossRef] [PubMed]
- Hostalek, U. Global Epidemiology of Prediabetes-Present and Future Perspectives. Clin. Diabetes Endocrinol. 2019, 5, 5. [Google Scholar] [CrossRef] [PubMed]
- Ford, E.S.; Zhao, G.; Li, C. Pre-Diabetes and the Risk for Cardiovascular Disease: A Systematic Review of the Evidence. J. Am. Coll. Cardiol. 2010, 55, 1310–1317. [Google Scholar] [CrossRef]
- Halban, P.A.; Polonsky, K.S.; Bowden, D.W.; Hawkins, M.A.; Ling, C.; Mather, K.J.; Powers, A.C.; Rhodes, C.J.; Sussel, L.; Weir, G.C. β-Cell Failure in Type 2 Diabetes: Postulated Mechanisms and Prospects for Prevention and Treatment. J. Clin. Endocrinol. Metab. 2014, 99, 1983–1992. [Google Scholar] [CrossRef]
- Selvin, E.; Lazo, M.; Chen, Y.; Shen, L.; Rubin, J.; McEvoy, J.W.; Hoogeveen, R.C.; Sharrett, A.R.; Ballantyne, C.M.; Coresh, J. Diabetes Mellitus, Prediabetes, and Incidence of Subclinical Myocardial Damage. Circulation 2014, 130, 1374–1382. [Google Scholar] [CrossRef]
- Yun, J.-S.; Jung, S.-H.; Shivakumar, M.; Xiao, B.; Khera, A.V.; Won, H.-H.; Kim, D. Polygenic Risk for Type 2 Diabetes, Lifestyle, Metabolic Health, and Cardiovascular Disease: A Prospective UK Biobank Study. Cardiovasc. Diabetol. 2022, 21, 131. [Google Scholar] [CrossRef]
- World Obesity Atlas. 2022. Available online: https://www.worldobesity.org/resources/resource-library/world-obesity-atlas-2022 (accessed on 15 August 2022).
- Sun, J.; Liu, Z.; Zhang, Z.; Zeng, Z.; Kang, W. The Correlation of Prediabetes and Type 2 Diabetes With Adiposity in Adults. Front. Nutr. 2022, 9, 818263. [Google Scholar] [CrossRef]
- Miao, Z.; Alvarez, M.; Ko, A.; Bhagat, Y.; Rahmani, E.; Jew, B.; Heinonen, S.; Muñoz-Hernandez, L.L.; Herrera-Hernandez, M.; Aguilar-Salinas, C.; et al. The Causal Effect of Obesity on Prediabetes and Insulin Resistance Reveals the Important Role of Adipose Tissue in Insulin Resistance. PLOS Genet. 2020, 16, e1009018. [Google Scholar] [CrossRef]
- Bycroft, C.; Freeman, C.; Petkova, D.; Band, G.; Elliott, L.T.; Sharp, K.; Motyer, A.; Vukcevic, D.; Delaneau, O.; O’Connell, J.; et al. The UK Biobank Resource with Deep Phenotyping and Genomic Data. Nature 2018, 562, 203–209. [Google Scholar] [CrossRef]
- Zeng, P. Statistical Analysis for Genome-Wide Association Study. J. Biomed. Res. 2015, 29, 285. [Google Scholar] [CrossRef]
- Golan, D.; Lander, E.S.; Rosset, S. Measuring Missing Heritability: Inferring the Contribution of Common Variants. Proc. Natl. Acad. Sci. USA 2014, 111, E5272–E5281. [Google Scholar] [CrossRef] [PubMed]
- Bailey, J.N.C.; Igo, R.P. Genetic Risk Scores. Curr. Protoc. Hum. Genet. 2016, 91, 1291–1299. [Google Scholar] [CrossRef]
- Ahlqvist, E.; Storm, P.; Käräjämäki, A.; Martinell, M.; Dorkhan, M.; Carlsson, A.; Vikman, P.; Prasad, R.B.; Aly, D.M.; Almgren, P.; et al. Novel Subgroups of Adult-Onset Diabetes and Their Association with Outcomes: A Data-Driven Cluster Analysis of Six Variables. Lancet Diabetes Endocrinol. 2018, 6, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Khera, A.V.; Chaffin, M.; Aragam, K.G.; Haas, M.E.; Roselli, C.; Choi, S.H.; Natarajan, P.; Lander, E.S.; Lubitz, S.A.; Ellinor, P.T.; et al. Genome-Wide Polygenic Scores for Common Diseases Identify Individuals with Risk Equivalent to Monogenic Mutations. Nat. Genet. 2018, 50, 1219–1224. [Google Scholar] [CrossRef] [PubMed]
- Padilla-Martínez, F.; Collin, F.; Kwasniewski, M.; Kretowski, A. Systematic Review of Polygenic Risk Scores for Type 1 and Type 2 Diabetes. Int. J. Mol. Sci. 2020, 21, 1703. [Google Scholar] [CrossRef] [PubMed]
- Li, J.H.; Szczerbinski, L.; Dawed, A.Y.; Kaur, V.; Todd, J.N.; Pearson, E.R.; Florez, J.C. A Polygenic Score for Type 2 Diabetes Risk Is Associated With Both the Acute and Sustained Response to Sulfonylureas. Diabetes 2021, 70, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Vassy, J.L.; Hivert, M.-F.; Porneala, B.; Dauriz, M.; Florez, J.C.; Dupuis, J.; Siscovick, D.S.; Fornage, M.; Rasmussen-Torvik, L.J.; Bouchard, C.; et al. Polygenic Type 2 Diabetes Prediction at the Limit of Common Variant Detection. Diabetes 2014, 63, 2172–2182. [Google Scholar] [CrossRef]
- Lall, K.; Mägi, R.; Morris, A.; Metspalu, A.; Fischer, K. Personalized Risk Prediction for Type 2 Diabetes: The Potential of Genetic Risk Scores. Genet. Med. 2016, 19, 322–329. [Google Scholar] [CrossRef]
- Tremblay, J.; Haloui, M.; Attaoua, R.; Tahir, R.; Hishmih, C.; Harvey, F.; Marois-Blanchet, F.-C.; Long, C.; Simon, P.; Santucci, L.; et al. Polygenic Risk Scores Predict Diabetes Complications and Their Response to Intensive Blood Pressure and Glucose Control. Diabetologia 2021, 64, 2012–2025. [Google Scholar] [CrossRef] [PubMed]
- Shaked, A.; Loza, B.; Olthoff, K.; Keating, B. Testing the Application of Polygenic Risk Scores in the Transplant Setting–Relevance for Precision Medicine. Clin. Transl. Med. 2022, 12, e1009. [Google Scholar] [CrossRef] [PubMed]
- Wagner, R.; Heni, M.; Tabák, A.G.; Machann, J.; Schick, F.; Randrianarisoa, E.; Hrabě de Angelis, M.; Birkenfeld, A.L.; Stefan, N.; Peter, A.; et al. Pathophysiology-Based Subphenotyping of Individuals at Elevated Risk for Type 2 Diabetes. Nat. Med. 2021, 27, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Szczerbinski, L. Polish Registry of Diabetes (PolReD). Identifier NCT04657367; 2020. Available online: https://clinicaltrials.gov/ct2/show/NCT04657367 (accessed on 2 November 2022).
- Sidorkiewicz, I.; Niemira, M.; Maliszewska, K.; Erol, A.; Bielska, A.; Szalkowska, A.; Adamska-Patruno, E.; Szczerbinski, L.; Gorska, M.; Kretowski, A. Circulating MiRNAs as a Predictive Biomarker of the Progression from Prediabetes to Diabetes: Outcomes of a 5-Year Prospective Observational Study. J. Clin. Med. 2020, 9, 2184. [Google Scholar] [CrossRef]
- Smushkin, G.; Vella, A. What Is Type 2 Diabetes? Med. Abingdon Engl. UK Ed 2010, 38, 597–601. [Google Scholar] [CrossRef] [PubMed]
- Panuganti, K.K.; Nguyen, M.; Kshirsagar, R.K. Obesity; StatPearls Publishing: Tampa, FL, USA, 2022. [Google Scholar]
- Choi, S.W.; Mak, T.S.-H.; O’Reilly, P.F. Tutorial: A Guide to Performing Polygenic Risk Score Analyses. Nat. Protoc. 2020, 15, 2759–2772. [Google Scholar] [CrossRef]
- Babb de Villiers, C.; Kroese, M.; Moorthie, S. Understanding Polygenic Models, Their Development and the Potential Application of Polygenic Scores in Healthcare. J. Med. Genet. 2020, 57, 725–732. [Google Scholar] [CrossRef]
- Type 2 Diabetes Knowledge Portal-Home. Available online: https://t2d.hugeamp.org/ (accessed on 21 June 2022).
- R Core Team R: A Language and Environment for Statistical Computing. R Found. Stat. Comput. Vienna, Austria, 2021. Available online: https://www.R-project.org/ (accessed on 2 November 2022).
- Aldossari, K.K.; Aldiab, A.; Al-Zahrani, J.M.; Al-Ghamdi, S.H.; Abdelrazik, M.; Batais, M.A.; Javad, S.; Nooruddin, S.; Razzak, H.A.; El-Metwally, A. Prevalence of Prediabetes, Diabetes, and Its Associated Risk Factors among Males in Saudi Arabia: A Population-Based Survey. J. Diabetes Res. 2018, 2018, 2194604. [Google Scholar] [CrossRef]
- White, K.A.M.; Daneshvari, S.; Lilyquist, J.; Luo, L.; Steffen, L.E.; Bivin, A.; Gurule, N.; Ducasa, G.M.; Torres, S.M.; Lindeman, R.; et al. Prediabetes: The Variation between HbA1c and Fasting Plasma Glucose. Int. J. Diabetol. Vasc. Dis. Res. 2015, Suppl. S2, 001. [Google Scholar] [CrossRef][Green Version]
- Hüls, A.; Krämer, U.; Carlsten, C.; Schikowski, T.; Ickstadt, K.; Schwender, H. Comparison of Weighting Approaches for Genetic Risk Scores in Gene-Environment Interaction Studies. BMC Genet. 2017, 18, 115. [Google Scholar] [CrossRef]
- Why Some Patients Don’t Take Prediabetes Seriously. Available online: https://www.ama-assn.org/delivering-care/diabetes/why-some-patients-dont-take-prediabetes-seriously (accessed on 14 September 2022).
- Ács, P.; Veress, R.; Rocha, P.; Dóczi, T.; Raposa, B.L.; Baumann, P.; Ostojic, S.; Pérmusz, V.; Makai, A. Criterion Validity and Reliability of the International Physical Activity Questionnaire–Hungarian Short Form against the RM42 Accelerometer. BMC Public Health 2021, 21, 381. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Siddiqui, W.J. Cholesterol Levels. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Pappan, N.; Rehman, A. Dyslipidemia. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Al Amri, T.; Bahijri, S.; Al-Raddadi, R.; Ajabnoor, G.; Al Ahmadi, J.; Jambi, H.; Borai, A.; Tuomilehto, J. The Association Between Prediabetes and Dyslipidemia Among Attendants of Primary Care Health Centers in Jeddah, Saudi Arabia. Diabetes Metab. Syndr. Obes. Targets Ther. 2019, 12, 2735–2743. [Google Scholar] [CrossRef] [PubMed]
- Hadi Alijanvand, M.; Aminorroaya, A.; Kazemi, I.; Amini, M.; Aminorroaya Yamini, S.; Mansourian, M. Prevalence and Predictors of Prediabetes and Its Coexistence with High Blood Pressure in First-Degree Relatives of Patients with Type 2 Diabetes: A 9-Year Cohort Study. J. Res. Med. Sci. Off. J. Isfahan Univ. Med. Sci. 2020, 25, 31. [Google Scholar] [CrossRef]
- Wagner, R.; Thorand, B.; Osterhoff, M.A.; Müller, G.; Böhm, A.; Meisinger, C.; Kowall, B.; Rathmann, W.; Kronenberg, F.; Staiger, H.; et al. Family History of Diabetes Is Associated with Higher Risk for Prediabetes: A Multicentre Analysis from the German Center for Diabetes Research. Diabetologia 2013, 56, 2176–2180. [Google Scholar] [CrossRef]
- Man, B.; Schwartz, A.; Pugach, O.; Xia, Y.; Gerber, B. A Clinical Diabetes Risk Prediction Model for Prediabetic Women with Prior Gestational Diabetes. PLoS ONE 2021, 16, e0252501. [Google Scholar] [CrossRef] [PubMed]
- Baker, C.F.; Overvad, K.; Dahm, C.C. Lean Body Mass and Risk of Type 2 Diabetes-a Danish Cohort Study. J. Diabetes Metab. Disord. 2019, 18, 445–451. [Google Scholar] [CrossRef]
- Al Hommos, N.A.; Ebenibo, S.; Edeoga, C.; Dagogo-Jack, S. Trajectories of Body Weight and Fat Mass in Relation to Incident Prediabetes in a Biracial Cohort of Free-Living Adults. J. Endocr. Soc. 2020, 5, bvaa164. [Google Scholar] [CrossRef]
- Luvuno, M.; Khathi, A.; Mabandla, M.V. Diet-Induced Prediabetes: Effects of Exercise Treatment on Risk Factors for Cardiovascular Complications. Nutr. Metab. 2021, 18, 45. [Google Scholar] [CrossRef]
- Scuteri, A.; Sanna, S.; Chen, W.-M.; Uda, M.; Albai, G.; Strait, J.; Najjar, S.; Nagaraja, R.; Orrú, M.; Usala, G.; et al. Genome-Wide Association Scan Shows Genetic Variants in the FTO Gene Are Associated with Obesity-Related Traits. PLoS Genet. 2007, 3, e115. [Google Scholar] [CrossRef] [PubMed]
- Adamska-Patruno, E.; Goscik, J.; Czajkowski, P.; Maliszewska, K.; Ciborowski, M.; Golonko, A.; Wawrusiewicz-Kurylonek, N.; Citko, A.; Waszczeniuk, M.; Kretowski, A.; et al. The MC4R Genetic Variants Are Associated with Lower Visceral Fat Accumulation and Higher Postprandial Relative Increase in Carbohydrate Utilization in Humans. Eur. J. Nutr. 2019, 58, 2929–2941. [Google Scholar] [CrossRef] [PubMed]
- Blaak, E.E.; Antoine, J.-M.; Benton, D.; Björck, I.; Bozzetto, L.; Brouns, F.; Diamant, M.; Dye, L.; Hulshof, T.; Holst, J.J.; et al. Impact of Postprandial Glycaemia on Health and Prevention of Disease. Obes. Rev. 2012, 13, 923–984. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.A.; Satheesh, G.; Vijayakumar, G.; Chandran, M.; Prabhu, P.R.; Simon, L.; Kutty, V.R.; Kartha, C.C.; Jaleel, A. Postprandial Metabolism Is Impaired in Overweight Normoglycemic Young Adults without Family History of Diabetes. Sci. Rep. 2020, 10, 353. [Google Scholar] [CrossRef]
- Ashenhurst, J.R.; Sazonova, O.V.; Svrchek, O.; Detweiler, S.; Kita, R.; Babalola, L.; McIntyre, M.; Aslibekyan, S.; Fontanillas, P.; Shringarpure, S.; et al. A Polygenic Score for Type 2 Diabetes Improves Risk Stratification Beyond Current Clinical Screening Factors in an Ancestrally Diverse Sample. Front. Genet. 2022, 13, 921. [Google Scholar] [CrossRef] [PubMed]
- Hahn, S.-J.; Kim, S.; Choi, Y.S.; Lee, J.; Kang, J. Prediction of Type 2 Diabetes Using Genome-Wide Polygenic Risk Score and Metabolic Profiles: A Machine Learning Analysis of Population-Based 10-Year Prospective Cohort Study. eBioMedicine 2022, 86, 104383. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Han, Y.; Jang, K.; Kim, M. Early Prediction for Prediabetes and Type 2 Diabetes Using the Genetic Risk Score and Oxidative Stress Score. Antioxidants 2022, 11, 1196. [Google Scholar] [CrossRef]
- Sullivan, G.M.; Feinn, R. Using Effect Size—Or Why the P Value Is Not Enough. J. Grad. Med. Educ. 2012, 4, 279–282. [Google Scholar] [CrossRef]
- Calin-Jageman, R.J.; Cumming, G. The New Statistics for Better Science: Ask How Much, How Uncertain, and What Else Is Known. Am. Stat. 2019, 73, 271–280. [Google Scholar] [CrossRef]
- Haupt, A.; Thamer, C.; Heni, M.; Machicao, F.; Machann, J.; Schick, F.; Stefan, N.; Fritsche, A.; Häring, H.-U.; Staiger, H. Novel Obesity Risk Loci Do Not Determine Distribution of Body Fat Depots: A Whole-Body MRI/MRS Study. Obesity 2010, 18, 1212–1217. [Google Scholar] [CrossRef] [PubMed]
- BCL-2 Family Proteins as Regulators of Mitochondria Metabolism-ScienceDirect. Available online: https://www.sciencedirect.com/science/article/pii/S0005272816300196?via%3Dihub (accessed on 2 November 2022).
- Fall, T.; Ärnlöv, J.; Berne, C.; Ingelsson, E. The Role of Obesity-Related Genetic Loci in Insulin Sensitivity. Diabet. Med. 2012, 29, e62–e66. [Google Scholar] [CrossRef]
- Speliotes, E.K.; Willer, C.J.; Berndt, S.I.; Monda, K.L.; Thorleifsson, G.; Jackson, A.U.; Lango Allen, H.; Lindgren, C.M.; Luan, J.; Mägi, R.; et al. Association Analyses of 249,796 Individuals Reveal 18 New Loci Associated with Body Mass Index. Nat. Genet. 2010, 42, 937–948. [Google Scholar] [CrossRef] [PubMed]
- Kulyté, A.; Rydén, M.; Mejhert, N.; Dungner, E.; Sjölin, E.; Arner, P.; Dahlman, I. MTCH2 in Human White Adipose Tissue and Obesity. J. Clin. Endocrinol. Metab. 2011, 96, E1661–E1665. [Google Scholar] [CrossRef] [PubMed]
- Lehner, B.; Semple, J.I.; Brown, S.E.; Counsell, D.; Campbell, R.D.; Sanderson, C.M. Analysis of a High-Throughput Yeast Two-Hybrid System and Its Use to Predict the Function of Intracellular Proteins Encoded within the Human MHC Class III Region. Genomics 2004, 83, 153–167. [Google Scholar] [CrossRef] [PubMed]
- Mariman, E.C.M.; Bouwman, F.G.; Aller, E.E.J.G.; van Baak, M.A.; Wang, P. Extreme Obesity Is Associated with Variation in Genes Related to the Circadian Rhythm of Food Intake and Hypothalamic Signaling. Physiol. Genomics 2015, 47, 225–231. [Google Scholar] [CrossRef]
- Chen, Y.; Shen, M.; Ji, C.; Huang, Y.; Shi, Y.; Ji, L.; Qin, Y.; Gu, Y.; Fu, Q.; Chen, H.; et al. Genome-Wide Identification of N6-Methyladenosine Associated SNPs as Potential Functional Variants for Type 1 Diabetes. Front. Endocrinol. 2022, 13, 913345. [Google Scholar] [CrossRef]
- McAllan, L.; Baranasic, D.; Villicaña, S.; Zhang, W.; Lehne, B.; Adamo, M.; Jenkinson, A.; Elkalaawy, M.; Mohammadi, B.; Hashemi, M.; et al. Integrative Genomic Analyses in Adipocytes Implicate DNA Methylation in Human Obesity and Diabetes; Genetic and Genomic Medicine. medRxiv 2021.
- Ofori, J.K.; Karagiannopoulos, A.; Nagao, M.; Westholm, E.; Ramadan, S.; Wendt, A.; Esguerra, J.L.S.; Eliasson, L. Human Islet MicroRNA-200c Is Elevated in Type 2 Diabetes and Targets the Transcription Factor ETV5 to Reduce Insulin Secretion. Diabetes 2022, 71, 275–284. [Google Scholar] [CrossRef]
- Gutierrez-Aguilar, R.; Kim, D.-H.; Casimir, M.; Dai, X.-Q.; Pfluger, P.T.; Park, J.; Haller, A.; Donelan, E.; Park, J.; D’Alessio, D.; et al. The Role of the Transcription Factor ETV5 in Insulin Exocytosis. Diabetologia 2014, 57, 383–391. [Google Scholar] [CrossRef]
- Plaza-Florido, A.; Pérez-Prieto, I.; Molina-Garcia, P.; Radom-Aizik, S.; Ortega, F.B.; Altmäe, S. Transcriptional and Epigenetic Response to Sedentary Behavior and Physical Activity in Children and Adolescents: A Systematic Review. Front. Pediatr. 2022, 10, 1312. [Google Scholar]
- KCTD15 Potassium Channel Tetramerization Domain Containing 15 [Homo Sapiens (Human)]-Gene-NCBI. Available online: https://www.ncbi.nlm.nih.gov/gene/79047#summary (accessed on 2 November 2022).
- Kotnik, P.; Knapič, E.; Kokošar, J.; Kovač, J.; Jerala, R.; Battelino, T.; Horvat, S. Identification of Novel Alleles Associated with Insulin Resistance in Childhood Obesity Using Pooled-DNA Genome-Wide Association Study Approach. Int. J. Obes. 2018, 42, 686–695. [Google Scholar] [CrossRef]

| Characteristics and Parameters | Baseline | Follow-Up | p-Value |
|---|---|---|---|
| Median (IR) | Median (IR) | ||
| Female (n%) | 245 (54.9%) | - | |
| Age (years) | 42.54 (30.33, 55.73) | 47.43 (35.63, 60.67) | 3.16 × 10−24 |
| BMI (kg/m2) | 26.87 (24.04, 30.85) | 27.51 (24.36, 31.780) | 7.64 × 10−13 |
| FFM (kg) | 53.92 (48.74, 61.52) | 50.74 (44.55, 62.16) | 1.13 × 10−21 |
| FM (kg) | 23.25 (20.06, 28.23) | 26.82 (21.22, 36.14) | 1.73 × 10−24 |
| MM (kg) | 24.98 (21.17, 30.3) | 24.85 (21.15, 32.58) | 0.0010 |
| VF (cm3) | 82.50 (65, 101) | 112 (72.25, 152) | 3.94 × 10−15 |
| SF (cm3) | 145.50 (117, 184) | 249 (180.25, 318.75) | 1.32 × 10−45 |
| VAT/SAT ratio | 0.54 (0.44, 0.64) | 0.42 (0.33, 0.56) | 3.82 × 10−13 |
| IPAQ (min/week) | 1344 (240, 4306) | 5368 (2530, 11546) | 3.90 × 10−24 |
| Fasting glucose (mg/dL) | 101 (95, 110) | 109 (98, 121) | 8.10 × 10−46 |
| 2 h glucose (mg/dL) | 120 (103, 131) | 129 (111.25, 139.75) | 1.50 × 10−14 |
| HbA1c (%) | 5.76 (5.51, 6.00) | 5.82 (5.59, 6.10) | 0.0002 |
| Fasting insulin (uU/mL) | 10.78 (8.50, 14.75) | 11.87 (9.40, 15.67) | 0.0006 |
| 2 h insulin (uU/mL) | 29.98 (17.17, 53.84) | 34.11 (22.23, 49.89) | 0.0154 |
| Chol (mg/dL) | 188 (165, 221) | 194 (169, 220) | 0.0726 |
| Tg (mg/dL) | 91 (67.25, 133.00) | 96.90 (71.25, 143.00) | 0.0918 |
| Hdl (mg/dL) | 59.70 (50.42, 68.00) | 57.5 (47, 70) | 0.2003 |
| Ldl (mg/dL) | 109.60 (83.25, 137.40) | 108.80 (87.40, 138.30) | 0.0760 |
| Metabolic Parameter | β (95% CI) | p-Value |
|---|---|---|
| Δ FFM (kg) | 0.0017 (−0.0029, 0.0063) | 0.462 |
| Δ FM (kg) | 0.0049 (−0.0006, 0.0091) | 0.025 |
| Δ MM (kg) | 0.0001 (−0.0004, 0.0002) | 0.548 |
| Δ VF (cm3) | 0.0001 (−0.0009, 0.0012) | 0.802 |
| Δ SF (cm3) | 0.0001 (−0.0004, 0.0006) | 0.738 |
| Δ VAT/SAT ratio | −0.0369 (−0.1955, 0.1216) | 0.647 |
| Δ IPAQ (min/week) | 0.0001 (−0.0004, 0.0002) | 0.269 |
| Δ Fasting glucose (mg/dL) | −0.0010 (−0.0034, 0.0013) | 0.394 |
| Δ 2 h glucose (mg/dL) | −0.0004 (−0.0015, 0.0007) | 0.467 |
| Δ HbA1c (%) | 0.0492 (−0.0242, 0.1226) | 0.188 |
| Δ Fasting insulin (uU/mL) | 0.0011 (−0.0024, 0.0045) | 0.548 |
| Δ 2 h insulin (uU/mL) | 0.0002 (−0.008, 0.0012) | 0.650 |
| Δ Chol (mg/dL) | 0.0063 (−0.0133, 0.0259) | 0.531 |
| Δ Tg (mg/dL) | −0.0013 (−0.0053, 0.0026) | 0.507 |
| Δ Hdl (mg/dL) | −0.0066 (−0.0261, 0.0130) | 0.511 |
| Δ Ldl (mg/dL) | −0.0065 (−0.0261, 0.0132) | 0.518 |
| Metabolic Parameter | β (95% CI) | p-Value |
|---|---|---|
| Δ FFM (kg) | 0.0021 (−0.0032, 0.0074) | 0.4383 |
| Δ FM (kg) | 0.0056 (−0.0008, 0.0105) | 0.0231 |
| Δ MM (kg) | 0.0002 (−0.0002, 0.0005) | 0.3184 |
| Δ VF (cm3) | 0.0002 (−0.0002, 0.0005) | 0.7600 |
| Δ SF (cm3) | 0.0001 (−0.006, 0.0006) | 0.8850 |
| Δ VAT/SAT ratio | 0.0205 (−0.1619, 0.2029) | 0.8252 |
| Δ IPAQ (min/week) | 0.0001 (−0.0004, 0.0002) | 0.4108 |
| Δ Glucose time 0 (mg/dl) | 0.0020 (−0.0007, 0.0047) | 0.1446 |
| Δ 2 h glucose (mg/dl) | 0.0013 (−0.0001, 0.0026) | 0.0341 |
| Δ HbA1c (%) | 0.0014 (−0.0830, 0.0859) | 0.9732 |
| Δ Fasting insulin (uU/mL) | −0.0024 (−0.0064, 0.0016) | 0.2316 |
| Δ 2 h insulin (uU/mL) | 0.0007 (−0.0004, 0.0018) | 0.2273 |
| Δ Chol (mg/dL) | 0.0168 (−0.0057, 0.0394) | 0.1434 |
| Δ Tg (mg/dL) | −0.0036 (−0.0081, 0.0009) | 0.1167 |
| Δ Hdl (mg/dL) | −0.0158 (−0.0383, 0.0067) | 0.1685 |
| Δ Ldl (mg/dL) | −0.0164 (−0.0390, 0.0061) | 0.1533 |
| rs10838738 (MTCH2) | A/A | A/G | G/G | p-value |
|---|---|---|---|---|
| N | 143 | 229 | 83 | |
| Δ 2 h glucose (mg/dL) | 19 (3, 35) | 13 (−6.75, 33) | 8 (−8, 23.5) | 0.017 |
| Δ 2 h insulin (uU/mL) | 5.88 (−5.85, 21.34) | 3.74 (−10.25, 22.49) | −1.68 (−26.02, 10.02) | 0.001 |
| rs2260000 (PRRC2A) | A/A | A/G | G/G | P§ |
| N | 151 | 223 | 81 | |
| Δ FM (kg) | 5.27 (0.50, 10.17) | 4.87 (1.32, 8.96) | 2.40 (−1.92, 6.45) | 0.051 |
| Δ VF (cm3) | 35.75 (−8.25, 73) | 29 (−6, 67) | 8 (−29.50, 53.50) | 0.068 |
| rs7647305 (ETV5) | C/C | C/T | T/T | P§ |
| N | 309 | 126 | 20 | |
| Δ 2 h glucose (mg/dl) | 13.3 (−1, 35) | 10.5 (−11.8, 25) | 9 (−30, 27.5) | 0.077 |
| Δ IPAQ (min/week) | 2712.5 (−604.9, 6962.2) | 2601 (−23.3, 9537.3) | 6463 (2470, 12,901) | 0.077 |
| rs29941 (KCTD15) | A/A | A/G | G/G | P§ |
| N | 38 | 204 | 213 | |
| Δ 2 h glucose (mg/dL) | 20.5 (12, 33) | 15 (0.75, 36.25) | 9.50 (−9.50, 26.25) | 0.068 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Padilla-Martinez, F.; Szczerbiński, Ł.; Citko, A.; Czajkowski, M.; Konopka, P.; Paszko, A.; Wawrusiewicz-Kurylonek, N.; Górska, M.; Kretowski, A. Testing the Utility of Polygenic Risk Scores for Type 2 Diabetes and Obesity in Predicting Metabolic Changes in a Prediabetic Population: An Observational Study. Int. J. Mol. Sci. 2022, 23, 16081. https://doi.org/10.3390/ijms232416081
Padilla-Martinez F, Szczerbiński Ł, Citko A, Czajkowski M, Konopka P, Paszko A, Wawrusiewicz-Kurylonek N, Górska M, Kretowski A. Testing the Utility of Polygenic Risk Scores for Type 2 Diabetes and Obesity in Predicting Metabolic Changes in a Prediabetic Population: An Observational Study. International Journal of Molecular Sciences. 2022; 23(24):16081. https://doi.org/10.3390/ijms232416081
Chicago/Turabian StylePadilla-Martinez, Felipe, Łukasz Szczerbiński, Anna Citko, Marcin Czajkowski, Paulina Konopka, Adam Paszko, Natalia Wawrusiewicz-Kurylonek, Maria Górska, and Adam Kretowski. 2022. "Testing the Utility of Polygenic Risk Scores for Type 2 Diabetes and Obesity in Predicting Metabolic Changes in a Prediabetic Population: An Observational Study" International Journal of Molecular Sciences 23, no. 24: 16081. https://doi.org/10.3390/ijms232416081
APA StylePadilla-Martinez, F., Szczerbiński, Ł., Citko, A., Czajkowski, M., Konopka, P., Paszko, A., Wawrusiewicz-Kurylonek, N., Górska, M., & Kretowski, A. (2022). Testing the Utility of Polygenic Risk Scores for Type 2 Diabetes and Obesity in Predicting Metabolic Changes in a Prediabetic Population: An Observational Study. International Journal of Molecular Sciences, 23(24), 16081. https://doi.org/10.3390/ijms232416081







