Neural Isoforms of Agrin Are Generated by Reduced PTBP1−RNA Interaction Network Spanning the Neuron−Specific Splicing Regions in AGRN
Abstract
1. Introduction
2. Results
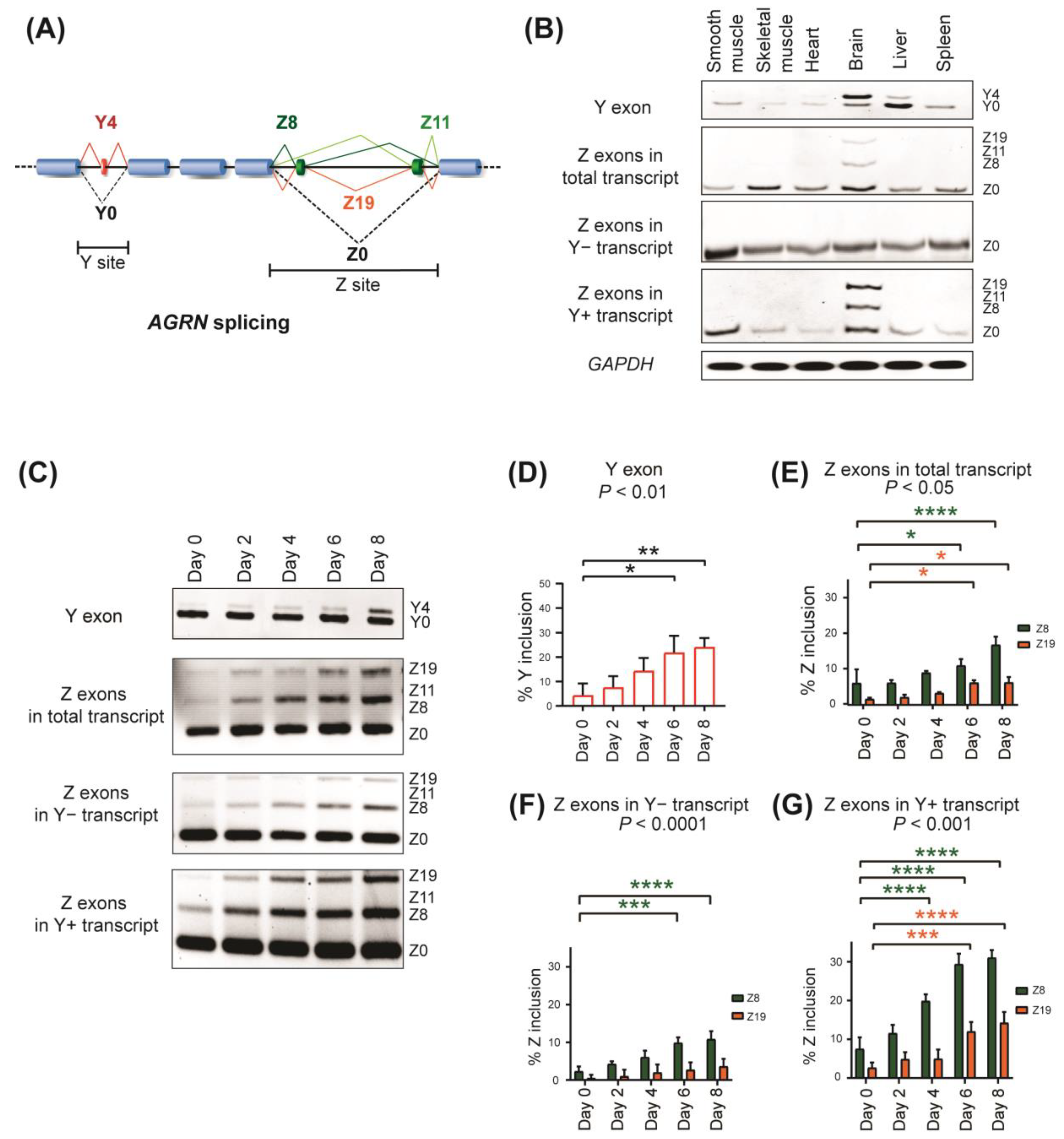
2.1. Alternative Splicing of AGRN Y and Z Exons in Human Cells and Tissues
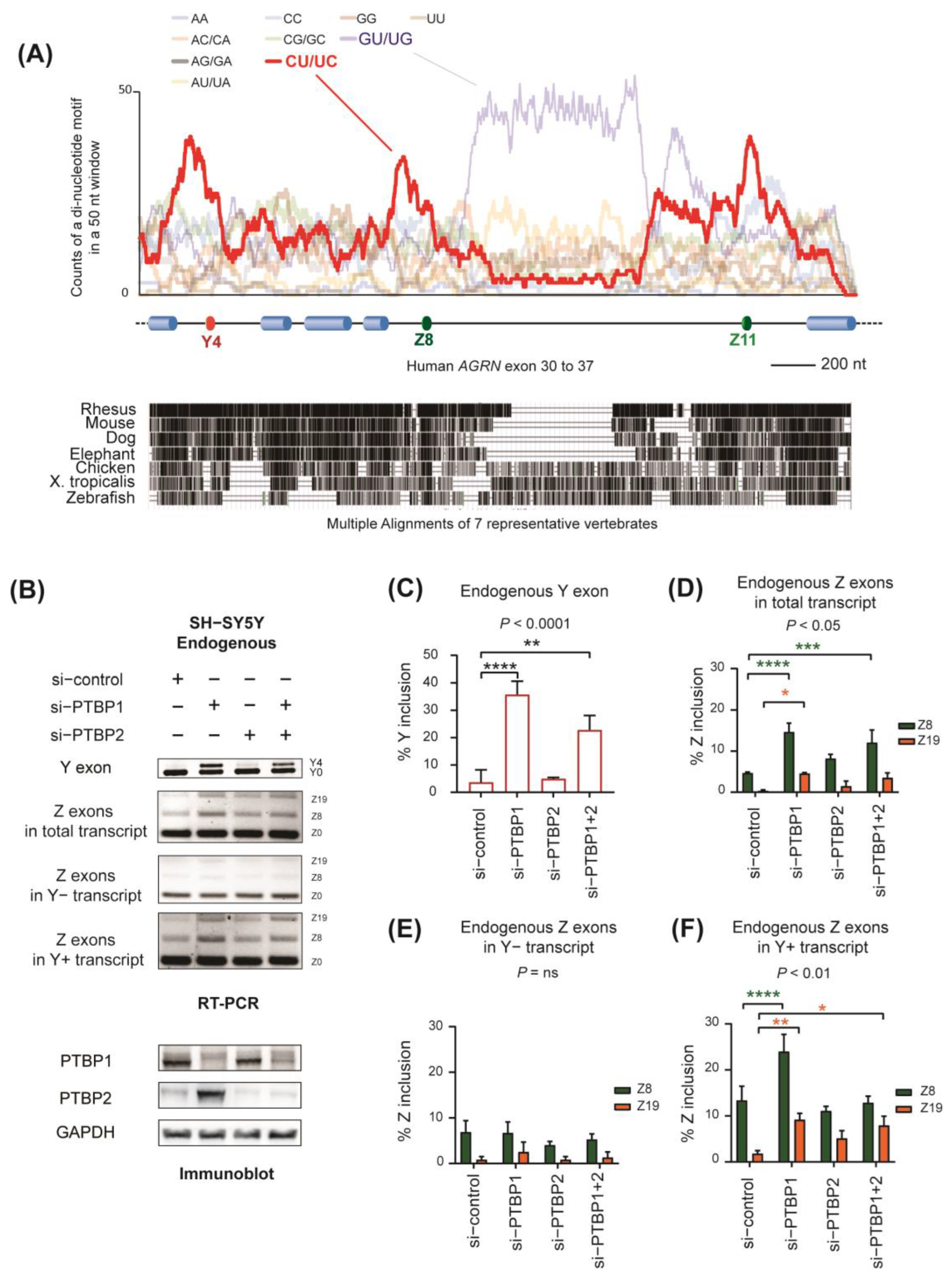
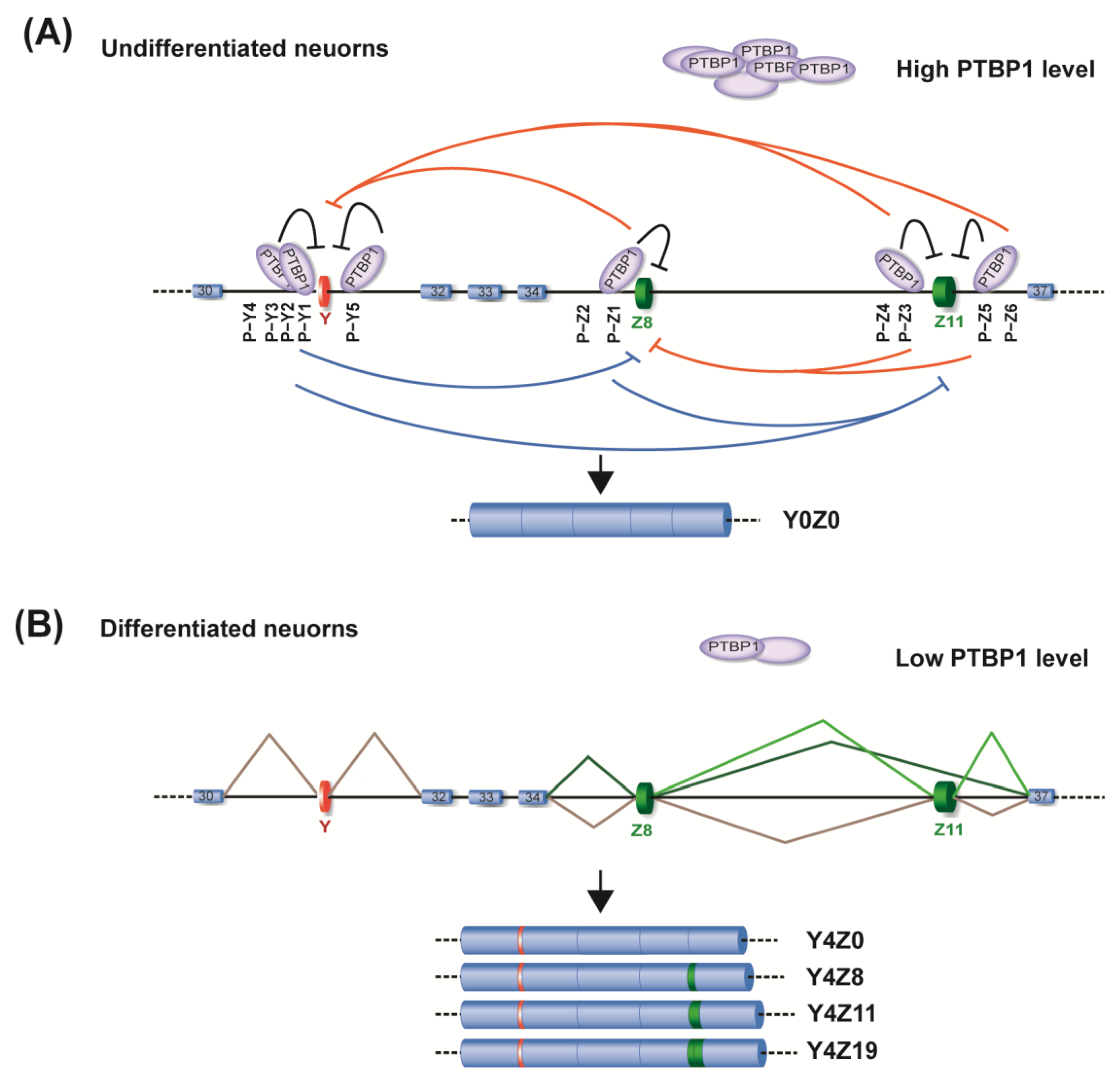
2.2. Identification of PTBP1 as a Repressor of Alternative Splicing of Y and Z Exons
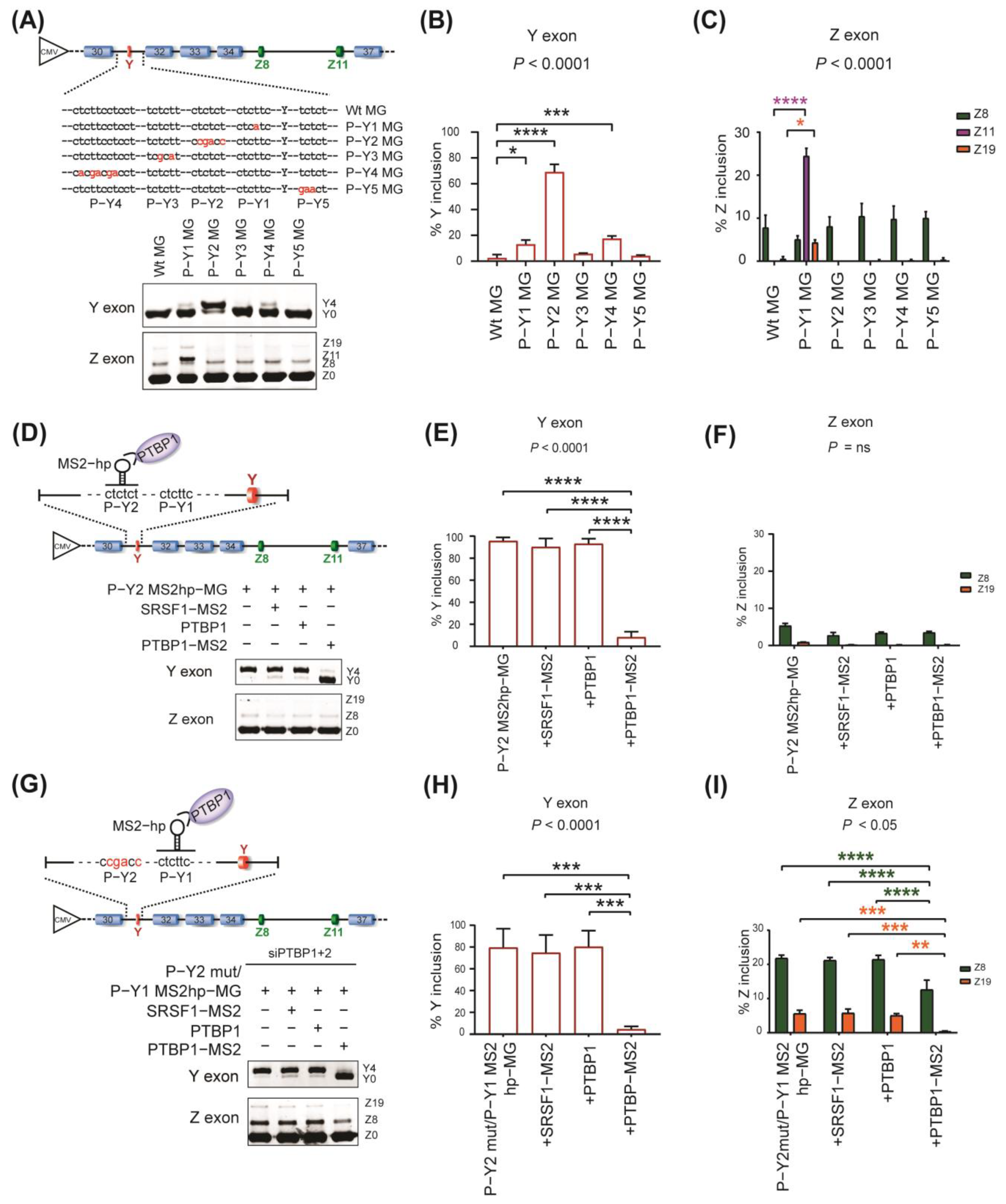
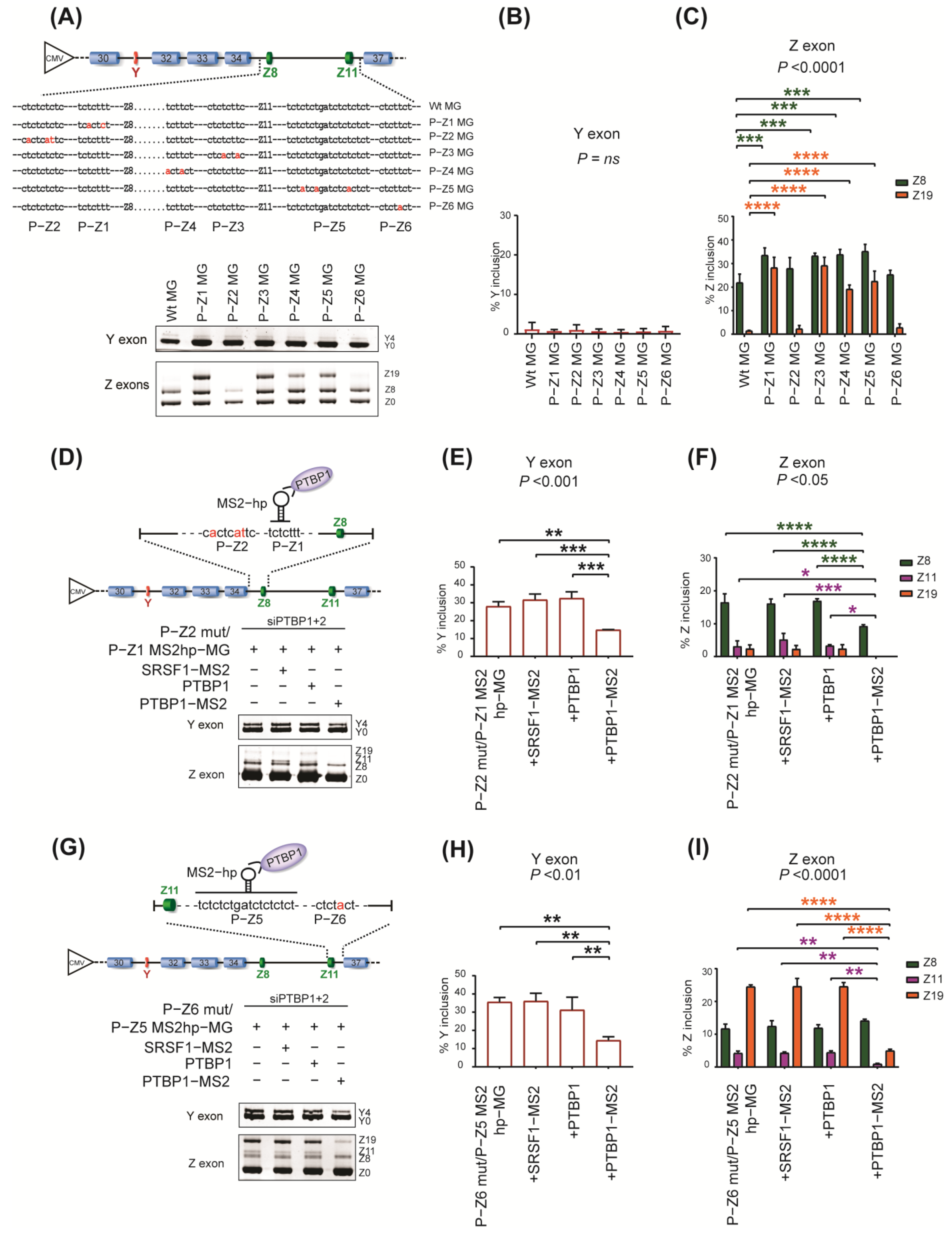
2.3. Specific Roles of PTBP1−Binding Sites around Y and Z Exons
2.4. The Essential Role of RRM4 in PTBP1 for the Repression of Both Y and Z Exons
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Construction of Minigenes and Expression Vectors for Splicing Analysis
4.3. Transfection of siRNAs and Expression Vectors
4.4. RNA Extraction and RT−PCR
4.5. Immunoblotting
4.6. Tethered Function Assay of PTBP1 and PTBP2
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Burgess, R.W.; Skarnes, W.C.; Sanes, J.R. Agrin isoforms with distinct amino termini: Differential expression, localization, and function. J. Cell Biol. 2000, 151, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Guarino, S.R.; Canciani, A.; Forneris, F. Dissecting the Extracellular Complexity of Neuromuscular Junction Organizers. Front. Mol. Biosci. 2020, 6, 156. [Google Scholar] [CrossRef] [PubMed]
- Reist, N.E.; Werle, M.J.; McMahan, U.J. Agrin released by motor neurons induces the aggregation of acetylcholine receptors at neuromuscular junctions. Neuron 1992, 8, 865–868. [Google Scholar] [CrossRef] [PubMed]
- Ferns, M.J.; Campanelli, J.T.; Hoch, W.; Scheller, R.H.; Hall, Z. The ability of agrin to cluster AChRs depends on alternative splicing and on cell surface proteoglycans. Neuron 1993, 11, 491–502. [Google Scholar] [CrossRef] [PubMed]
- Ohno, K.; Ohkawara, B.; Shen, X.-M.; Selcen, D.; Engel, A.G. Clinical and Pathologic Features of Congenital Myasthenic Syndromes Caused by 35 Genes−A Comprehensive Review. Int. J. Mol. Sci. 2023, 24, 3730. [Google Scholar] [CrossRef]
- Ohkawara, B.; Shen, X.-M.; Selcen, D.; Nazim, M.; Bril, V.; Tarnopolsky, A.M.; Brady, L.; Fukami, S.; Amato, A.A.; Yis, U.; et al. Congenital myasthenic syndrome−associated agrin variants affect clustering of acetylcholine receptors in a domain−specific manner. J. Clin. Investig. 2020, 5, e132023. [Google Scholar] [CrossRef]
- Zhang, Z.; Pinto, A.; Wan, L.; Wang, W.; Berg, M.; Oliva, I.; Singh, L.; Dengler, C.; Wei, Z.; Dreyfuss, G. Dysregulation of synaptogenesis genes antecedes motor neuron pathology in spinal muscular atrophy. Proc. Natl. Acad. Sci. USA 2013, 110, 19348–19353. [Google Scholar] [CrossRef]
- Boido, M.; Amicis, E.; Valsecchi, V.; Trevisan, M.; Ala, U.; Ruegg, M.A.; Hettwer, S.; Vercelli, A. Increasing agrin function antagonizes muscle atrophy and motor impairment in spinal muscular atrophy. Front. Cell. Neurosci. 2018, 12, 17. [Google Scholar] [CrossRef]
- Kim, J.-K.; Caine, C.; Awano, T.; Herbst, R.; Monani, U.R. Motor neuronal repletion of the NMJ organizer, Agrin, modulates the severity of the spinal muscular atrophy disease phenotype in model mice. Hum. Mol. Genet. 2017, 26, 2377–2385. [Google Scholar] [CrossRef]
- Hoch, W.; Ferns, M.; Campanelli, J.T.; Hall, Z.W.; Scheller, R.H. Developmental regulation of highly active alternatively spliced forms of agrin. Neuron 1993, 11, 479–490. [Google Scholar] [CrossRef]
- Ohno, K.; Rahman, M.A.; Nazim, M.; Nasrin, F.; Lin, Y.; Takeda, J.I.; Masuda, A. Splicing regulation and dysregulation of cholinergic genes expressed at the neuromuscular junction. J. Neurochem. 2017, 142, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Ruggiu, M.; Herbst, R.; Kim, N.; Jevesk, M.; Fak, J.J.; Mann, M.; Fischbach, G.; Burden, S.; Darnell, R. Rescuing Z+ agrin splicing in Nova null mice restores synapse formation and unmasks a physiologic defect in motor neuron firing. Proc. Natl. Acad. Sci. USA 2009, 106, 3513–3518. [Google Scholar] [CrossRef] [PubMed]
- Burgess, R.W.; Nguyen, Q.T.; Son, Y.-J.; Lichtman, J.W.; Sanes, J.R. Alternatively Spliced Isoforms of Nerve− and Muscle−Derived Agrin. Neuron 1999, 23, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Ma, E.; Morgan, R.; Godfrey, E. Distribution of agrin mRNAs in the chick embryo nervous system. J. Neurosci. 1994, 14, 2943–2952. [Google Scholar] [CrossRef]
- Nikolaou, N.; Gordon, P.M.; Hamid, F.; Taylor, R.; Lloyd, J.-J.; Makeyev, E.V.; Houart, C. Cytoplasmic pool of U1 spliceosome protein SNRNP70 shapes the axonal transcriptome and regulates motor connectivity. Curr. Biol. 2022, 32, 5099–5115. [Google Scholar] [CrossRef]
- Gesemann, M.; Denzer, A.J.; Ruegg, M.A. Acetylcholine receptor−aggregating activity of agrin isoforms and mapping of the active site. J. Cell Biol. 1995, 128, 625–636. [Google Scholar] [CrossRef]
- Wei, N.; Lin, C.Q.; Modafferi, E.F.; Gomes, W.A.; Black, D.L. A unique intronic splicing enhancer controls the inclusion of the agrin Y exon. RNA 1997, 3, 1275–1288. [Google Scholar]
- Saulière, J.; Sureau, A.; Expert−Bezançon, A.; Marie, J. The polypyrimidine tract binding protein (PTB) represses splicing of exon 6B from the beta−tropomyosin pre−mRNA by directly interfering with the binding of the U2AF65 subunit. J. Cell Biol. 2006, 26, 8755–8769. [Google Scholar] [CrossRef]
- Lamichhane, R.; Daubner, G.M.; Thomas−Crusells, J.; Auweter, S.D.; Manatschal, C.; Austin, K.S.; Valniuk, O.; Allain, F.H.-T.; Rueda, D. RNA looping by PTB: Evidence using FRET and NMR spectroscopy for a role in splicing repression. Proc. Natl. Acad. Sci. USA 2010, 107, 4105–4110. [Google Scholar] [CrossRef]
- Wagner, E.J.; Garcia−Blanco, M.A. Polypyrimidine tract binding protein antagonizes exon definition. Mol. Cell. Biol. 2001, 21, 3281–3288. [Google Scholar] [CrossRef]
- Ashiya, M.; Grabowski, P.J. A neuron−specific splicing switch mediated by an array of pre−mRNA repressor sites: Evidence of a regulatory role for the polypyrimidine tract binding protein and a brain−specific PTB counterpart. RNA 1997, 3, 996–1015. [Google Scholar] [PubMed]
- Boutz, P.L.; Stoilov, P.; Li, Q.; Lin, C.; Chawla, G.; Ostrow, K.; Shiue, L.; Ares, M.; Black, D.L. A post−transcriptional regulatory switch in polypyrimidine tract−binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 2007, 21, 1636–1652. [Google Scholar] [CrossRef] [PubMed]
- Coutinho−Mansfield, G.C.; Xue, Y.; Zhang, Y.; Fu, X.-D. PTB/nPTB switch: A post−transcriptional mechanism for programming neuronal differentiation. Genes Dev. 2007, 21, 1573–1577. [Google Scholar] [CrossRef] [PubMed]
- Lonsdale, J.; Thomas, J.; Salvatore, M.; Phillips, R.; Lo, E.; Shad, S. The Genotype−Tissue Expression (GTEx) project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef] [PubMed]
- Mallet, J.; Houhou, L.; Pajak, F.; Oda, Y.; Cervini, R.; Bejanin, S.; Berrard, S. The cholinergic locus: ChAT and VAChT genes. J. Physiol. 1998, 92, 145–147. [Google Scholar] [CrossRef] [PubMed]
- Takase, K.; Mitsushima, D.; Funabashi, T.; Kimura, F. Sex difference in the 24−h acetylcholine release profile in the premotor/supplementary motor area of behaving rats. Brain Res. 2007, 1154, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Amir−Ahmady, B.; Boutz, P.L.; Markovtsov, V.; Phillips, M.L.; Black, D.L. Exon repression by polypyrimidine tract binding protein. RNA 2005, 11, 699–716. [Google Scholar] [CrossRef]
- Luo, Y.; Hitz, B.C.; Gabdank, I.; A Hilton, J.; Kagda, M.S.; Lam, B.; Myers, Z.; Sud, P.; Jou, J.; Lin, K.; et al. New developments on the Encyclopedia of DNA Elements (ENCODE) data portal. Nucleic Acids Res. 2019, 48, D882–D889. [Google Scholar] [CrossRef]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef]
- Van Nostrand, E.L.; Freese, P.; Pratt, G.A.; Wang, X.; Wei, X.; Xiao, R.; Blue, S.M.; Chen, J.-Y.; Cody, N.A.L.; Dominguez, D.; et al. A large−scale binding and functional map of human RNA−binding proteins. Nature 2020, 583, 711–719. [Google Scholar] [CrossRef]
- Piva, F.; Giulietti, M.; Burini, A.B.; Principato, G. SpliceAid 2: A database of human splicing factors expression data and RNA target motifs. Hum. Mutat. 2011, 33, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Spellman, R.; Llorian, M.; Smith, C.W.J. Crossregulation and Functional Redundancy between the Splicing Regulator PTB and Its Paralogs nPTB and ROD1. Mol. Cell 2007, 27, 420–434. [Google Scholar] [CrossRef] [PubMed]
- Oberstrass, F.C.; Auweter, S.D.; Erat, M.; Hargous, Y.; Henning, A.; Wenter, P.; Reymond, L.; Amir−Ahmady, B.; Pitsch, S.; Black, D.L.; et al. Structure of PTB bound to RNA: Specific binding and implications for splicing regulation. Science 2005, 309, 2054–2057. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhang, W.; Reed, R.B.; Liu, W.; Grabowski, P.J. Mutations in RRM4 uncouple the splicing repression and RNA−binding activities of polypyrimidine tract binding protein. RNA 2002, 8, 137–149. [Google Scholar] [CrossRef]
- Zhang, H.; Sathyamurthy, A.; Liu, F.; Li, L.; Zhang, L.; Dong, Z.; Cui, W.; Sun, X.; Zhao, K.; Wang, H.; et al. Agrin−Lrp4−Ror2 signaling regulates adult hippocampal neurogenesis in mice. Elife 2019, 8, e45303. [Google Scholar] [CrossRef]
- Kim, M.J.; Liu, I.; Song, Y.; Lee, J.; Halfter, W.; Balice−Gordon, R.J.; Linney, E.; Cole, G.J. Agrin is required for posterior development and motor axon outgrowth and branching in embryonic zebrafish. Glycobiology 2006, 17, 231–247. [Google Scholar] [CrossRef]
- Irimia, M.; Weatheritt, R.J.; Ellis, J.D.; Parikshak, N.N.; Gonatopoulos−Pournatzis, T.; Babor, M.; Quesnel−Vallières, M.; Tapial, J.; Raj, B.; O’hanlon, D.; et al. A Highly Conserved Program of Neuronal Microexons Is Misregulated in Autistic Brains. Cell 2014, 159, 1511–1523. [Google Scholar] [CrossRef] [PubMed]
- Porter, R.S.; Jaamour, F.; Iwase, S. Neuron−specific alternative splicing of transcriptional machineries: Implications for neurodevelopmental disorders. Mol. Cell. Neurosci. 2018, 87, 35–45. [Google Scholar] [CrossRef]
- Li, Y.I.; Sanchez−Pulido, L.; Haerty, W.; Ponting, C.P. RBFOX and PTBP1 proteins regulate the alternative splicing of micro−exons in human brain transcripts. Genome Res. 2014, 25, 1–13. [Google Scholar] [CrossRef]
- Cereda, M.; Pozzoli, U.; Rot, G.; Juvan, P.; Schweitzer, A.; Clark, T.; Ule, J. RNAmotifs: Prediction of multivalent RNA motifs that control alternative splicing. Genome Biol. 2014, 15, R20. [Google Scholar] [CrossRef]
- Pina, J.M.; Hernandez, L.A.; Keppetipola, N.M. Polypyrimidine tract binding proteins PTBP1 and PTBP2 interact with distinct proteins under splicing conditions. PLoS ONE 2022, 17, e0263287. [Google Scholar] [CrossRef] [PubMed]
- Hamid, F.M.; Makeyev, E.V. A mechanism underlying position−specific regulation of alternative splicing. Nucleic Acids Res. 2017, 45, 12455–12468. [Google Scholar] [CrossRef] [PubMed]
- Ye, R.; Hu, N.; Cao, C.; Su, R.; Xu, S.; Yang, C.; Zhou, X.; Xue, Y. Capture RIC−seq reveals positional rules of PTBP1−associated RNA loops in splicing regulation. Mol. Cell 2023, 83, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Beusch, I.; Barraud, P.; Moursy, A.; Cléry, A.; Allain, F.H.-T. Tandem hnRNP A1 RNA recognition motifs act in concert to repress the splicing of survival motor neuron exon 7. Elife 2017, 6, e25736. [Google Scholar] [CrossRef] [PubMed]
- Spendiff, S.; Howarth, R.; McMacken, G.; Davey, T.; Quinlan, K.; O’Connor, E.; Slater, C.; Hettwer, S.; Mäder, A.; Roos, A.; et al. Modulation of the Acetylcholine Receptor Clustering Pathway Improves Neuromuscular Junction Structure and Muscle Strength in a Mouse Model of Congenital Myasthenic Syndrome. Front. Mol. Neurosci. 2020, 13, 594220. [Google Scholar] [CrossRef]
- Li, Z.; Li, M.; Wood, K.; Hettwer, S.; Muley, S.; Shi, F.; Liu, Q.; Ladha, S. Engineered agrin attenuates the severity of experimental autoimmune myasthenia gravis. Muscle Nerve 2017, 57, 814–820. [Google Scholar] [CrossRef]
- Havens, M.A.; Hastings, M.L. Splice−switching antisense oligonucleotides as therapeutic drugs. Nucleic Acids Res. 2016, 44, 6549–6563. [Google Scholar] [CrossRef]
- Masuda, A.; Shen, X.-M.; Ito, M.; Matsuura, T.; Engel, A.G.; Ohno, K. hnRNP H enhances skipping of a nonfunctional exon P3A in CHRNA1 and a mutation disrupting its binding causes congenital myasthenic syndrome. Hum. Mol. Genet. 2008, 17, 4022–4035. [Google Scholar] [CrossRef]
- Rahman, M.A.; Masuda, A.; Ohe, K.; Ito, M.; Hutchinson, D.O.; Mayeda, A.; Engel, A.G.; Ohno, K. HnRNP L and hnRNP LL antagonistically modulate PTB−mediated splicing suppression of CHRNA1 pre−Mrna. Sci. Rep. 2013, 3, srep02931. [Google Scholar] [CrossRef]
- Rahman, M.A.; Azuma, Y.; Nasrin, F.; Takeda, J.I.; Nazim, M.; Ahsan, K.B.; Masuda, A.; Engel, A.G.; Ohno, K. SRSF1 and hnRNP H antagonistically regulate splicing of COLQ exon 16 in a congenital myasthenic syndrome. Sci. Rep. 2015, 5, 13208. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bushra, S.; Lin, Y.-N.; Joudaki, A.; Ito, M.; Ohkawara, B.; Ohno, K.; Masuda, A. Neural Isoforms of Agrin Are Generated by Reduced PTBP1−RNA Interaction Network Spanning the Neuron−Specific Splicing Regions in AGRN. Int. J. Mol. Sci. 2023, 24, 7420. https://doi.org/10.3390/ijms24087420
Bushra S, Lin Y-N, Joudaki A, Ito M, Ohkawara B, Ohno K, Masuda A. Neural Isoforms of Agrin Are Generated by Reduced PTBP1−RNA Interaction Network Spanning the Neuron−Specific Splicing Regions in AGRN. International Journal of Molecular Sciences. 2023; 24(8):7420. https://doi.org/10.3390/ijms24087420
Chicago/Turabian StyleBushra, Samira, Ying-Ni Lin, Atefeh Joudaki, Mikako Ito, Bisei Ohkawara, Kinji Ohno, and Akio Masuda. 2023. "Neural Isoforms of Agrin Are Generated by Reduced PTBP1−RNA Interaction Network Spanning the Neuron−Specific Splicing Regions in AGRN" International Journal of Molecular Sciences 24, no. 8: 7420. https://doi.org/10.3390/ijms24087420
APA StyleBushra, S., Lin, Y.-N., Joudaki, A., Ito, M., Ohkawara, B., Ohno, K., & Masuda, A. (2023). Neural Isoforms of Agrin Are Generated by Reduced PTBP1−RNA Interaction Network Spanning the Neuron−Specific Splicing Regions in AGRN. International Journal of Molecular Sciences, 24(8), 7420. https://doi.org/10.3390/ijms24087420





