Abstract
Multiple metabolic pathways are utilized to maintain cellular homeostasis. Given the evidence that altered cell metabolism significantly contributes to glioma biology, the current research efforts aim to improve our understanding of metabolic rewiring between glioma’s complex genotype and tissue context. In addition, extensive molecular profiling has revealed activated oncogenes and inactivated tumor suppressors that directly or indirectly impact the cellular metabolism that is associated with the pathogenesis of gliomas. The mutation status of isocitrate dehydrogenases (IDHs) is one of the most important prognostic factors in adult-type diffuse gliomas. This review presents an overview of the metabolic alterations in IDH-mutant gliomas and IDH-wildtype glioblastoma (GBM). A particular focus is placed on targeting metabolic vulnerabilities to identify new therapeutic strategies for glioma.
1. Introduction
Cellular metabolism generates energy (ATP), biosynthetic precursors, cofactors, reducing equivalents (NADPH and NADH), and macromolecules, such as nucleic acids, proteins, and lipids, which are crucial for biological functions. As a result, the metabolic pathways in normal and malignant cells play a pivotal role in cell growth and survival. Experimental studies have shown that altered cellular metabolism supports sustained proliferation and enhances tumor development and progression [1]. Increased glycolytic activity is required for the rapid division of cancer cells, leading to glucose accumulation in glycolytic tumor tissues [2]. To find evidence that cellular metabolism contributes to pathogenesis, non-invasive metabolic screening has been developed, including 18F- fluorodeoxyglucose positron emission tomography (FDG-PET) [2]. Deregulating cellular energy metabolism is now recognized as a fundamental hallmark of cancer, as proposed by Hanahan and Weinberg [3].
Normal and transformed cells that are proliferating rely on glycolysis instead of oxidative phosphorylation (OXPHOS) and produce lactate from glucose, even in the presence of adequate oxygen, known as the Warburg effect [4,5]. Unlike the intense dependence on glycolytic metabolism in growing and dividing cells, most differentiated cells use OXPHOS for ATP production rather than glycolysis [6]. It has been established that rapidly dividing cells undergo metabolic adaptation without mitochondrial function defects [7]. For example, the inhibition of lactate efflux adversely affects the glioma invasion, indicating that accelerated lactate efflux is required for invasive abilities in tumor cells [8]. In addition to the invasive phenotype, glycolytic metabolism can regulate and delay programmed cell death [9].
Gliomas are thought to originate from glial or precursor cells. Several lines of evidence reveal significant differences in the metabolic profiles between isocitrate dehydrogenase (IDH)-mutant gliomas and IDH-wildtype glioblastomas [10,11,12]. For example, increased reactive oxygen species (ROS) are detected in IDH1-mutant cells [10]. Additionally, IDH1-mutant gliomas rely on oxidative phosphorylation, while IDH1-wildtype gliomas primarily rely on glycolysis for ATP production [11,12]. Similarly, a patient-derived oligodendroglioma xenograft model showed an increased mitochondrial activity compared to IDH1-wildtype xenografts [13]. This review presents the metabolic differences between IDH-mutant and IDH-wildtype gliomas. Furthermore, it reviews the novel therapeutic strategies that target metabolic alterations in preclinical and clinical studies.
1.1. Glioma
Gliomas account for 80% of malignant brain tumors [14] and have been classified and graded based on histological features [15]. However, this has resulted in inter-observer variability and minimal clinical efficacy. Despite the standard treatments, including surgical resection and chemoradiotherapy, the clinical outcomes have been unfavorable over the previous decades due to the development of drug resistance and tumor recurrence [15]. Next-generation sequencing provides information on genetically distinct alterations in adult and pediatric gliomas [16,17,18,19]. For example, mutations in IDH and histone variants frequently occur in adult and pediatric gliomas, respectively [15]. Oligodendroglioma is characterized by the co-deletion of chromosome arms 1p and 19q (1p/19q codel) along with IDH mutations (Table 1), while astrocytoma is 1p/19q non-codel with loss of alpha-thalassemia/mental retardation syndrome X-linked (ATRX). BRAF is the most commonly altered molecular driver in pediatric low-grade gliomas [20], with BRAF V600E mutations being more frequent in pleomorphic xanthoastrocytoma (PXA) and ganglioglioma (GG) [20,21].

Table 1.
Molecular-based classification of adult-type diffuse gliomas.
1.2. Glycolysis
The brain consumes approximately 25% of the glucose in the human body [22]. Specifically, astrocytes rely on glycolytic pathways, while neurons exhibit higher rates of the oxidative metabolism [23]. The brain uses alternative energy substrates such as lactate and ketone bodies in harsh environmental conditions. Cells take up glucose via specific glucose transporters (GLUTs), and hexokinase (HK) converts glucose to glucose-6-phosphate, which is the first step in glucose metabolism. HK1 is highly expressed in normal brain and low-grade gliomas, whereas HK2 is overexpressed in developing embryos and glioblastoma (GBM) tissue [24,25]. A study by Wolf and colleagues suggested that the demethylation of HK2 intron1 triggers its expression in human GBM [25]. The pentose phosphate pathway (PPP) is closely linked with glycolysis and is essential for NADPH regeneration synthesis [26].
The last committed step in glycolysis is the conversion of phosphoenolpyruvate to pyruvate, which is catalyzed by pyruvate kinase. Variable expressions and heterozygous mutations of pyruvate kinase type M2 (PKM2) have been reported in human breast tumors [27]. Similarly, PKM2 is upregulated in GBM specimens, and the siRNA-mediated downregulation of PKM2 leads to decreased levels of ATP and glutathione [28]. Mechanistically, PKM2 interacts with β-catenin upon the stimulation of the epidermal growth factor (EGF), and PKM2-dependent β-catenin transactivation is required for GBM development [29]. A similar but independent study reported that PKM2 physically interacts with Histone H3, and PKM2-dependent phosphorylation is essential for cell proliferation and tumor growth [30].
1.3. Mitochondrial Metabolism
The mitochondrial pyruvate carrier (MPC), which consists of two proteins, MPC1 and MPC2, is essential for efficient pyruvate uptake in the inner mitochondrial membrane. Impaired mitochondrial transport has been observed in many cancers, including gliomas. In particular, mitochondrial pyruvate carrier1 (MPC1) is under-expressed in gliomas, which is correlated with reduced survival in patients [31]. It has also been demonstrated that MPC restoration forms a mitochondrial complex and reduces tumor growth in vivo [31]. Interestingly, high MPC1 expression is strongly associated with a better prognosis in IDH-mutant and 1p/19q codel gliomas, but not GBM [32].
Other mitochondrial enzymes involved in oxidative metabolism include pyruvate dehydrogenase (PDH), which irreversibly converts pyruvate into acetyl-CoA. PDH is phosphorylated and inactivated by pyruvate dehydrogenase kinase (PDK), leading to decreased pyruvate oxidation in mitochondria and accelerated lactate production in the cytosol. The activity of PDH can be increased by PDH phosphatase (PDP) expression, which is repressed in patient-derived GBM samples [33]. In line with this, PDP1 restoration reduces GBM tumor growth [33].
Acetyl-CoA combines with oxaloacetate to form citrate. Citrate can be exported from the mitochondria and cleaved by ATP-citrate lyase (ACLY) to generate acetyl-CoA for fatty acid synthesis and histone acetylation. Within the mitochondria, several reactions enable citrate to be decarboxylated to oxaloacetate, producing CO2 and converting nicotinamide (NAD) and flavin adenine dinucleotides (FAD) to NADH and FADH2. These reducing equivalents are oxidized in the mitochondrial electron transport chain (ETC) to generate an electrochemical gradient, which is necessary for ATP synthase. Interestingly, mutations in the mitochondrial complex III and IV, ETC components, have been found in GBM [34]. Cytochrome c oxidase (complex IV) and ATP synthase (complex V) in the ETC can be repressed by D-2-hydroxyglutate [35,36].
1.4. Glutamine Metabolism
Glutamine metabolism serves as a nitrogen and carbon source for the biosynthesis of nucleotides and amino acids and it can replenish the carbon backbone as an anaplerotic substrate for the tricarboxylic acid (TCA) cycle function. Additionally, glutamine metabolism supports NADPH production for fatty acid synthesis. During glutaminolysis, glutamine is first converted to glutamate by glutaminase (GLS). Glutamine-derived glutamate can be metabolized by glutamate dehydrogenase (GDH) to produce α-ketoglutarate (α-KG) for maintaining cellular homeostasis (Figure 1). Glutamate can also be converted to glutamine by glutamine synthetase (GS), which is an astrocytic enzyme.
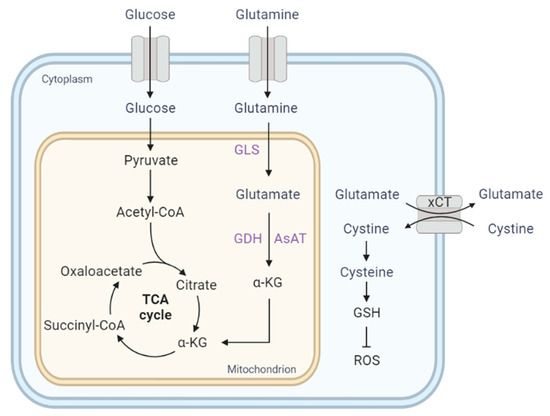
Figure 1.
Glutamine metabolism supplies TCA cycle intermediates (anaplerosis). The cystine/glutamate antiporter xCT (SLC7A11) promotes extracellular cystine uptake for glutathione biosynthesis. α-KG, α-ketoglutarate; AsAT, aspartate aminotransferase; GDH, glutamate dehydrogenase; GLS, glutaminase; GSH, glutathione; ROS, reactive oxygen species.
Transformed cells exhibit a high rate of glutamine metabolism during rapid proliferation [37], and increased glutamine uptake within tumors has been observed in human gliomas but not in normal brains [38]. ASCT2 (Slc1a5), the key glutamine importer, is strongly expressed in a rat astrocytoma-derived glioma model [39]. It has been demonstrated that MYC-dependent metabolic alteration or glutaminolysis renders cells addicted to glutamine for protein and nucleotide biosynthesis [40], and consequently, the transaminase inhibitor aminooxyacetate (AOA) induces selective toxicity in MYC-transformed cells [40]. In glutamine-starved GBM cells, GS sustains de novo purine biosynthesis [41].
Glutamine metabolism also promotes drug resistance to mTOR kinase inhibitors in an α-KG-dependent manner [42]. This compensatory upregulation of GLS and glutamate by mTOR inhibitors confers survival advantages [42]. It has been shown that the pharmacologic inhibition of mTOR kinase promotes cystine uptake and glutamate secretion via the cysteine/glutamate antiporter (xCT) encoded by the SLC7A11 gene [43]. Particularly, cysteine uptake is important for glioma cells to maintain cellular redox balance by producing glutathione (GSH) [43] (Figure 1). Gu et al. found that mTORC2 specifically binds to xCT, reducing xCT activity by phosphorylation on serine 26 [43]. In addition, xCT is responsible for glioma-mediated neuronal toxicity via the glutamate release [44,45]. Importantly, the treatment of sulfasalazine (SAS), an FDA-approved xCT inhibitor, reduces peritumoral glutamate in glioma patients [44]. Like the glutamine importer, glioma cells upregulate the expression of glutamatergic receptors, such as NMDA and AMPA [46,47,48]. Interestingly, IDH-wildtype gliomas utilize glutamine and glucose for metabolic pathways, whereas IDH1-mutant gliomas depend on glutamate and lactate [49]. This is discussed further in the section on metabolic reprogramming in IDH-mutant glioma.
1.5. Lipid Metabolism
Fatty acid and cholesterol biosynthesis are essential for the basic structure of cellular membranes in proliferating tumor cells. The evidence shows that fatty acids can cross the blood–brain barrier (BBB) via fatty acid transport proteins [50]. In contrast, dietary cholesterol cannot enter the central nervous system due to the BBB. The evidence suggests that GBM cells depend on cholesterol metabolism and exhibit selective vulnerability to liver X receptor (LXR) ligands [51]. Moreover, GBM with constitutively active epidermal growth factor receptor (EGFR) signaling is particularly susceptible to the depletion of sterol regulatory element-binding protein 1 (SREBP-1), which is a transcription factor for fatty acid and cholesterol synthesis [52]. A recent study reported that triglycerides (TG) are prominently formed in GBM tissues and are required for GBM survival via autophagy or glucose deprivation-mediated TG hydrolysis [53]. Kant and colleagues demonstrated that fatty acid β-oxidation (FAO) plays a central role during gliomagenesis [54]. Comprehensive metabolic profiling of patient-derived gliomas revealed that the biological function of FAO depends on the diverse tumor microenvironment [54]. Surprisingly, the antidepressant fluoxetine inhibits sphingomyelin phosphodiesterase 1 (SMPD1), the key enzyme for sphingolipid biosynthesis, and tumor progression in patient-derived GBM orthotopic xenograft models [55]. Consistent with this finding, clinical observations show that fluoxetine significantly prolongs the survival of GBM patients [55].
2. Metabolic Reprogramming in IDH-Mutant Glioma
Under physiological and pathological conditions, IDH enzymes play an important role in cellular metabolism as a critical component of the TCA (also known as the citric acid or Krebs cycle). IDH1 and IDH2 use nicotinamide adenine dinucleotide phosphate (NADP+) as a cofactor to generate NADPH, as illustrated in Figure 1 [56]. NADPH is vital for buffering ROS and lipid metabolism [57,58]. IDH1 is a cytosolic NADP+-dependent metabolic enzyme that mediates the oxidative decarboxylation of isocitrate to produce α-ketoglutarate (α-KG) or 2-oxoglutarate (2-OG). Additionally, IDH is essential for antioxidant defense through glutathione recycling [59].
Isocitrate dehydrogenase (IDH) mutations are prevalent in astrocytoma and oligodendroglioma [60,61]. IDH2, which is an analog of IDH1, is predominantly localized in mitochondria but mutated in gliomas at much lower frequencies [62]. IDH3 is also a mitochondrial enzyme that uses NAD+ instead of NADP+. Hotspot mutations of IDH1/2 have been identified in various human malignancies, including diffuse gliomas, myelodysplastic syndrome (MDS), chondrosarcoma, intrahepatic cholangiocarcinoma (ICC), and acute myeloid leukemia (AML) [17,61,63,64,65,66]. IDH1 mutations work in tandem with other oncogenic events to promote astrocyte proliferation and glioma development [67,68]. After a telomere-induced crisis, IDH mutations induce telomerase reverse transcriptase (TERT) reactivation, which is linked to astrocyte immortalization and transformation [69].
Heterozygous mutations at codon R132 or R172, respectively, lead to the production of D-2-hydroxyglutate (2-HG) with neomorphic enzyme activity in IDH1 and IDH2 (Figure 2) [66,70]. Moreover, IDH mutations significantly impact components of the TCA cycle intermediates and amino acids [71]. In IDH1- or IDH2-mutant cells, the levels of tyrosine, serine, threonine, methionine, tryptophan, phenylalanine, asparagine, and glycine are increased, while glutamate, aspartate, and N-acetylated amino acids are depleted [71]. Due to the limited capacity to metabolize 2-HG, intracellular 2-HG can accumulate up to 30 mM and impede α-KG as an antagonist [70]. The production of 2-HG alters redox metabolism and triggers oxidative stress in cell culture conditions, leading to the dependency of IDH-mutant cells on exogenous lipid sources [72]. The mitochondrial production of proline is enhanced to maintain redox homeostasis in IDH1-mutant glioma cells [73].
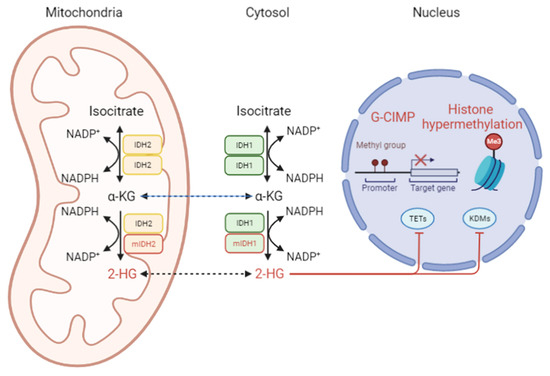
Figure 2.
The pathophysiological role of the IDH enzyme. As homodimers, IDH1 or IDH2 produce α-KG in the cytosol or mitochondria, respectively. The prevalence of the IDH mutation is high in adult-type low-grade gliomas. mIDH gains a neomorphic enzymatic activity, leading to 2-HG accumulation. Consequently, 2-HG competitively binds to ten-eleven translocation (TET) enzymes and histone lysine demethylase (KDMs). 2-HG, D-2-hydroxyglutarate; α-KG, α-ketoglutarate; G-CIMP, glioma-CpG island methylator phenotype; H3, KDM, histone lysine demethylase; mIDH1/2, mutant IDH1/2; TET, ten-eleven translocation protein.
2-HG affects the α-KG-dependent dioxygenases family of enzymes, including the TET family of DNA hydroxylases and JumonjiC (JmjC) domain-containing histone lysine demethylases (KDMs) (Figure 2) [74,75]. Recently, 2-HG-induced overexpression of stearyl-CoA desaturase (SCD) was found to cause significant alterations in the phospholipids and morphology of the endoplasmic reticulum (ER) and Golgi [76]. IDH1-mutant glioma lines are more susceptible to oleic acid-induced apoptosis than wildtype counterparts [76]. Fack et al. showed that orthotopic patient-derived xenografts of IDH-mutants exhibit remarkable differences in phospholipid composition, reduced glucose turnover, and lower energy potential [11]. These authors further revealed that cystathionine-β-synthase (CBS) is highly expressed in IDH-mutant gliomas, and its expression correlates with prolonged survival in oligodendroglioma patients [11]. 2-HG also inhibits 2-OG-dependent branched-chain amino acid (BCAA) transaminases (BCATs) and glutamate synthesis, increasing glutaminase dependence [77]. McBrayer and colleagues demonstrated that the GLS inhibitor CB-839 impairs cell proliferation by depleting glutathione, particularly in IDH1-mutant HOG cells under oxidative stress but not in their IDH-wildtype counterparts [77]. Interestingly, Zaprinast, a phosphodiesterase type 5 inhibitor, reduces the levels of 2-HG and inhibits the glutaminase (GLS) enzyme in IDH1-mutant cells [78].
An earlier study showed that the prolyl hydroxylase domain (PHD)-containing enzymes are inhibited by 2-HG treatment [74]. However, unlike the initial observation, several studies have shown that 2-HG promotes prolyl hydroxylase activity, leading to low levels of the hypoxia-inducible factor subunit HIF-1alpha in IDH-mutant gliomas [79,80]. IDH-mutant tumors grow slowly under hypoxic conditions in vivo, which is possibly due to altered metabolic consequences [12]. In particular, increased OXPHOS and decreased glutamine metabolism are observed in IDH1-mutant cells. As a result, the selective inhibition of glutaminase slows down IDH-mutant cells [81]. Conversely, the overexpression of glutamate dehydrogenase 2 (GLUD2) rescues the growth-inhibitory effect of IDH1 mutation in murine glioma progenitor cells [82].
3. Metabolic Reprogramming in IDH-Wildtype Glioblastoma
Due to its highly aggressive nature, GBM was the first tumor type to be sequenced by The Cancer Genome Atlas [16]. More than 90% of GBMs exhibit genetic aberrations in the RTK/RAS/PI3K pathway [16]. Constitutive Akt activation is sufficient to promote glucose consumption, exhibiting high rates of glycolysis [83]. GLUT3 is highly expressed as a glucose transporter, particularly in classical and proneural GBM subtypes, via PAK4-YAP/TAZ signaling [84]. Additionally, shRNA-mediated knockdown targeting integrin β3 strongly reduces the expression of GLUT3, glucose uptake, and lactate production [84]. Phosphatase and tensin homolog (PTEN) mutations are associated with high levels of HK2, enabling GBM cells to proliferate in a distinctly unique microenvironment [24].
As one of the major downstream targets of the PI3K/AKT signaling pathway, the mammalian target of rapamycin (mTOR) signaling is dysregulated in many cancers, including glioblastoma. The experimental evidence demonstrates that Nf1 deficiency promotes astrocyte proliferation in an mTOR-dependent manner [85]. It is becoming increasingly apparent that α-KG or 2-HG inhibits ATP synthase and mTOR signaling [35,36,86]. IDH-wildtype GBM cells selectively produce 2-HG under hypoxic conditions [87]. Intlekofer et al. reported that lactate dehydrogenase A (LDHA) profoundly affects hypoxia-induced 2-HG [87].
EGFR gene amplification is found in about half of GBM patients [16], frequently harboring the EGFR gene rearrangement-induced constitutively activated mutant, EGFRvIII. It is becoming evident that the EGFRvIII mutation promotes glycolytic gene expression via MYC-dependent tumor cell metabolism [88]. Delta MAX, a truncated MAX protein, enhances the glycolytic gene expression and tumorigenic potential in EGFRvIII GBM cells [88]. MYC activation induces the expression of genes involved in glycolysis and glutaminolysis [89]. Using metabolic imaging in an orthotopic xenograft mouse model, Mair and colleagues found that lactate labeling positively correlates with the c-MYC-mediated expression of HK2, monocarboxylate transporters, and lactate dehydrogenase A (LDHA) [90]. Patient-derived GBM lines activated by MYC show glucose dependency, and these cell lines are selectively responsive to glycolytic inhibition with nicotinamide phosphoribosyl-transferase (NAMPT) inhibitors [91].
p53 balances glycolysis and oxidative phosphorylation, reducing ROS for cell survival under normal conditions. Under severe metabolic stress, p53 also has a pro-oxidant activity that can remove impaired cells [92]. Mai et al. found that cytoplasmic p53 is required for erlotinib-induced apoptosis. The combined targeting of EGFR-driven glucose utilization and pharmacological p53 stabilization suppresses tumor growth in orthotopic GBM xenograft models [93]. It is important to note that Costunolide-induced ROS production has selective toxicity, particularly in glioma cells A172 and the U87MG bearing wildtype p53, but not in p53-mutant T98G [94]. TERT regulates PPP and glycogen accumulation [94]. The TERT-promoter mutations C228T and C250T are commonly found in GBM and oligodendroglioma [95]. TERT-promoter mutant GBM tumors show high fatty acid synthase (FASN) levels and lipid accumulation [96]. In addition, TERT downregulation by TERT siRNA results in the decreased expression of peroxisome proliferator-activated receptor gamma co-activator 1-alpha (PGC-1α) [96].
4. Crosstalk of Metabolic and Epigenetic Signaling in Glioma
The interplay between epigenetic modifications and metabolic alterations affects tumor cell heterogeneity and plasticity in gliomas. For example, the forced expression of Enhancer of zeste homolog 2 (EZH2), the catalytic subunit of the polycomb repressive complex 2 (PRC2), enhances glycolytic metabolism instead of mitochondrial respiration [97]. In addition, HIF1α is required for EZH2-mediated metabolic adaptation in gliomas [97]. MiR-215 is post-transcriptionally induced by HIF1α bound to Drosha/DGCR8 complexes under hypoxia [98]. Interestingly, the inhibition of MiR-215 attenuates the sphere-forming ability and the tumorigenic capability of glioma stem cells [98]. The lysine methyltransferase G9a and G9a-like protein (GLP) methylate HIF-1α at K674 and inhibit its transcriptional activity under hypoxia [99]. Importantly, G9a is downregulated by chronic hypoxic conditions in GBM [99].
As described above, SREBP-1 plays a central role in lipogenesis. Glucose enhances the stability of SREBP cleavage-activating protein (SCAP) by N-glycosylation, leading to SREBP-1 activation [100]. In this sense, EGFR signaling can trigger SREBP-1 activation via increasing the glucose uptake [100]. On the contrary, defects in the N-glycosylation of SCAP reduce the orthotopic tumor growth in GBM-bearing mice [100]. As a negative feedback regulator, miR-29 expression is induced by the SCAP and SREBP-1 complex, and subsequently, miR-29 inhibits SCAP and SREBP-1 by targeting their 3′-untranslated region (3′-UTR) [101].
Kelch-like ECH-associated protein 1 (KEAP1) plays a crucial role in the ubiquitin-mediated degradation of NF-E2-related factor 2 (NRF2) [102]. In many human cancers, somatic mutations of KEAP1 or NRF2 disrupt the interaction of these two proteins, consequently leading to the accumulation of NRF2 and upregulation of its target genes, even under stressed conditions [102]. It has been reported that KIAA0132, the human homolog of INrf2, inhibits the ubiquitin–proteasome pathway for NRF2 degradation [103].
5. Tumor Microenvironment
Nutrient availability in the tissue context and cell-autonomous mechanisms, such as genomic alterations and oncogenic signaling, can modulate metabolic needs in brain tumors. Recent investigations have demonstrated that low tumor oxygenation, also known as hypoxia, correlates with glioma cell spreading and worse patient survival [104]. Lactate efflux into the extracellular space creates an acidic tumor microenvironment (TME), resulting in drug resistance and blocking the cytotoxic function of T cells [105]. Low oxygen tension also alters the levels of NAD and NADP, enhancing the catabolism of proteins to fulfill the bioenergetic demand [106]. A recent study showed that monocyte-derived macrophages are more abundant in IDH-wildtype gliomas, while microglia are enriched in IDH-mutant gliomas [107].
2-HG inhibits complement activation in a dose-dependent manner and diminishes complement-mediated phagocytosis [108]. T cells can uptake tumor cell-derived 2-HG via the sodium-dependent dicarboxylate transporter 3 (SLC13A3), leading to impaired T-cell antitumor immunity [109]. It is therefore becoming apparent that a decreased number of infiltrating T cells is observed in IDH-mutant gliomas [107,108,109]. Additionally, 2-HG inhibits T-cell proliferation, migration, and cytokine secretion [108]. The computational characterization approach found that MHC-I subunit human leukocyte antigen (HLA) genes are significantly methylated in IDH-mutant glioma cell lines compared with IDH-wildtype GBM lines, suggesting that the MHC-I-mediated antigen presentation is impaired in IDH-mutant gliomas [110].
6. Therapeutic Approaches for Targeting Metabolic Vulnerabilities
Altered cell metabolism in cancer cells offers metabolic vulnerabilities that could be exploited therapeutically. Thus, a comprehensive understanding of glioma metabolism involved in tumor heterogeneity and drug resistance mechanisms can target metabolic vulnerability and translate into the clinic to benefit patients (Figure 3). As a glycolytic inhibitor, 2-deoxy-D-glucose (2-DG) has been shown to potentiate radiation-induced ER stress in GSCs [111]. 2-DG also protects normal brain tissue from radiation damage in the clinic [112]. Dietary restrictions have been shown to sensitize gliomas to radiation therapy [113]. An in silico super-enhancer screen identified ELOVL Fatty Acid Elongase 2 (ELOVL2) as critical for GBM stem cell proliferation [114]. The combined targeting of EGFR signaling and polyunsaturated fatty acid synthesis displays a synergistic effect on glioma stem cells (GSCs) [114]. Although the mTOR inhibitor rapamycin has not been effective in the clinic, ATP-competitive mTOR kinase inhibitors CC214-1 and CC214-2 provide promising therapeutic efficacy in orthotopic xenografts [115]. Gini and colleagues found that a preferential effect of CC214-1 is more pronounced in glioma cells with EGFRvIII expression and PTEN loss [115]. Importantly, the combined inhibition of mTOR kinase and glutaminase (GLS) profoundly reduces tumor growth in a GBM xenograft model [42].
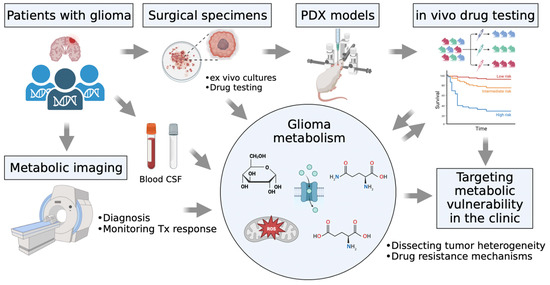
Figure 3.
Metabolic profiles of glioma characterized by plasma and the cerebrospinal fluid (CSF) of the patients, patient-derived cell lines, intracranial mouse models, and preclinical drug testing.
The accumulated evidence suggests that maintaining 2-HG levels or IDH1 mutations may not be essential for glioma growth, as the loss of 2-HG or mutant IDH1 expression alone is insufficient to prevent tumor propagation [116,117]. Interestingly, IDH mutations or the epigenetic effect of 2-HG can decrease the level of NAD+, and inhibiting nicotinamide phosphoribosyltransferase (NAMPT) can induce cytotoxicity in endogenous IDH1/2-mutant cancer cells [116]. Additionally, the increased enzymatic activity of SIRT1, an NAD+-consuming enzyme, in combination with NAMPT inhibition, can enhance the antiproliferative effect in patient-derived IDH-mutant cells [118].
GSCs show reduced mitochondrial respiration, and they are resistant to conventional alkylating chemotherapy drugs, such as temozolomide (TMZ) and 1,3-bis(2-chloroethyl)-N-nitrosourea (BCNU) [119]. The glycolytic inhibition mediated by 3-bromo-2-oxopropionate-1-propyl ester (3-BrOP), along with carmustine (BCNU), shows a dramatic effect in killing GSCs [120]. Ritonavir (RTV), a non-specific GLUT antagonist, shows low BBB permeability [121]. Dual treatment with RTV plus BCNU increases the overall survival in the GL261 murine tumor model [121]. Interestingly, high glucose transporter isoform 1 (GLUT1) expression is detected in quiescent endothelial cells [122]. Surprisingly, the loss of GLUT1 in endothelial cells impairs brain angiogenesis in vivo without altering BBB physical functions [122].
Glioma cells utilize both glycolysis and mitochondrial oxidation in vivo [123]. Pharmacological inhibition of mitochondrial components, mutant IDHs, and lipids is also being tested in ongoing clinical studies (Table 2). Dichloroacetate (DCA) induces ROS production, showing antitumor and antiangiogenic effects in C6 glioma cells in vivo [124]. As a pyruvate dehydrogenase inhibitor, DCA penetrates the BBB and normalizes the mitochondrial functions in three of five GBM patients [125]. Mechanistically, DCA contributes to p53 activation and mitochondrial ROS generation [125]. Oliva et al. found that mitochondrial DNA is susceptible to damage by sustained TMZ treatment [126]. In addition, the pharmacological and genetic intervention of cytochrome c oxidase (COX) restores TMZ sensitivity in TMZ-resistant glioma cells [126]. The small molecule IACS-010759, an inhibitor of mitochondria complex I of the ETC, reduces tumor growth in mouse models of brain cancers and acute myeloid leukemia (AML) [127].

Table 2.
Ongoing clinical trials of metabolic agents in adult-type diffuse gliomas.
Ivermectin, an antiparasitic drug, markedly suppresses GBM tumor growth via mitochondrial respiration inhibition [128]. Given that tumor hypoxia is a key contributor to radioresistance, targeting tumor hypoxia by antiparasitic agents (ivermectin, proguanil, mefloquine, quinacrine, and atovaquone) is a promising approach for enhancing radiosensitivity [129]. Imipridone, which is also called ONC201, suppresses glucose metabolism and OXPHOS-dependent ATP production in stem-like GBM cells [130]. High levels of oxaloacetate have been shown to reduce glioma growth in animal models [131], and anhydrous Enol-Oxaloacetate (AEO) is being utilized to evaluate clinical response. A recent study reported that targeting GLS enzymatic activity by CB-839 eradicates stem-like GBM cells [132]. Based on the preclinical finding, CB-839, combined with radiation and temozolomide, is being utilized in phase 1 clinical trial of IDH-mutant astrocytoma (Table 2; ClinicalTrials.gov NCT03528642).
7. Conclusions
Oncogene-directed reprogramming provides a unique metabolic adaptation to anabolic growth or elevated rate requirements. On the other hand, unique dependencies expose metabolic vulnerabilities that can be exploited as a metabolically targeted therapeutic approach. However, several challenges need to be considered, such as the permeability of the BBB, glioma heterogeneity, and the dynamic tumor microenvironment, particularly in terms of the nutrient and oxygen availability. An improved understanding of metabolism in normal physiology and brain tumors is needed to revolutionize the treatment of these intractable diseases. Given that alternative therapeutic options can be combined with epigenetic drugs or immunotherapy, further studies are required to tackle heterogeneous glioma metabolism.
Funding
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (2022R1F1A1063909).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The figures were created using BioRender.com.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Venneti, S.; Thompson, C.B. Metabolic Reprogramming in Brain Tumors. Annu. Rev. Pathol. 2017, 12, 515–545. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.M.; Parolia, A.; Dunphy, M.P.; Venneti, S. Non-invasive metabolic imaging of brain tumours in the era of precision medicine. Nat. Rev. Clin. Oncol. 2016, 13, 725–739. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed]
- Warburg, O. On the origin of cancer cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef]
- Ward, P.S.; Thompson, C.B. Metabolic reprogramming: A cancer hallmark even warburg did not anticipate. Cancer Cell 2012, 21, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Lunt, S.Y.; Vander Heiden, M.G. Aerobic glycolysis: Meeting the metabolic requirements of cell proliferation. Annu. Rev. Cell Dev. Biol. 2011, 27, 441–464. [Google Scholar] [CrossRef]
- Colen, C.B.; Shen, Y.; Ghoddoussi, F.; Yu, P.; Francis, T.B.; Koch, B.J.; Monterey, M.D.; Galloway, M.P.; Sloan, A.E.; Mathupala, S.P. Metabolic targeting of lactate efflux by malignant glioma inhibits invasiveness and induces necrosis: An in vivo study. Neoplasia 2011, 13, 620–632. [Google Scholar] [CrossRef]
- Plas, D.R.; Thompson, C.B. Cell metabolism in the regulation of programmed cell death. Trends Endocrinol. Metab. 2002, 13, 75–78. [Google Scholar] [CrossRef]
- Garrett, M.; Sperry, J.; Braas, D.; Yan, W.; Le, T.M.; Mottahedeh, J.; Ludwig, K.; Eskin, A.; Qin, Y.; Levy, R.; et al. Metabolic characterization of isocitrate dehydrogenase (IDH) mutant and IDH wildtype gliomaspheres uncovers cell type-specific vulnerabilities. Cancer Metab. 2018, 6, 4. [Google Scholar] [CrossRef]
- Fack, F.; Tardito, S.; Hochart, G.; Oudin, A.; Zheng, L.; Fritah, S.; Golebiewska, A.; Nazarov, P.V.; Bernard, A.; Hau, A.C.; et al. Altered metabolic landscape in IDH-mutant gliomas affects phospholipid, energy, and oxidative stress pathways. EMBO Mol. Med. 2017, 9, 1681–1695. [Google Scholar] [CrossRef] [PubMed]
- Grassian, A.R.; Parker, S.J.; Davidson, S.M.; Divakaruni, A.S.; Green, C.R.; Zhang, X.; Slocum, K.L.; Pu, M.; Lin, F.; Vickers, C.; et al. IDH1 mutations alter citric acid cycle metabolism and increase dependence on oxidative mitochondrial metabolism. Cancer Res. 2014, 74, 3317–3331. [Google Scholar] [CrossRef] [PubMed]
- Navis, A.C.; Niclou, S.P.; Fack, F.; Stieber, D.; van Lith, S.; Verrijp, K.; Wright, A.; Stauber, J.; Tops, B.; Otte-Holler, I.; et al. Increased mitochondrial activity in a novel IDH1-R132H mutant human oligodendroglioma xenograft model: In situ detection of 2-HG and alpha-KG. Acta Neuropathol. Commun. 2013, 1, 18. [Google Scholar] [CrossRef] [PubMed]
- Ostrom, Q.T.; Patil, N.; Cioffi, G.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2013–2017. Neuro. Oncol. 2020, 22, iv1–iv96. [Google Scholar] [CrossRef] [PubMed]
- Weller, M.; Wick, W.; Aldape, K.; Brada, M.; Berger, M.; Pfister, S.M.; Nishikawa, R.; Rosenthal, M.; Wen, P.Y.; Stupp, R.; et al. Glioma. Nat. Rev. Dis. Prim. 2015, 1, 15017. [Google Scholar] [CrossRef] [PubMed]
- Brennan, C.W.; Verhaak, R.G.; McKenna, A.; Campos, B.; Noushmehr, H.; Salama, S.R.; Zheng, S.; Chakravarty, D.; Sanborn, J.Z.; Berman, S.H.; et al. The somatic genomic landscape of glioblastoma. Cell 2013, 155, 462–477. [Google Scholar] [CrossRef]
- Parsons, D.W.; Jones, S.; Zhang, X.; Lin, J.C.; Leary, R.J.; Angenendt, P.; Mankoo, P.; Carter, H.; Siu, I.M.; Gallia, G.L.; et al. An integrated genomic analysis of human glioblastoma multiforme. Science 2008, 321, 1807–1812. [Google Scholar] [CrossRef]
- Suzuki, H.; Aoki, K.; Chiba, K.; Sato, Y.; Shiozawa, Y.; Shiraishi, Y.; Shimamura, T.; Niida, A.; Motomura, K.; Ohka, F.; et al. Mutational landscape and clonal architecture in grade II and III gliomas. Nat. Genet. 2015, 47, 458–468. [Google Scholar] [CrossRef]
- Wu, G.; Broniscer, A.; McEachron, T.A.; Lu, C.; Paugh, B.S.; Becksfort, J.; Qu, C.; Ding, L.; Huether, R.; Parker, M.; et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat. Genet. 2012, 44, 251–253. [Google Scholar] [CrossRef]
- Ryall, S.; Zapotocky, M.; Fukuoka, K.; Nobre, L.; Guerreiro Stucklin, A.; Bennett, J.; Siddaway, R.; Li, C.; Pajovic, S.; Arnoldo, A.; et al. Integrated Molecular and Clinical Analysis of 1,000 Pediatric Low-Grade Gliomas. Cancer Cell 2020, 37, 569–583.e565. [Google Scholar] [CrossRef]
- Dias-Santagata, D.; Lam, Q.; Vernovsky, K.; Vena, N.; Lennerz, J.K.; Borger, D.R.; Batchelor, T.T.; Ligon, K.L.; Iafrate, A.J.; Ligon, A.H.; et al. BRAF V600E mutations are common in pleomorphic xanthoastrocytoma: Diagnostic and therapeutic implications. PLoS ONE 2011, 6, e17948. [Google Scholar] [CrossRef] [PubMed]
- Goyal, M.S.; Raichle, M.E. Glucose Requirements of the Developing Human Brain. J. Pediatr. Gastroenterol. Nutr. 2018, 66, S46–S49. [Google Scholar] [CrossRef] [PubMed]
- Belanger, M.; Allaman, I.; Magistretti, P.J. Brain energy metabolism: Focus on astrocyte-neuron metabolic cooperation. Cell Metab. 2011, 14, 724–738. [Google Scholar] [CrossRef] [PubMed]
- Wolf, A.; Agnihotri, S.; Micallef, J.; Mukherjee, J.; Sabha, N.; Cairns, R.; Hawkins, C.; Guha, A. Hexokinase 2 is a key mediator of aerobic glycolysis and promotes tumor growth in human glioblastoma multiforme. J. Exp. Med. 2011, 208, 313–326. [Google Scholar] [CrossRef]
- Wolf, A.; Agnihotri, S.; Munoz, D.; Guha, A. Developmental profile and regulation of the glycolytic enzyme hexokinase 2 in normal brain and glioblastoma multiforme. Neurobiol. Dis. 2011, 44, 84–91. [Google Scholar] [CrossRef]
- Ros, S.; Schulze, A. Balancing glycolytic flux: The role of 6-phosphofructo-2-kinase/fructose 2,6-bisphosphatases in cancer metabolism. Cancer Metab. 2013, 1, 8. [Google Scholar] [CrossRef] [PubMed]
- Israelsen, W.J.; Dayton, T.L.; Davidson, S.M.; Fiske, B.P.; Hosios, A.M.; Bellinger, G.; Li, J.; Yu, Y.; Sasaki, M.; Horner, J.W.; et al. PKM2 isoform-specific deletion reveals a differential requirement for pyruvate kinase in tumor cells. Cell 2013, 155, 397–409. [Google Scholar] [CrossRef] [PubMed]
- Kefas, B.; Comeau, L.; Erdle, N.; Montgomery, E.; Amos, S.; Purow, B. Pyruvate kinase M2 is a target of the tumor-suppressive microRNA-326 and regulates the survival of glioma cells. Neuro. Oncol. 2010, 12, 1102–1112. [Google Scholar] [CrossRef]
- Yang, W.; Xia, Y.; Ji, H.; Zheng, Y.; Liang, J.; Huang, W.; Gao, X.; Aldape, K.; Lu, Z. Nuclear PKM2 regulates beta-catenin transactivation upon EGFR activation. Nature 2011, 480, 118–122. [Google Scholar] [CrossRef]
- Yang, W.; Xia, Y.; Hawke, D.; Li, X.; Liang, J.; Xing, D.; Aldape, K.; Hunter, T.; Alfred Yung, W.K.; Lu, Z. PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell 2012, 150, 685–696. [Google Scholar] [CrossRef]
- Schell, J.C.; Olson, K.A.; Jiang, L.; Hawkins, A.J.; Van Vranken, J.G.; Xie, J.; Egnatchik, R.A.; Earl, E.G.; DeBerardinis, R.J.; Rutter, J. A role for the mitochondrial pyruvate carrier as a repressor of the Warburg effect and colon cancer cell growth. Mol. Cell 2014, 56, 400–413. [Google Scholar] [CrossRef] [PubMed]
- Karsy, M.; Guan, J.; Huang, L.E. Prognostic role of mitochondrial pyruvate carrier in isocitrate dehydrogenase-mutant glioma. J. Neurosurg. 2018, 130, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Prabhu, A.; Sarcar, B.; Miller, C.R.; Kim, S.H.; Nakano, I.; Forsyth, P.; Chinnaiyan, P. Ras-mediated modulation of pyruvate dehydrogenase activity regulates mitochondrial reserve capacity and contributes to glioblastoma tumorigenesis. Neuro. Oncol. 2015, 17, 1220–1230. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, R.E.; Keatley, K.; Littlewood, D.T.; Meunier, B.; Holt, W.V.; An, Q.; Higgins, S.C.; Polyzoidis, S.; Stephenson, K.F.; Ashkan, K.; et al. Identification and functional prediction of mitochondrial complex III and IV mutations associated with glioblastoma. Neuro. Oncol. 2015, 17, 942–952. [Google Scholar] [CrossRef]
- Latini, A.; da Silva, C.G.; Ferreira, G.C.; Schuck, P.F.; Scussiato, K.; Sarkis, J.J.; Dutra Filho, C.S.; Wyse, A.T.; Wannmacher, C.M.; Wajner, M. Mitochondrial energy metabolism is markedly impaired by D-2-hydroxyglutaric acid in rat tissues. Mol. Genet. Metab. 2005, 86, 188–199. [Google Scholar] [CrossRef] [PubMed]
- Kolker, S.; Pawlak, V.; Ahlemeyer, B.; Okun, J.G.; Horster, F.; Mayatepek, E.; Krieglstein, J.; Hoffmann, G.F.; Kohr, G. NMDA receptor activation and respiratory chain complex V inhibition contribute to neurodegeneration in d-2-hydroxyglutaric aciduria. Eur. J. Neurosci. 2002, 16, 21–28. [Google Scholar] [CrossRef]
- DeBerardinis, R.J.; Mancuso, A.; Daikhin, E.; Nissim, I.; Yudkoff, M.; Wehrli, S.; Thompson, C.B. Beyond aerobic glycolysis: Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc. Natl. Acad. Sci. USA 2007, 104, 19345–19350. [Google Scholar] [CrossRef] [PubMed]
- Venneti, S.; Dunphy, M.P.; Zhang, H.; Pitter, K.L.; Zanzonico, P.; Campos, C.; Carlin, S.D.; La Rocca, G.; Lyashchenko, S.; Ploessl, K.; et al. Glutamine-based PET imaging facilitates enhanced metabolic evaluation of gliomas in vivo. Sci. Transl. Med. 2015, 7, 274ra217. [Google Scholar] [CrossRef]
- Dolinska, M.; Dybel, A.; Zablocka, B.; Albrecht, J. Glutamine transport in C6 glioma cells shows ASCT2 system characteristics. Neurochem. Int. 2003, 43, 501–507. [Google Scholar] [CrossRef]
- Wise, D.R.; DeBerardinis, R.J.; Mancuso, A.; Sayed, N.; Zhang, X.Y.; Pfeiffer, H.K.; Nissim, I.; Daikhin, E.; Yudkoff, M.; McMahon, S.B.; et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc. Natl. Acad. Sci. USA 2008, 105, 18782–18787. [Google Scholar] [CrossRef]
- Tardito, S.; Oudin, A.; Ahmed, S.U.; Fack, F.; Keunen, O.; Zheng, L.; Miletic, H.; Sakariassen, P.O.; Weinstock, A.; Wagner, A.; et al. Glutamine synthetase activity fuels nucleotide biosynthesis and supports growth of glutamine-restricted glioblastoma. Nat. Cell Biol. 2015, 17, 1556–1568. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Sasayama, T.; Irino, Y.; Takata, K.; Nagashima, H.; Satoh, N.; Kyotani, K.; Mizowaki, T.; Imahori, T.; Ejima, Y.; et al. Compensatory glutamine metabolism promotes glioblastoma resistance to mTOR inhibitor treatment. J. Clin. Investig. 2015, 125, 1591–1602. [Google Scholar] [CrossRef]
- Gu, Y.; Albuquerque, C.P.; Braas, D.; Zhang, W.; Villa, G.R.; Bi, J.; Ikegami, S.; Masui, K.; Gini, B.; Yang, H.; et al. mTORC2 Regulates Amino Acid Metabolism in Cancer by Phosphorylation of the Cystine-Glutamate Antiporter xCT. Mol. Cell 2017, 67, 128–138.e127. [Google Scholar] [CrossRef] [PubMed]
- Robert, S.M.; Buckingham, S.C.; Campbell, S.L.; Robel, S.; Holt, K.T.; Ogunrinu-Babarinde, T.; Warren, P.P.; White, D.M.; Reid, M.A.; Eschbacher, J.M.; et al. SLC7A11 expression is associated with seizures and predicts poor survival in patients with malignant glioma. Sci. Transl. Med. 2015, 7, 289ra286. [Google Scholar] [CrossRef] [PubMed]
- Park, J.W.; Kilic, O.; Deo, M.; Jimenez-Cowell, K.; Demirdizen, E.; Kim, H.; Turcan, S. CIC reduces xCT/SLC7A11 expression and glutamate release in glioma. Acta Neuropathol. Commun. 2023, 11, 13. [Google Scholar] [CrossRef]
- Venkataramani, V.; Tanev, D.I.; Strahle, C.; Studier-Fischer, A.; Fankhauser, L.; Kessler, T.; Korber, C.; Kardorff, M.; Ratliff, M.; Xie, R.; et al. Glutamatergic synaptic input to glioma cells drives brain tumour progression. Nature 2019, 573, 532–538. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, H.S.; Morishita, W.; Geraghty, A.C.; Silverbush, D.; Gillespie, S.M.; Arzt, M.; Tam, L.T.; Espenel, C.; Ponnuswami, A.; Ni, L.; et al. Electrical and synaptic integration of glioma into neural circuits. Nature 2019, 573, 539–545. [Google Scholar] [CrossRef]
- Zeng, Q.; Michael, I.P.; Zhang, P.; Saghafinia, S.; Knott, G.; Jiao, W.; McCabe, B.D.; Galvan, J.A.; Robinson, H.P.C.; Zlobec, I.; et al. Synaptic proximity enables NMDAR signalling to promote brain metastasis. Nature 2019, 573, 526–531. [Google Scholar] [CrossRef]
- Lenting, K.; Khurshed, M.; Peeters, T.H.; van den Heuvel, C.; van Lith, S.A.M.; de Bitter, T.; Hendriks, W.; Span, P.N.; Molenaar, R.J.; Botman, D.; et al. Isocitrate dehydrogenase 1-mutated human gliomas depend on lactate and glutamate to alleviate metabolic stress. FASEB J. 2019, 33, 557–571. [Google Scholar] [CrossRef]
- Bruce, K.D.; Zsombok, A.; Eckel, R.H. Lipid Processing in the Brain: A Key Regulator of Systemic Metabolism. Front. Endocrinol. 2017, 8, 60. [Google Scholar] [CrossRef]
- Villa, G.R.; Hulce, J.J.; Zanca, C.; Bi, J.; Ikegami, S.; Cahill, G.L.; Gu, Y.; Lum, K.M.; Masui, K.; Yang, H.; et al. An LXR-Cholesterol Axis Creates a Metabolic Co-Dependency for Brain Cancers. Cancer Cell 2016, 30, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.; Prins, R.M.; Dang, J.; Kuga, D.; Iwanami, A.; Soto, H.; Lin, K.Y.; Huang, T.T.; Akhavan, D.; Hock, M.B.; et al. EGFR signaling through an Akt-SREBP-1-dependent, rapamycin-resistant pathway sensitizes glioblastomas to antilipogenic therapy. Sci. Signal. 2009, 2, ra82. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Geng, F.; Cheng, X.; Guo, Q.; Zhong, Y.; Cloughesy, T.F.; Yong, W.H.; Chakravarti, A.; Guo, D. Lipid Droplets Maintain Energy Homeostasis and Glioblastoma Growth via Autophagic Release of Stored Fatty Acids. iScience 2020, 23, 101569. [Google Scholar] [CrossRef] [PubMed]
- Kant, S.; Kesarwani, P.; Prabhu, A.; Graham, S.F.; Buelow, K.L.; Nakano, I.; Chinnaiyan, P. Enhanced fatty acid oxidation provides glioblastoma cells metabolic plasticity to accommodate to its dynamic nutrient microenvironment. Cell Death Dis. 2020, 11, 253. [Google Scholar] [CrossRef]
- Bi, J.; Khan, A.; Tang, J.; Armando, A.M.; Wu, S.; Zhang, W.; Gimple, R.C.; Reed, A.; Jing, H.; Koga, T.; et al. Targeting glioblastoma signaling and metabolism with a re-purposed brain-penetrant drug. Cell Rep. 2021, 37, 109957. [Google Scholar] [CrossRef] [PubMed]
- Hurley, J.H.; Dean, A.M.; Koshland, D.E., Jr.; Stroud, R.M. Catalytic mechanism of NADP(+)-dependent isocitrate dehydrogenase: Implications from the structures of magnesium-isocitrate and NADP+ complexes. Biochemistry 1991, 30, 8671–8678. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.H.; Son, M.K.; Koh, H.J.; Lee, S.M.; Song, I.H.; Kim, Y.O.; Lee, Y.S.; Jeong, K.S.; Kim, W.B.; Park, J.W.; et al. Control of mitochondrial redox balance and cellular defense against oxidative damage by mitochondrial NADP+-dependent isocitrate dehydrogenase. J. Biol. Chem. 2001, 276, 16168–16176. [Google Scholar] [CrossRef] [PubMed]
- Koh, H.J.; Lee, S.M.; Son, B.G.; Lee, S.H.; Ryoo, Z.Y.; Chang, K.T.; Park, J.W.; Park, D.C.; Song, B.J.; Veech, R.L.; et al. Cytosolic NADP+-dependent isocitrate dehydrogenase plays a key role in lipid metabolism. J. Biol. Chem. 2004, 279, 39968–39974. [Google Scholar] [CrossRef]
- Lee, S.M.; Koh, H.J.; Park, D.C.; Song, B.J.; Huh, T.L.; Park, J.W. Cytosolic NADP(+)-dependent isocitrate dehydrogenase status modulates oxidative damage to cells. Free. Radic. Biol. Med. 2002, 32, 1185–1196. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research, N.; Brat, D.J.; Verhaak, R.G.; Aldape, K.D.; Yung, W.K.; Salama, S.R.; Cooper, L.A.; Rheinbay, E.; Miller, C.R.; Vitucci, M.; et al. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N. Engl. J. Med. 2015, 372, 2481–2498. [Google Scholar] [CrossRef]
- Yan, H.; Parsons, D.W.; Jin, G.; McLendon, R.; Rasheed, B.A.; Yuan, W.; Kos, I.; Batinic-Haberle, I.; Jones, S.; Riggins, G.J.; et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 2009, 360, 765–773. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, C.; Meyer, J.; Balss, J.; Capper, D.; Mueller, W.; Christians, A.; Felsberg, J.; Wolter, M.; Mawrin, C.; Wick, W.; et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: A study of 1,010 diffuse gliomas. Acta Neuropathol. 2009, 118, 469–474. [Google Scholar] [CrossRef] [PubMed]
- Mardis, E.R.; Ding, L.; Dooling, D.J.; Larson, D.E.; McLellan, M.D.; Chen, K.; Koboldt, D.C.; Fulton, R.S.; Delehaunty, K.D.; McGrath, S.D.; et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N. Engl. J. Med. 2009, 361, 1058–1066. [Google Scholar] [CrossRef] [PubMed]
- Amary, M.F.; Bacsi, K.; Maggiani, F.; Damato, S.; Halai, D.; Berisha, F.; Pollock, R.; O’Donnell, P.; Grigoriadis, A.; Diss, T.; et al. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J. Pathol. 2011, 224, 334–343. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Dong, Q.; Zhang, C.; Kuan, P.F.; Liu, Y.; Jeck, W.R.; Andersen, J.B.; Jiang, W.; Savich, G.L.; Tan, T.X.; et al. Mutations in isocitrate dehydrogenase 1 and 2 occur frequently in intrahepatic cholangiocarcinomas and share hypermethylation targets with glioblastomas. Oncogene 2013, 32, 3091–3100. [Google Scholar] [CrossRef]
- Ward, P.S.; Patel, J.; Wise, D.R.; Abdel-Wahab, O.; Bennett, B.D.; Coller, H.A.; Cross, J.R.; Fantin, V.R.; Hedvat, C.V.; Perl, A.E.; et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 2010, 17, 225–234. [Google Scholar] [CrossRef]
- Bettegowda, C.; Agrawal, N.; Jiao, Y.; Sausen, M.; Wood, L.D.; Hruban, R.H.; Rodriguez, F.J.; Cahill, D.P.; McLendon, R.; Riggins, G.; et al. Mutations in CIC and FUBP1 contribute to human oligodendroglioma. Science 2011, 333, 1453–1455. [Google Scholar] [CrossRef]
- Philip, B.; Yu, D.X.; Silvis, M.R.; Shin, C.H.; Robinson, J.P.; Robinson, G.L.; Welker, A.E.; Angel, S.N.; Tripp, S.R.; Sonnen, J.A.; et al. Mutant IDH1 Promotes Glioma Formation In Vivo. Cell Rep. 2018, 23, 1553–1564. [Google Scholar] [CrossRef]
- Ohba, S.; Mukherjee, J.; Johannessen, T.C.; Mancini, A.; Chow, T.T.; Wood, M.; Jones, L.; Mazor, T.; Marshall, R.E.; Viswanath, P.; et al. Mutant IDH1 Expression Drives TERT Promoter Reactivation as Part of the Cellular Transformation Process. Cancer Res. 2016, 76, 6680–6689. [Google Scholar] [CrossRef] [PubMed]
- Dang, L.; White, D.W.; Gross, S.; Bennett, B.D.; Bittinger, M.A.; Driggers, E.M.; Fantin, V.R.; Jang, H.G.; Jin, S.; Keenan, M.C.; et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 2009, 462, 739–744. [Google Scholar] [CrossRef]
- Reitman, Z.J.; Jin, G.; Karoly, E.D.; Spasojevic, I.; Yang, J.; Kinzler, K.W.; He, Y.; Bigner, D.D.; Vogelstein, B.; Yan, H. Profiling the effects of isocitrate dehydrogenase 1 and 2 mutations on the cellular metabolome. Proc. Natl. Acad. Sci. USA 2011, 108, 3270–3275. [Google Scholar] [CrossRef] [PubMed]
- Badur, M.G.; Muthusamy, T.; Parker, S.J.; Ma, S.; McBrayer, S.K.; Cordes, T.; Magana, J.H.; Guan, K.L.; Metallo, C.M. Oncogenic R132 IDH1 Mutations Limit NADPH for De Novo Lipogenesis through (D)2-Hydroxyglutarate Production in Fibrosarcoma Sells. Cell Rep. 2018, 25, 1018–1026.e1014. [Google Scholar] [CrossRef] [PubMed]
- Hollinshead, K.E.R.; Munford, H.; Eales, K.L.; Bardella, C.; Li, C.; Escribano-Gonzalez, C.; Thakker, A.; Nonnenmacher, Y.; Kluckova, K.; Jeeves, M.; et al. Oncogenic IDH1 Mutations Promote Enhanced Proline Synthesis through PYCR1 to Support the Maintenance of Mitochondrial Redox Homeostasis. Cell Rep. 2018, 22, 3107–3114. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Yang, H.; Liu, Y.; Yang, Y.; Wang, P.; Kim, S.H.; Ito, S.; Yang, C.; Wang, P.; Xiao, M.T.; et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell 2011, 19, 17–30. [Google Scholar] [CrossRef]
- Zhao, S.; Lin, Y.; Xu, W.; Jiang, W.; Zha, Z.; Wang, P.; Yu, W.; Li, Z.; Gong, L.; Peng, Y.; et al. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science 2009, 324, 261–265. [Google Scholar] [CrossRef]
- Lita, A.; Pliss, A.; Kuzmin, A.; Yamasaki, T.; Zhang, L.; Dowdy, T.; Burks, C.; de Val, N.; Celiku, O.; Ruiz-Rodado, V.; et al. IDH1 mutations induce organelle defects via dysregulated phospholipids. Nat. Commun. 2021, 12, 614. [Google Scholar] [CrossRef] [PubMed]
- McBrayer, S.K.; Mayers, J.R.; DiNatale, G.J.; Shi, D.D.; Khanal, J.; Chakraborty, A.A.; Sarosiek, K.A.; Briggs, K.J.; Robbins, A.K.; Sewastianik, T.; et al. Transaminase Inhibition by 2-Hydroxyglutarate Impairs Glutamate Biosynthesis and Redox Homeostasis in Glioma. Cell 2018, 175, 101–116.e125. [Google Scholar] [CrossRef]
- Elhammali, A.; Ippolito, J.E.; Collins, L.; Crowley, J.; Marasa, J.; Piwnica-Worms, D. A high-throughput fluorimetric assay for 2-hydroxyglutarate identifies Zaprinast as a glutaminase inhibitor. Cancer Discov. 2014, 4, 828–839. [Google Scholar] [CrossRef]
- Koivunen, P.; Lee, S.; Duncan, C.G.; Lopez, G.; Lu, G.; Ramkissoon, S.; Losman, J.A.; Joensuu, P.; Bergmann, U.; Gross, S.; et al. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature 2012, 483, 484–488. [Google Scholar] [CrossRef]
- Kickingereder, P.; Sahm, F.; Radbruch, A.; Wick, W.; Heiland, S.; Deimling, A.; Bendszus, M.; Wiestler, B. IDH mutation status is associated with a distinct hypoxia/angiogenesis transcriptome signature which is non-invasively predictable with rCBV imaging in human glioma. Sci. Rep. 2015, 5, 16238. [Google Scholar] [CrossRef]
- Seltzer, M.J.; Bennett, B.D.; Joshi, A.D.; Gao, P.; Thomas, A.G.; Ferraris, D.V.; Tsukamoto, T.; Rojas, C.J.; Slusher, B.S.; Rabinowitz, J.D.; et al. Inhibition of glutaminase preferentially slows growth of glioma cells with mutant IDH1. Cancer Res. 2010, 70, 8981–8987. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Nishimura, M.C.; Kharbanda, S.; Peale, F.; Deng, Y.; Daemen, A.; Forrest, W.F.; Kwong, M.; Hedehus, M.; Hatzivassiliou, G.; et al. Hominoid-specific enzyme GLUD2 promotes growth of IDH1R132H glioma. Proc. Natl. Acad. Sci. USA 2014, 111, 14217–14222. [Google Scholar] [CrossRef] [PubMed]
- Elstrom, R.L.; Bauer, D.E.; Buzzai, M.; Karnauskas, R.; Harris, M.H.; Plas, D.R.; Zhuang, H.; Cinalli, R.M.; Alavi, A.; Rudin, C.M.; et al. Akt stimulates aerobic glycolysis in cancer cells. Cancer Res. 2004, 64, 3892–3899. [Google Scholar] [CrossRef] [PubMed]
- Cosset, E.; Ilmjarv, S.; Dutoit, V.; Elliott, K.; von Schalscha, T.; Camargo, M.F.; Reiss, A.; Moroishi, T.; Seguin, L.; Gomez, G.; et al. Glut3 Addiction Is a Druggable Vulnerability for a Molecularly Defined Subpopulation of Glioblastoma. Cancer Cell 2017, 32, 856–868.e855. [Google Scholar] [CrossRef]
- Banerjee, S.; Crouse, N.R.; Emnett, R.J.; Gianino, S.M.; Gutmann, D.H. Neurofibromatosis-1 regulates mTOR-mediated astrocyte growth and glioma formation in a TSC/Rheb-independent manner. Proc. Natl. Acad. Sci. USA 2011, 108, 15996–16001. [Google Scholar] [CrossRef]
- Fu, X.; Chin, R.M.; Vergnes, L.; Hwang, H.; Deng, G.; Xing, Y.; Pai, M.Y.; Li, S.; Ta, L.; Fazlollahi, F.; et al. 2-Hydroxyglutarate Inhibits ATP Synthase and mTOR Signaling. Cell Metab. 2015, 22, 508–515. [Google Scholar] [CrossRef]
- Intlekofer, A.M.; Dematteo, R.G.; Venneti, S.; Finley, L.W.; Lu, C.; Judkins, A.R.; Rustenburg, A.S.; Grinaway, P.B.; Chodera, J.D.; Cross, J.R.; et al. Hypoxia Induces Production of L-2-Hydroxyglutarate. Cell Metab. 2015, 22, 304–311. [Google Scholar] [CrossRef]
- Babic, I.; Anderson, E.S.; Tanaka, K.; Guo, D.; Masui, K.; Li, B.; Zhu, S.; Gu, Y.; Villa, G.R.; Akhavan, D.; et al. EGFR mutation-induced alternative splicing of Max contributes to growth of glycolytic tumors in brain cancer. Cell Metab. 2013, 17, 1000–1008. [Google Scholar] [CrossRef]
- Dang, C.V. MYC, metabolism, cell growth, and tumorigenesis. Cold Spring Harb Perspect Med. 2013, 3, a014217. [Google Scholar] [CrossRef]
- Mair, R.; Wright, A.J.; Ros, S.; Hu, D.E.; Booth, T.; Kreis, F.; Rao, J.; Watts, C.; Brindle, K.M. Metabolic Imaging Detects Low Levels of Glycolytic Activity That Vary with Levels of c-Myc Expression in Patient-Derived Xenograft Models of Glioblastoma. Cancer Res. 2018, 78, 5408–5418. [Google Scholar] [CrossRef]
- Tateishi, K.; Iafrate, A.J.; Ho, Q.; Curry, W.T.; Batchelor, T.T.; Flaherty, K.T.; Onozato, M.L.; Lelic, N.; Sundaram, S.; Cahill, D.P.; et al. Myc-Driven Glycolysis Is a Therapeutic Target in Glioblastoma. Clin. Cancer Res. 2016, 22, 4452–4465. [Google Scholar] [CrossRef] [PubMed]
- Kruiswijk, F.; Labuschagne, C.F.; Vousden, K.H. p53 in survival, death and metabolic health: A lifeguard with a licence to kill. Nat. Rev. Mol. Cell Biol. 2015, 16, 393–405. [Google Scholar] [CrossRef] [PubMed]
- Mai, W.X.; Gosa, L.; Daniels, V.W.; Ta, L.; Tsang, J.E.; Higgins, B.; Gilmore, W.B.; Bayley, N.A.; Harati, M.D.; Lee, J.T.; et al. Cytoplasmic p53 couples oncogene-driven glucose metabolism to apoptosis and is a therapeutic target in glioblastoma. Nat. Med. 2017, 23, 1342–1351. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, F.; Dixit, D.; Sharma, V.; Kumar, A.; Joshi, S.D.; Sarkar, C.; Sen, E. Nrf2-driven TERT regulates pentose phosphate pathway in glioblastoma. Cell Death Dis. 2016, 7, e2213. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Koh, J.; Kim, S.I.; Won, J.K.; Park, C.K.; Choi, S.H.; Park, S.H. The frequency and prognostic effect of TERT promoter mutation in diffuse gliomas. Acta Neuropathol. Commun. 2017, 5, 62. [Google Scholar] [CrossRef]
- Ahmad, F.; Patrick, S.; Sheikh, T.; Sharma, V.; Pathak, P.; Malgulwar, P.B.; Kumar, A.; Joshi, S.D.; Sarkar, C.; Sen, E. Telomerase reverse transcriptase (TERT)—Enhancer of zeste homolog 2 (EZH2) network regulates lipid metabolism and DNA damage responses in glioblastoma. J. Neurochem. 2017, 143, 671–683. [Google Scholar] [CrossRef]
- Pang, B.; Zheng, X.R.; Tian, J.X.; Gao, T.H.; Gu, G.Y.; Zhang, R.; Fu, Y.B.; Pang, Q.; Li, X.G.; Liu, Q. EZH2 promotes metabolic reprogramming in glioblastomas through epigenetic repression of EAF2-HIF1alpha signaling. Oncotarget 2016, 7, 45134–45143. [Google Scholar] [CrossRef]
- Hu, J.; Sun, T.; Wang, H.; Chen, Z.; Wang, S.; Yuan, L.; Liu, T.; Li, H.R.; Wang, P.; Feng, Y.; et al. MiR-215 Is Induced Post-transcriptionally via HIF-Drosha Complex and Mediates Glioma-Initiating Cell Adaptation to Hypoxia by Targeting KDM1B. Cancer Cell 2016, 29, 49–60. [Google Scholar] [CrossRef]
- Bao, L.; Chen, Y.; Lai, H.T.; Wu, S.Y.; Wang, J.E.; Hatanpaa, K.J.; Raisanen, J.M.; Fontenot, M.; Lega, B.; Chiang, C.M.; et al. Methylation of hypoxia-inducible factor (HIF)-1alpha by G9a/GLP inhibits HIF-1 transcriptional activity and cell migration. Nucleic. Acids Res. 2018, 46, 6576–6591. [Google Scholar] [CrossRef]
- Cheng, C.; Ru, P.; Geng, F.; Liu, J.; Yoo, J.Y.; Wu, X.; Cheng, X.; Euthine, V.; Hu, P.; Guo, J.Y.; et al. Glucose-Mediated N-glycosylation of SCAP Is Essential for SREBP-1 Activation and Tumor Growth. Cancer Cell 2015, 28, 569–581. [Google Scholar] [CrossRef]
- Ru, P.; Hu, P.; Geng, F.; Mo, X.; Cheng, C.; Yoo, J.Y.; Cheng, X.; Wu, X.; Guo, J.Y.; Nakano, I.; et al. Feedback Loop Regulation of SCAP/SREBP-1 by miR-29 Modulates EGFR Signaling-Driven Glioblastoma Growth. Cell Rep. 2016, 16, 1527–1535. [Google Scholar] [CrossRef] [PubMed]
- Taguchi, K.; Motohashi, H.; Yamamoto, M. Molecular mechanisms of the Keap1-Nrf2 pathway in stress response and cancer evolution. Genes Cells 2011, 16, 123–140. [Google Scholar] [CrossRef] [PubMed]
- Sekhar, K.R.; Yan, X.X.; Freeman, M.L. Nrf2 degradation by the ubiquitin proteasome pathway is inhibited by KIAA0132, the human homolog to INrf2. Oncogene 2002, 21, 6829–6834. [Google Scholar] [CrossRef]
- Monteiro, A.R.; Hill, R.; Pilkington, G.J.; Madureira, P.A. The Role of Hypoxia in Glioblastoma Invasion. Cells 2017, 6, 45. [Google Scholar] [CrossRef]
- Jing, X.; Yang, F.; Shao, C.; Wei, K.; Xie, M.; Shen, H.; Shu, Y. Role of hypoxia in cancer therapy by regulating the tumor microenvironment. Mol. Cancer 2019, 18, 157. [Google Scholar] [CrossRef]
- Kucharzewska, P.; Christianson, H.C.; Belting, M. Global profiling of metabolic adaptation to hypoxic stress in human glioblastoma cells. PLoS ONE 2015, 10, e0116740. [Google Scholar] [CrossRef] [PubMed]
- Klemm, F.; Maas, R.R.; Bowman, R.L.; Kornete, M.; Soukup, K.; Nassiri, S.; Brouland, J.P.; Iacobuzio-Donahue, C.A.; Brennan, C.; Tabar, V.; et al. Interrogation of the Microenvironmental Landscape in Brain Tumors Reveals Disease-Specific Alterations of Immune Cells. Cell 2020, 181, 1643–1660.e1617. [Google Scholar] [CrossRef]
- Zhang, L.; Sorensen, M.D.; Kristensen, B.W.; Reifenberger, G.; McIntyre, T.M.; Lin, F. D-2-Hydroxyglutarate Is an Intercellular Mediator in IDH-Mutant Gliomas Inhibiting Complement and T Cells. Clin. Cancer Res. 2018, 24, 5381–5391. [Google Scholar] [CrossRef]
- Bunse, L.; Pusch, S.; Bunse, T.; Sahm, F.; Sanghvi, K.; Friedrich, M.; Alansary, D.; Sonner, J.K.; Green, E.; Deumelandt, K.; et al. Suppression of antitumor T cell immunity by the oncometabolite (R)-2-hydroxyglutarate. Nat. Med. 2018, 24, 1192–1203. [Google Scholar] [CrossRef] [PubMed]
- Luoto, S.; Hermelo, I.; Vuorinen, E.M.; Hannus, P.; Kesseli, J.; Nykter, M.; Granberg, K.J. Computational Characterization of Suppressive Immune Microenvironments in Glioblastoma. Cancer Res. 2018, 78, 5574–5585. [Google Scholar] [CrossRef]
- Shah, S.S.; Rodriguez, G.A.; Musick, A.; Walters, W.M.; de Cordoba, N.; Barbarite, E.; Marlow, M.M.; Marples, B.; Prince, J.S.; Komotar, R.J.; et al. Targeting Glioblastoma Stem Cells with 2-Deoxy-D-Glucose (2-DG) Potentiates Radiation-Induced Unfolded Protein Response (UPR). Cancers 2019, 11, 159. [Google Scholar] [CrossRef] [PubMed]
- Dwarakanath, B.S.; Singh, D.; Banerji, A.K.; Sarin, R.; Venkataramana, N.K.; Jalali, R.; Vishwanath, P.N.; Mohanti, B.K.; Tripathi, R.P.; Kalia, V.K.; et al. Clinical studies for improving radiotherapy with 2-deoxy-D-glucose: Present status and future prospects. J. Cancer Res. Ther. 2009, 5, S21–S26. [Google Scholar] [CrossRef]
- Taylor, S.R.; Falcone, J.N.; Cantley, L.C.; Goncalves, M.D. Developing dietary interventions as therapy for cancer. Nat. Rev. Cancer 2022, 22, 452–466. [Google Scholar] [CrossRef] [PubMed]
- Gimple, R.C.; Kidwell, R.L.; Kim, L.J.Y.; Sun, T.; Gromovsky, A.D.; Wu, Q.; Wolf, M.; Lv, D.; Bhargava, S.; Jiang, L.; et al. Glioma Stem Cell-Specific Superenhancer Promotes Polyunsaturated Fatty-Acid Synthesis to Support EGFR Signaling. Cancer Discov. 2019, 9, 1248–1267. [Google Scholar] [CrossRef]
- Gini, B.; Zanca, C.; Guo, D.; Matsutani, T.; Masui, K.; Ikegami, S.; Yang, H.; Nathanson, D.; Villa, G.R.; Shackelford, D.; et al. The mTOR kinase inhibitors, CC214-1 and CC214-2, preferentially block the growth of EGFRvIII-activated glioblastomas. Clin. Cancer Res. 2013, 19, 5722–5732. [Google Scholar] [CrossRef] [PubMed]
- Tateishi, K.; Wakimoto, H.; Iafrate, A.J.; Tanaka, S.; Loebel, F.; Lelic, N.; Wiederschain, D.; Bedel, O.; Deng, G.; Zhang, B.; et al. Extreme Vulnerability of IDH1 Mutant Cancers to NAD+ Depletion. Cancer Cell 2015, 28, 773–784. [Google Scholar] [CrossRef]
- Turcan, S.; Makarov, V.; Taranda, J.; Wang, Y.; Fabius, A.W.M.; Wu, W.; Zheng, Y.; El-Amine, N.; Haddock, S.; Nanjangud, G.; et al. Mutant-IDH1-dependent chromatin state reprogramming, reversibility, and persistence. Nat. Genet. 2018, 50, 62–72. [Google Scholar] [CrossRef]
- Miller, J.J.; Fink, A.; Banagis, J.A.; Nagashima, H.; Subramanian, M.; Lee, C.K.; Melamed, L.; Tummala, S.S.; Tateishi, K.; Wakimoto, H.; et al. Sirtuin activation targets IDH-mutant tumors. Neuro. Oncol. 2021, 23, 53–62. [Google Scholar] [CrossRef]
- Kang, M.K.; Kang, S.K. Tumorigenesis of chemotherapeutic drug-resistant cancer stem-like cells in brain glioma. Stem Cells Dev. 2007, 16, 837–847. [Google Scholar] [CrossRef]
- Yuan, S.; Wang, F.; Chen, G.; Zhang, H.; Feng, L.; Wang, L.; Colman, H.; Keating, M.J.; Li, X.; Xu, R.H.; et al. Effective elimination of cancer stem cells by a novel drug combination strategy. Stem Cells 2013, 31, 23–34. [Google Scholar] [CrossRef]
- Azzalin, A.; Nato, G.; Parmigiani, E.; Garello, F.; Buffo, A.; Magrassi, L. Inhibitors of GLUT/SLC2A Enhance the Action of BCNU and Temozolomide against High-Grade Gliomas. Neoplasia 2017, 19, 364–373. [Google Scholar] [CrossRef] [PubMed]
- Veys, K.; Fan, Z.; Ghobrial, M.; Bouche, A.; Garcia-Caballero, M.; Vriens, K.; Conchinha, N.V.; Seuwen, A.; Schlegel, F.; Gorski, T.; et al. Role of the GLUT1 Glucose Transporter in Postnatal CNS Angiogenesis and Blood-Brain Barrier Integrity. Circ. Res. 2020, 127, 466–482. [Google Scholar] [CrossRef] [PubMed]
- Marin-Valencia, I.; Yang, C.; Mashimo, T.; Cho, S.; Baek, H.; Yang, X.L.; Rajagopalan, K.N.; Maddie, M.; Vemireddy, V.; Zhao, Z.; et al. Analysis of tumor metabolism reveals mitochondrial glucose oxidation in genetically diverse human glioblastomas in the mouse brain in vivo. Cell Metab. 2012, 15, 827–837. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Zhao, X.; Ren, W.; Wang, X.; Yu, K.F.; Li, D.; Zhang, X.; Zhang, Q. Antitumor activity of dichloroacetate on C6 glioma cell: In vitro and in vivo evaluation. OncoTargets Ther. 2013, 6, 189–198. [Google Scholar] [CrossRef]
- Michelakis, E.D.; Sutendra, G.; Dromparis, P.; Webster, L.; Haromy, A.; Niven, E.; Maguire, C.; Gammer, T.L.; Mackey, J.R.; Fulton, D.; et al. Metabolic modulation of glioblastoma with dichloroacetate. Sci. Transl. Med. 2010, 2, 31ra34. [Google Scholar] [CrossRef] [PubMed]
- Oliva, C.R.; Nozell, S.E.; Diers, A.; McClugage, S.G., 3rd; Sarkaria, J.N.; Markert, J.M.; Darley-Usmar, V.M.; Bailey, S.M.; Gillespie, G.Y.; Landar, A.; et al. Acquisition of temozolomide chemoresistance in gliomas leads to remodeling of mitochondrial electron transport chain. J. Biol. Chem. 2010, 285, 39759–39767. [Google Scholar] [CrossRef]
- Molina, J.R.; Sun, Y.; Protopopova, M.; Gera, S.; Bandi, M.; Bristow, C.; McAfoos, T.; Morlacchi, P.; Ackroyd, J.; Agip, A.A.; et al. An inhibitor of oxidative phosphorylation exploits cancer vulnerability. Nat. Med. 2018, 24, 1036–1046. [Google Scholar] [CrossRef]
- Liu, Y.; Fang, S.; Sun, Q.; Liu, B. Anthelmintic drug ivermectin inhibits angiogenesis, growth and survival of glioblastoma through inducing mitochondrial dysfunction and oxidative stress. Biochem. Biophys. Res. Commun. 2016, 480, 415–421. [Google Scholar] [CrossRef]
- Mudassar, F.; Shen, H.; O’Neill, G.; Hau, E. Targeting tumor hypoxia and mitochondrial metabolism with anti-parasitic drugs to improve radiation response in high-grade gliomas. J. Exp. Clin. Cancer Res. 2020, 39, 208. [Google Scholar] [CrossRef]
- Ishida, C.T.; Zhang, Y.; Bianchetti, E.; Shu, C.; Nguyen, T.T.T.; Kleiner, G.; Sanchez-Quintero, M.J.; Quinzii, C.M.; Westhoff, M.A.; Karpel-Massler, G.; et al. Metabolic Reprogramming by Dual AKT/ERK Inhibition through Imipridones Elicits Unique Vulnerabilities in Glioblastoma. Clin. Cancer Res. 2018, 24, 5392–5406. [Google Scholar] [CrossRef]
- Ruban, A.; Berkutzki, T.; Cooper, I.; Mohar, B.; Teichberg, V.I. Blood glutamate scavengers prolong the survival of rats and mice with brain-implanted gliomas. Invesitig. New Drugs 2012, 30, 2226–2235. [Google Scholar] [CrossRef] [PubMed]
- Koch, K.; Hartmann, R.; Tsiampali, J.; Uhlmann, C.; Nickel, A.C.; He, X.; Kamp, M.A.; Sabel, M.; Barker, R.A.; Steiger, H.J.; et al. A comparative pharmaco-metabolomic study of glutaminase inhibitors in glioma stem-like cells confirms biological effectiveness but reveals differences in target-specificity. Cell Death Discov. 2020, 6, 20. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).