A Practical Method for Amino Acid Analysis by LC-MS Using Precolumn Derivatization with Urea
Abstract
1. Introduction
2. Results and Discussion
2.1. Feasibility of Urea Derivatization for Amino Acid Analysis
2.2. Method Optimization
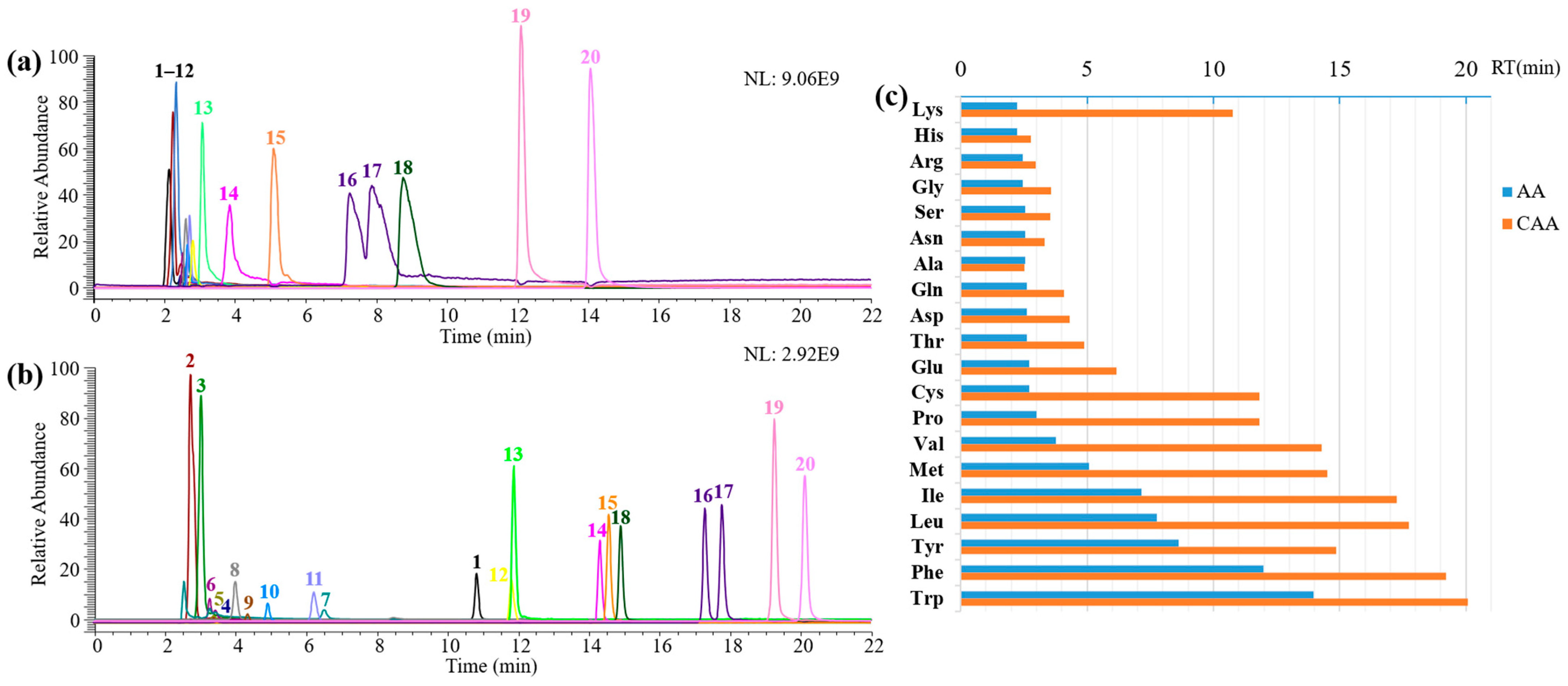
2.3. Derivatization of 20 Amino Acid Mixtures and Real Samples
2.4. Derivatization of Peptides
3. Materials and Methods
3.1. Materials
3.2. Preparation of Standard Curve
3.3. Derivatization Condition Optimization
3.4. Derivatization Reaction with AA Mixture, Real Samples, and Peptides
3.5. LC-ESI-MS Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kaspar, H.; Dettmer, K.; Gronwald, W.; Oefner, P.J. Advances in amino acid analysis. Anal. Bioanal. Chem. 2009, 393, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Zhong, C.; Zou, C.; Wang, B.; Zhang, N. Analytical methods for amino acid determination in organisms. Amino Acids 2020, 52, 1071–1088. [Google Scholar] [CrossRef]
- Shah, A.J.; Crespi, F.; Heidbreder, C. Amino acid neurotransmitters: Separation approaches and diagnostic value. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2002, 781, 151–163. [Google Scholar] [CrossRef] [PubMed]
- Munz, E.; Jakob, P.M.; Borisjuk, L. The potential of nuclear magnetic resonance to track lipids in planta. Biochimie 2016, 130, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Yuan, B.; Ding, Y.; Kamal, G.M.; Shao, L.; Zhou, Z.; Jiang, B.; Sun, P.; Zhang, X.; Liu, M. Reconstructing diffusion ordered NMR spectroscopy by simultaneous inversion of Laplace transform. J. Magn. Reson. 2017, 278, 1–7. [Google Scholar] [CrossRef]
- Poinsot, V.; Ong-Meang, V.; Ric, A.; Gavard, P.; Perquis, L.; Couderc, F. Recent advances in amino acid analysis by capillary electromigration methods: June 2015–May 2017. Electrophoresis 2018, 39, 190–208. [Google Scholar] [CrossRef]
- Lu, T.; Olesik, S.V. Electrospun polyvinyl alcohol ultra-thin layer chromatography of amino acids. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2013, 912, 98–104. [Google Scholar] [CrossRef]
- Gałęzowska, G.; Ratajczyk, J.; Wolska, L. Determination of amino acids in human biological fluids by high-performance liquid chromatography: Critical review. Amino Acids 2021, 53, 993–1009. [Google Scholar] [CrossRef]
- MacKenzie, S.L. Recent developments in amino acid analysis by gas-liquid chromatography. Methods Biochem. Anal. 1981, 27, 1–88. [Google Scholar]
- Wang, H.; McNeil, Y.R.; Yeo, T.W.; Anstey, N.M. Simultaneous determination of multiple amino acids in plasma in critical illness by high performance liquid chromatography with ultraviolet and fluorescence detection. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2013, 940, 53–58. [Google Scholar] [CrossRef]
- Song, Y.; Funatsu, T.; Tsunoda, M. Amino acid analysis using core-shell particle column. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2013, 927, 214–217. [Google Scholar] [CrossRef] [PubMed]
- Tasakis, R.N.; Touraki, M. Identification of bacteriocins secreted by the probiotic Lactococcus lactis following microwave-assisted acid hydrolysis (MAAH), amino acid content analysis, and bioinformatics. Anal. Bioanal. Chem. 2018, 410, 1299–1310. [Google Scholar] [CrossRef] [PubMed]
- Tsai, T.H.; Wang, M.; Ressom, H.W. Preprocessing and analysis of LC-MS-based proteomic data. Methods Mol. Biol. 2016, 1362, 63–76. [Google Scholar] [PubMed]
- Le, A.; Ng, A.; Kwan, T.; Cusmano-Ozog, K.; Cowan, T.M. A rapid, sensitive method for quantitative analysis of underivatized amino acids by liquid chromatography–tandem mass spectrometry (LC–MS/MS). J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2014, 944, 166–174. [Google Scholar] [CrossRef] [PubMed]
- Mochizuki, T.; Todoroki, K.; Inoue, K.; Min, J.Z.; Toyo’oka, T. Isotopic variants of light and heavy L-pyroglutamic acid succinimidyl esters as the derivatization reagents for DL-amino acid chiral metabolomics identification by liquid chromatography and electrospray ionization mass spectrometry. Anal. Chim. Acta 2014, 811, 51–59. [Google Scholar] [CrossRef]
- van Bentum, M.; Selbach, M. An Introduction to Advanced Targeted Acquisition Methods. Mol. Cell Proteom. 2021, 20, 100165. [Google Scholar] [CrossRef]
- Ziegler, J.; Abel, S. Analysis of amino acids by HPLC/electrospray negative ion tandem mass spectrometry using 9-fluorenylmethoxycarbonyl chloride (Fmoc-Cl) derivatization. Amino Acids 2014, 46, 2799–2808. [Google Scholar] [CrossRef]
- Han, M.; Xie, M.; Han, J.; Yuan, D.; Yang, T.; Xie, Y. Development and validation of a rapid, selective, and sensitive LC–MS/MS method for simultaneous determination of d-and l-amino acids in human serum: Application to the study of hepatocellular carcinoma. Anal. Bioanal. Chem. 2018, 410, 2517–2531. [Google Scholar] [CrossRef]
- Zanetta, J.; Vincendon, G.; Mandel, P.; Combos, G. The utilisation of I-dimethylaminonaphthalene-5-sulphonyl chloride for quantitative determination of free amino acids and partial analysis of primary structure of proteins. J. Chromatogr. 1970, 51, 441–458. [Google Scholar] [CrossRef]
- Roth, M. Fluorescence reaction for amino acids. Anal. Chem. 1971, 43, 880–882. [Google Scholar] [CrossRef]
- Cuchiaro, H.; Laurens, L.M.L. Total Protein Analysis in Algae via Bulk Amino Acid Detection: Optimization of Amino Acid Derivatization after Hydrolysis with O-Phthalaldehyde 3-Mercaptopropionic Acid (OPA-3MPA). J. Agric. Food Chem. 2019, 67, 5672–5679. [Google Scholar] [CrossRef]
- Watanabe, Y.; Imai, K. Pre-column labelling for high-performance liquid chromatography of amino acids with 7-fluoro-4-nitrobenzo-2-oxa-1, 3-diazole and its application to protein hydrolysates. J. Chromatogr. A 1982, 239, 723–732. [Google Scholar] [CrossRef]
- Einarsson, S.; Josefsson, B.; Lagerkvist, S. Determination of amino acids with 9-fluorenylmethyl chloroformate and reversed-phase high-performance liquid chromatography. J. Chromatogr. A 1983, 282, 609–618. [Google Scholar] [CrossRef]
- Cohen, S.A.; Michaud, D.P. Synthesis of a fluorescent derivatizing reagent, 6-aminoquinolyl-N-hydroxysate amino acids via high-performance liquid chromatography. Anal. Biochem. 1993, 211, 279–287. [Google Scholar] [CrossRef] [PubMed]
- Shimbo, K.; Yahashi, A.; Hirayama, K.; Nakazawa, M.; Miyano, H. Multifunctional and Highly Sensitive Precolumn Reagents for Amino Acids in Liquid Chromatography/Tandem Mass Spectrometry. Anal. Chem. 2009, 81, 5172–5179. [Google Scholar] [CrossRef]
- Cohen, S.A.; Bidlingmeyer, B.A.; Tarvin, T.L. PITC derivatives in amino acid analysis. Nature 1986, 320, 769–770. [Google Scholar] [CrossRef]
- Verardo, G.; Geatti, P.; Strazzolini, P. Rapid and Efficient Microwave-Assisted Synthesis of N-Carbamoyl-L-amino Acids. Synth. Commun. 2007, 37, 1833–1844. [Google Scholar] [CrossRef]
- Zapata Flores, E.J.; Bùi, N.K.N.; Selberg, S.; Herodes, K.; Leito, I. Comparison of two azobenzene-based amino acid derivatization reagents for LC-MS/MS analysis in positive and negative ESI modes. Talanta 2023, 252, 123803. [Google Scholar] [CrossRef]
- Rebane, R.; Oldekop, M.L.; Herodes, K. Comparison of amino acid derivatization reagents for LC-ESI-MS analysis. Introducing a novel phosphazene-based derivatization reagent. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2012, 904, 99–106. [Google Scholar] [CrossRef]
- Rebane, R.; Herodes, K. Comparison of three buffer solutions for amino acid derivatization and following analysis by liquid chromatography electrospray mass spectrometry. J. Chromatogr. A 2012, 1245, 134–142. [Google Scholar] [CrossRef]
- Uutela, P.I.; Ketola, R.A.; Piepponen, P.; Kostiainen, R.J.A.C.A. Comparison of different amino acid derivatives and analysis of rat brain microdialysates by liquid chromatography tandem mass spectrometry. Anal. Chim. Acta 2009, 633, 223–231. [Google Scholar] [CrossRef] [PubMed]
- Jámbor, A.; Molnár-Perl, I. Amino acid analysis by high-performance liquid chromatography after derivatization with 9-fluorenylmethyloxycarbonyl chloride Literature overview and further study. J. Chromatogr. A 2009, 1216, 3064–3077. [Google Scholar] [CrossRef] [PubMed]
- Rebane, R.; Herodes, K. A sensitive method for free amino acids analysis by liquid chromatography with ultraviolet and mass spectrometric detection using precolumn derivatization with diethyl ethoxymethylenemalonate: Application to the honey analysis. Anal. Chim. Acta 2010, 672, 79–84. [Google Scholar] [CrossRef] [PubMed]




| Analyte | Linear Equation | Linear Range (nM) | R2 | LOD (nM) a | LOD (nM) b | LOQ (nM) c | LOQ (nM) d |
|---|---|---|---|---|---|---|---|
| Ala | y = 7.5905 × 1011x + 672444175.81 | 103–107 | 0.9939 | 130.78 | 58.60 | 435.50 | 195.40 |
| Phe | y =1.10681 × 1012x + 1634677812.7 | 103–107 | 0.9923 | 19.96 | 11.64 | 66.47 | 38.80 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, R.; Huang, B.; Lu, G.; Fu, S.; Ying, J.; Zhao, Y. A Practical Method for Amino Acid Analysis by LC-MS Using Precolumn Derivatization with Urea. Int. J. Mol. Sci. 2023, 24, 7332. https://doi.org/10.3390/ijms24087332
Zhao R, Huang B, Lu G, Fu S, Ying J, Zhao Y. A Practical Method for Amino Acid Analysis by LC-MS Using Precolumn Derivatization with Urea. International Journal of Molecular Sciences. 2023; 24(8):7332. https://doi.org/10.3390/ijms24087332
Chicago/Turabian StyleZhao, Runjin, Biling Huang, Gang Lu, Songsen Fu, Jianxi Ying, and Yufen Zhao. 2023. "A Practical Method for Amino Acid Analysis by LC-MS Using Precolumn Derivatization with Urea" International Journal of Molecular Sciences 24, no. 8: 7332. https://doi.org/10.3390/ijms24087332
APA StyleZhao, R., Huang, B., Lu, G., Fu, S., Ying, J., & Zhao, Y. (2023). A Practical Method for Amino Acid Analysis by LC-MS Using Precolumn Derivatization with Urea. International Journal of Molecular Sciences, 24(8), 7332. https://doi.org/10.3390/ijms24087332






