Longitudinal Variations in Antibody Responses against SARS-CoV-2 Spike Epitopes upon Serial Vaccinations
Abstract
1. Introduction
2. Results
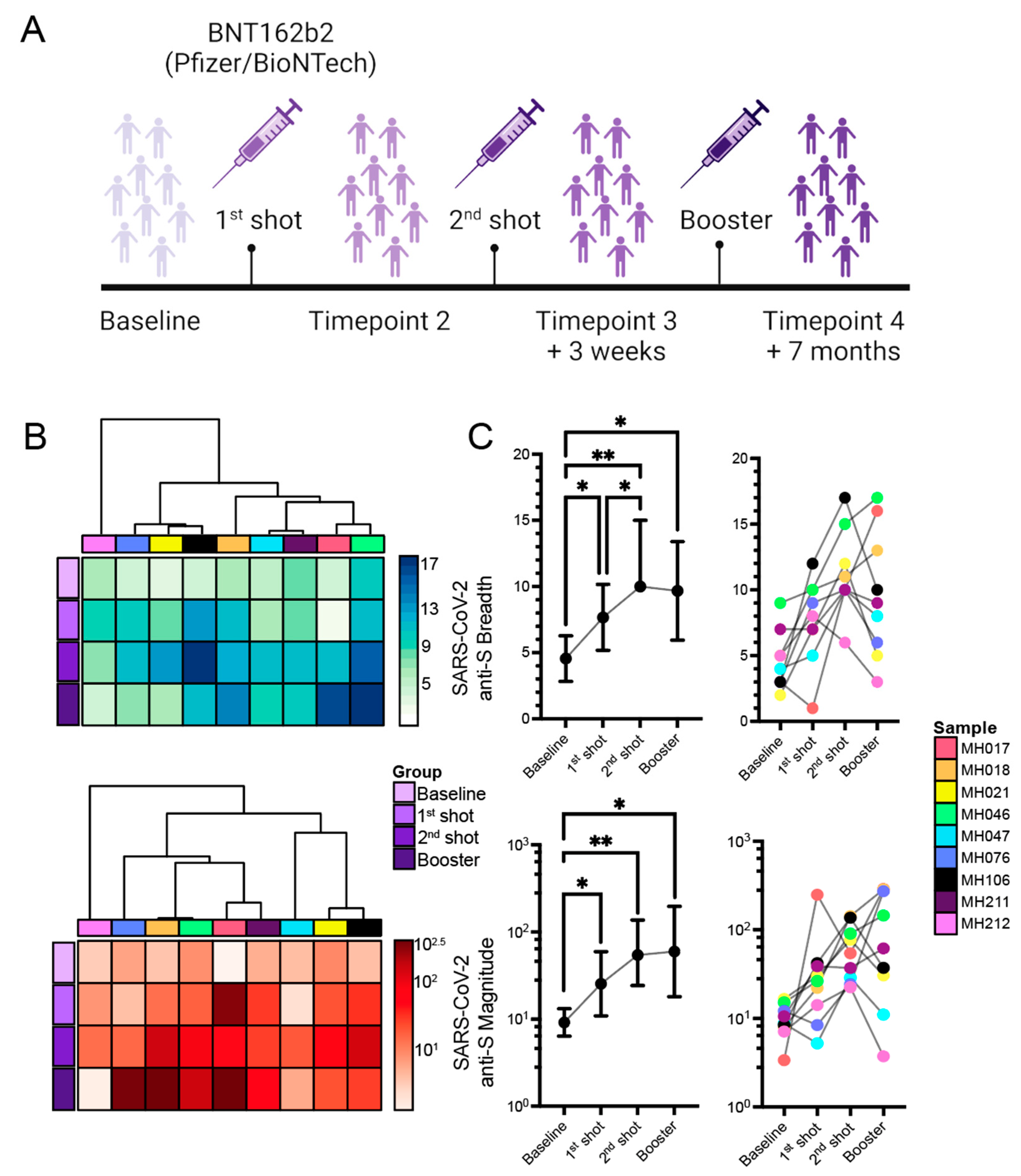
2.1. Ab Responses to SARS-CoV-2 mRNA Vaccination
Association of Differences of Vaccination Time or Sample Collection with Changes in Responses against the S Protein
2.2. Cross-Reactivities of the Anti-SARS-CoV-2 Ab Responses against Other CoVs
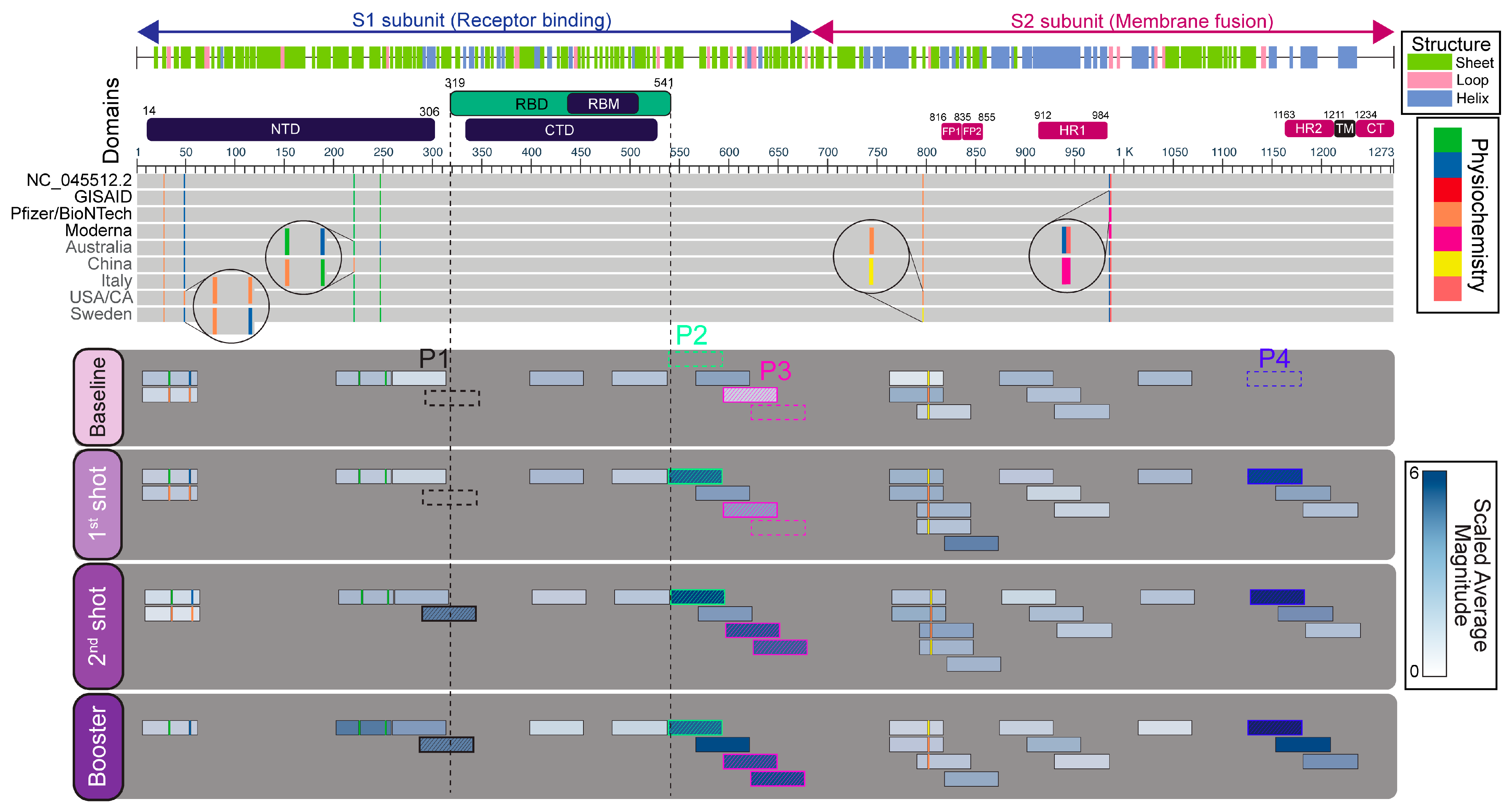
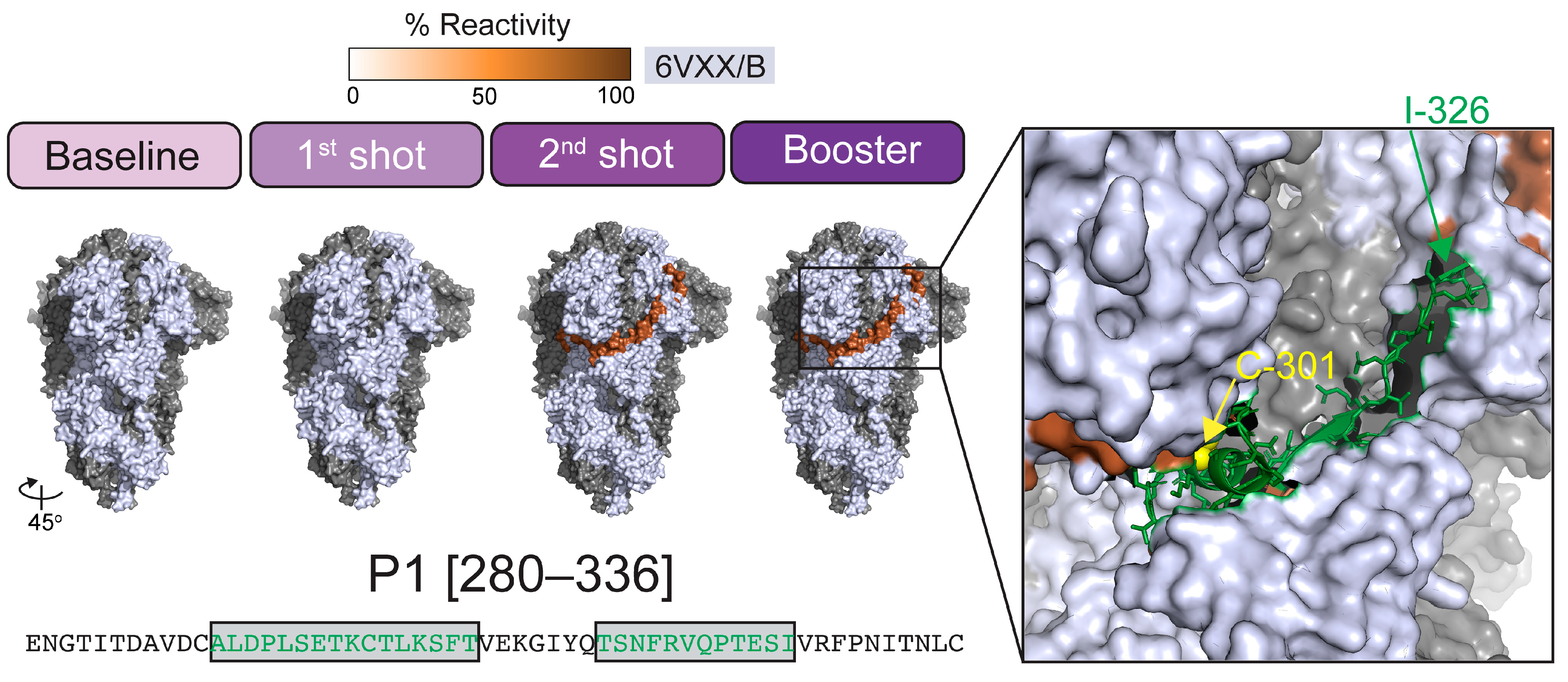
2.3. Immunogenicity Map and Annotation of SARS-CoV-2 Spike (S) Glycoprotein-Reactive Peptides
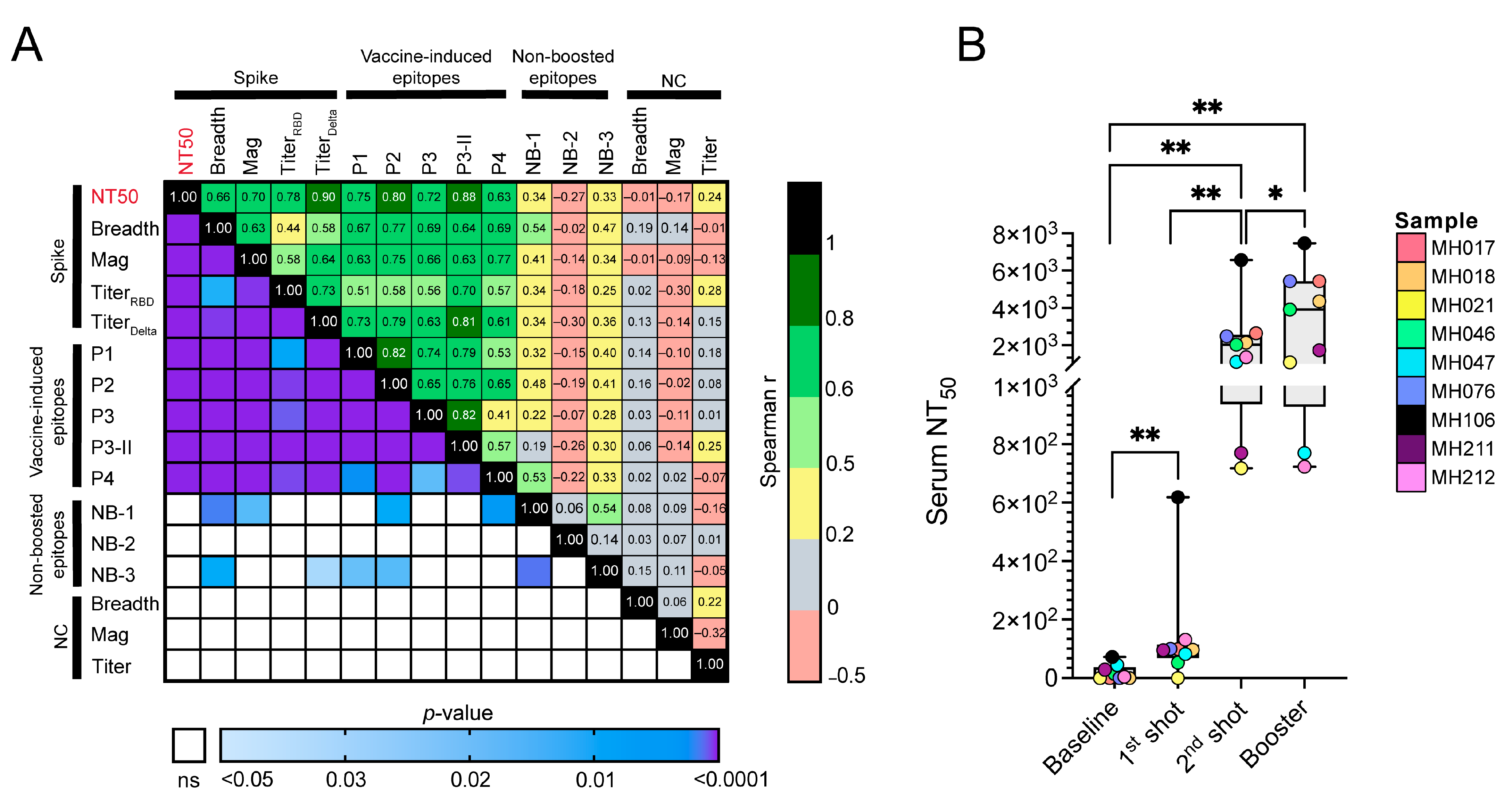
2.4. Associations of Vaccine-Induced and Non-Boosted Responses with Titer, Neutralization, and the Breadth and Magnitude of Anti-S Responses
3. Discussion
4. Materials and Methods
4.1. Subject Recruitment/COVID-19 Serological Analysis
4.2. Phage Immunoprecipitation and Sequencing (PhIP-Seq)
4.3. Pseudovirus Production and Neutralization Assays
4.4. Data Processing and Statistical Analyses
4.5. Epitope and Structural Mapping
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ryan, E.T. The Cholera Pandemic, Still with Us after Half a Century: Time to Rethink. PLoS Negl. Trop. Dis. 2011, 5, e1003. [Google Scholar] [CrossRef] [PubMed]
- Quinn, T.C. Global burden of the HIV pandemic. Lancet 1996, 348, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, Y.C.; Wu, T.Z.; Liu, D.P.; Shao, P.L.; Chang, L.Y.; Lu, C.Y.; Lee, C.Y.; Huang, F.Y.; Huang, L.M. Influenza Pandemics: Past, Present and Future. J. Formos. Med. Assoc. 2006, 105, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Dong, E.; Du, H.; Gardner, L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020, 20, 533–534. [Google Scholar] [CrossRef] [PubMed]
- Gorbalenya, A.E.; Baker, S.C.; Baric, R.S.; de Groot, R.J.; Drosten, C.; Gulyaeva, A.A.; Haagmans, B.L.; Lauber, C.; Leontovich, A.M.; Neuman, B.W.; et al. The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar] [CrossRef]
- Wu, A.; Peng, Y.; Huang, B.; Ding, X.; Wang, X.; Niu, P.; Meng, J.; Zhu, Z.; Zhang, Z.; Wang, J.; et al. Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China. Cell Host Microbe 2020, 27, 325–328. [Google Scholar] [CrossRef]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef]
- Liu, L.; Wang, P.; Nair, M.S.; Yu, J.; Rapp, M.; Wang, Q.; Luo, Y.; Chan, J.F.W.; Sahi, V.; Figueroa, A.; et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature 2020, 584, 450–456. [Google Scholar] [CrossRef]
- Cao, Y.; Su, B.; Guo, X.; Sun, W.; Deng, Y.; Bao, L.; Zhu, Q.; Zhang, X.; Zheng, Y.; Geng, C.; et al. Potent Neutralizing Antibodies against SARS-CoV-2 Identified by High-Throughput Single-Cell Sequencing of Convalescent Patients’ B Cells. Cell 2020, 182, 73–84.e16. [Google Scholar] [CrossRef]
- Brouwer, P.J.M.; Caniels, T.G.; van der Straten, K.; Snitselaar, J.L.; Aldon, Y.; Bangaru, S.; Torres, J.L.; Okba, N.M.A.; Claireaux, M.; Kerster, G.; et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science 2020, 369, 643–650. [Google Scholar] [CrossRef]
- Khoury, D.S.; Cromer, D.; Reynaldi, A.; Schlub, T.E.; Wheatley, A.K.; Juno, J.A.; Subbarao, K.; Kent, S.J.; Triccas, J.A.; Davenport, M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021, 27, 1205–1211. [Google Scholar] [CrossRef]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA COVID-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef]
- Corbett, K.S.; Edwards, D.K.; Leist, S.R.; Abiona, O.M.; Boyoglu-Barnum, S.; Gillespie, R.A.; Himansu, S.; Schäfer, A.; Ziwawo, C.T.; DiPiazza, A.T.; et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature 2020, 586, 567–571. [Google Scholar] [CrossRef]
- Wrapp, D.; Wang, N.; Corbett, K.; Goldsmith, J.; Hsieh, C.L.; Abiona, O.; Graham, B.S.; McLellan, J.S. undefined Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef]
- Lopez Bernal, J.; Andrews, N.; Gower, C.; Gallagher, E.; Simmons, R.; Thelwall, S.; Stowe, J.; Tessier, E.; Groves, N.; Dabrera, G.; et al. Effectiveness of COVID-19 Vaccines against the B.1.617.2 (Delta) Variant. N. Engl. J. Med. 2021, 385, 585–594. [Google Scholar] [CrossRef]
- Nemet, I.; Kliker, L.; Lustig, Y.; Zuckerman, N.; Erster, O.; Cohen, C.; Kreiss, Y.; Alroy-Preis, S.; Regev-Yochay, G.; Mendelson, E.; et al. Third BNT162b2 Vaccination Neutralization of SARS-CoV-2 Omicron Infection. N. Engl. J. Med. 2022, 386, 492–494. [Google Scholar] [CrossRef]
- Saciuk, Y.; Kertes, J.; Shamir Stein, N.; Ekka Zohar, A. Effectiveness of a Third Dose of BNT162b2 mRNA Vaccine. J. Infect. Dis. 2022, 225, 30–33. [Google Scholar] [CrossRef]
- Arbel, R.; Hammerman, A.; Sergienko, R.; Friger, M.; Peretz, A.; Netzer, D.; Yaron, S. BNT162b2 Vaccine Booster and Mortality Due to COVID-19. N. Engl. J. Med. 2021, 385, 2413–2420. [Google Scholar] [CrossRef]
- Buckner, C.M.; Kardava, L.; El Merhebi, O.; Narpala, S.R.; Serebryannyy, L.; Lin, B.C.; Wang, W.; Zhang, X.; Lopes de Assis, F.; Kelly, S.E.M.; et al. Interval between prior SARS-CoV-2 infection and booster vaccination impacts magnitude and quality of antibody and B cell responses. Cell 2022, 185, 4333–4346.e14. [Google Scholar] [CrossRef]
- Bowen, J.E.; Addetia, A.; Dang, H.V.; Stewart, C.; Brown, J.T.; Sharkey, W.K.; Sprouse, K.R.; Walls, A.C.; Mazzitelli, I.G.; Logue, J.K.; et al. Omicron spike function and neutralizing activity elicited by a comprehensive panel of vaccines. Science 2022, 377, 890–894. [Google Scholar] [CrossRef]
- Sullivan, D.J.; Franchini, M.; Joyner, M.J.; Casadevall, A.; Focosi, D. Analysis of anti-SARS-CoV-2 Omicron-neutralizing antibody titers in different vaccinated and unvaccinated convalescent plasma sources. Nat. Commun. 2022, 13, 6478. [Google Scholar] [CrossRef] [PubMed]
- Bates, T.A.; McBride, S.K.; Leier, H.C.; Guzman, G.; Lyski, Z.L.; Schoen, D.; Winders, B.; Lee, J.Y.; Lee, D.X.; Messer, W.B.; et al. Vaccination before or after SARS-CoV-2 infection leads to robust humoral response and antibodies that effectively neutralize variants. Sci. Immunol. 2022, 7, 8014. [Google Scholar] [CrossRef] [PubMed]
- Shrotri, M.; Fragaszy, E.; Nguyen, V.; Navaratnam, A.M.D.; Geismar, C.; Beale, S.; Kovar, J.; Byrne, T.E.; Fong, W.L.E.; Patel, P.; et al. Spike-antibody responses to COVID-19 vaccination by demographic and clinical factors in a prospective community cohort study. Nat. Commun. 2022, 13, 5780. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ma, M.L.; Lei, Q.; Wang, F.; Hong, W.; Lai, D.Y.; Hou, H.; Xu, Z.W.; Zhang, B.; Chen, H.; et al. Linear epitope landscape of the SARS-CoV-2 Spike protein constructed from 1051 COVID-19 patients. Cell Rep. 2021, 34, 108915. [Google Scholar] [CrossRef]
- Bartsch, Y.C.; Cizmeci, D.; Kang, J.; Zohar, T.; Periasamy, S.; Mehta, N.; Tolboom, J.; Van der Fits, L.; Sadoff, J.; Comeaux, C.; et al. Antibody effector functions are associated with protection from respiratory syncytial virus. Cell 2022, 185, 4873–4886.e10. [Google Scholar] [CrossRef]
- Xu, G.J.; Kula, T.; Xu, Q.; Li, M.Z.; Vernon, S.D.; Ndung’u, T.; Ruxrungtham, K.; Sanchez, J.; Brander, C.; Chung, R.T.; et al. Comprehensive serological profiling of human populations using a synthetic human virome. Science 2015, 348, aaa0698. [Google Scholar] [CrossRef]
- Mohan, D.; Wansley, D.L.; Sie, B.M.; Noon, M.S.; Baer, A.N.; Laserson, U.; Larman, H.B. PhIP-Seq characterization of serum antibodies using oligonucleotide-encoded peptidomes. Nat. Protoc. 2018, 13, 1958–1978. [Google Scholar] [CrossRef]
- Haynes, W.A.; Kamath, K.; Bozekowski, J.; Baum-Jones, E.; Campbell, M.; Casanovas-Massana, A.; Daugherty, P.S.; Dela Cruz, C.S.; Dhal, A.; Farhadian, S.F.; et al. High-resolution epitope mapping and characterization of SARS-CoV-2 antibodies in large cohorts of subjects with COVID-19. Commun. Biol. 2021, 4, 1317. [Google Scholar] [CrossRef]
- Shrock, E.; Fujimura, E.; Kula, T.; Timms, R.T.; Lee, I.H.; Leng, Y.; Robinson, M.L.; Sie, B.M.; Li, M.Z.; Chen, Y.; et al. Viral epitope profiling of COVID-19 patients reveals cross-reactivity and correlates of severity. Science 2020, 370, eabd4250. [Google Scholar] [CrossRef]
- Lee, J.-S.; Poo, H.; Han, D.P.; Hong, S.-P.; Kim, K.; Cho, M.W.; Kim, E.; Sung, M.-H.; Kim, C.-J. Mucosal immunization with surface-displayed severe acute respiratory syndrome coronavirus spike protein on Lactobacillus casei induces neutralizing antibodies in mice. J. Virol. 2006, 80, 4079–4087. [Google Scholar] [CrossRef]
- Amrun, S.N.; Lee, C.Y.P.; Lee, B.; Fong, S.W.; Young, B.E.; Chee, R.S.L.; Yeo, N.K.W.; Torres-Ruesta, A.; Carissimo, G.; Poh, C.M.; et al. Linear B-cell epitopes in the spike and nucleocapsid proteins as markers of SARS-CoV-2 exposure and disease severity. EBioMedicine 2020, 58, 102911. [Google Scholar] [CrossRef]
- Farrera-Soler, L.; Daguer, J.P.; Barluenga, S.; Vadas, O.; Cohen, P.; Pagano, S.; Yerly, S.; Kaiser, L.; Vuilleumier, N.; Winssinger, N. Identification of immunodominant linear epitopes from SARS-CoV-2 patient plasma. PLoS ONE 2020, 15, e0238089. [Google Scholar] [CrossRef]
- Li, Y.; Lai, D.Y.; Zhang, H.N.; Jiang, H.W.; Tian, X.; Ma, M.L.; Qi, H.; Meng, Q.F.; Guo, S.J.; Wu, Y.; et al. Linear epitopes of SARS-CoV-2 spike protein elicit neutralizing antibodies in COVID-19 patients. Cell Mol. Immunol. 2020, 17, 1095–1097. [Google Scholar] [CrossRef]
- Bhattacharya, M.; Sharma, A.R.; Patra, P.; Ghosh, P.; Sharma, G.; Patra, B.C.; Lee, S.S.; Chakraborty, C. Development of epitope-based peptide vaccine against novel coronavirus 2019 (SARS-COV-2): Immunoinformatics approach. J. Med. Virol. 2020, 92, 618–631. [Google Scholar] [CrossRef]
- He, Y.; Zhou, Y.; Wu, H.; Luo, B.; Chen, J.; Li, W.; Jiang, S. Identification of Immunodominant Sites on the Spike Protein of Severe Acute Respiratory Syndrome (SARS) Coronavirus: Implication for Developing SARS Diagnostics and Vaccines. J. Immunol. 2004, 173, 4050–4057. [Google Scholar] [CrossRef]
- Singh Slathia, P.; Sharma, P. Prediction of T and B Cell Epitopes in the Proteome of SARS-CoV-2 for Potential Use in Diagnostics and Vaccine Design. ChemRxiv 2020. [Google Scholar] [CrossRef]
- Ahmed, S.F.; Quadeer, A.A.; McKay, M.R. Preliminary Identification of Potential Vaccine Targets for the COVID-19 Coronavirus (SARS-CoV-2) Based on SARS-CoV Immunological Studies. Viruses 2020, 12, 254. [Google Scholar] [CrossRef]
- Poh, C.M.; Carissimo, G.; Wang, B.; Amrun, S.N.; Lee, C.Y.P.; Chee, R.S.L.; Fong, S.W.; Yeo, N.K.W.; Lee, W.H.; Torres-Ruesta, A.; et al. Two linear epitopes on the SARS-CoV-2 spike protein that elicit neutralising antibodies in COVID-19 patients. Nat. Commun. 2020, 11, 2806. [Google Scholar] [CrossRef]
- Wisnewski, A.V.; Redlich, C.A.; Liu, J.; Kamath, K.; Abad, Q.A.; Smith, R.F.; Fazen, L.; Santiago, R.; Luna, J.C.; Martinez, B.; et al. Immunogenic amino acid motifs and linear epitopes of COVID-19 mRNA vaccines. PLoS ONE 2021, 16, e0252849. [Google Scholar] [CrossRef]
- Karachaliou, M.; Moncunill, G.; Espinosa, A.; Castaño-Vinyals, G.; Rubio, R.; Vidal, M.; Jiménez, A.; Prados, E.; Carreras, A.; Cortés, B.; et al. SARS-CoV-2 infection, vaccination, and antibody response trajectories in adults: A cohort study in Catalonia. BMC Med. 2022, 20, 1–16. [Google Scholar] [CrossRef]
- Bellusci, L.; Grubbs, G.; Zahra, F.T.; Forgacs, D.; Golding, H.; Ross, T.M.; Khurana, S. Antibody affinity and cross-variant neutralization of SARS-CoV-2 Omicron BA.1, BA.2 and BA.3 following third mRNA vaccination. Nat. Commun. 2022, 13, 347. [Google Scholar] [CrossRef]
- Bowen, J.E.; Park, Y.-J.; Stewart, C.; Brown, J.T.; Sharkey, W.K.; Walls, A.C.; Joshi, A.; Sprouse, K.R.; McCallum, M.; Tortorici, M.A.; et al. SARS-CoV-2 spike conformation determines plasma neutralizing activity elicited by a wide panel of human vaccines. Sci. Immunol. 2022, 7, eadf1421. [Google Scholar] [CrossRef] [PubMed]
- Suryadevara, N.; Shrihari, S.; Gilchuk, P.; VanBlargan, L.A.; Binshtein, E.; Zost, S.J.; Nargi, R.S.; Sutton, R.E.; Winkler, E.S.; Chen, E.C.; et al. Neutralizing and protective human monoclonal antibodies recognizing the N-terminal domain of the SARS-CoV-2 spike protein. Cell 2021, 184, 2316–2331.e15. [Google Scholar] [CrossRef] [PubMed]
- Collier, D.A.; De Marco, A.; Ferreira, I.A.T.M.; Meng, B.; Datir, R.P.; Walls, A.C.; Kemp, S.A.; Bassi, J.; Pinto, D.; Silacci-Fregni, C.; et al. Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature 2021, 593, 136–141. [Google Scholar] [CrossRef]
- Vishwakarma, P.; Yadav, N.; Rizvi, Z.A.; Khan, N.A.; Chiranjivi, A.K.; Mani, S.; Bansal, M.; Dwivedi, P.; Shrivastava, T.; Kumar, R.; et al. Severe Acute Respiratory Syndrome Coronavirus 2 Spike Protein Based Novel Epitopes Induce Potent Immune Responses in vivo and Inhibit Viral Replication in vitro. Front. Immunol. 2021, 12, 513. [Google Scholar] [CrossRef]
- Voss, W.N.; Hou, Y.J.; Johnson, N.V.; Delidakis, G.; Kim, J.E.; Javanmardi, K.; Horton, A.P.; Bartzoka, F.; Paresi, C.J.; Tanno, Y.; et al. Prevalent, protective, and convergent IgG recognition of SARS-CoV-2 non-RBD spike epitopes. Science 2021, 372, 1108–1112. [Google Scholar] [CrossRef]
- Piccoli, L.; Park, Y.J.; Tortorici, M.A.; Czudnochowski, N.; Walls, A.C.; Beltramello, M.; Silacci-Fregni, C.; Pinto, D.; Rosen, L.E.; Bowen, J.E.; et al. Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology. Cell 2020, 183, 1024–1042.e21. [Google Scholar] [CrossRef]
- Bianchini, F.; Crivelli, V.; Abernathy, M.E.; Guerra, C.; Palus, M.; Muri, J.; Marcotte, H.; Piralla, A.; Pedotti, M.; De Gasparo, R.; et al. Human neutralizing antibodies to cold linear epitopes and subdomain 1 of the SARS-CoV-2 spike glycoprotein. Sci. Immunol. 2023, 8, eade0958. [Google Scholar] [CrossRef]
- To, K.K.W.; Tsang, O.T.Y.; Leung, W.S.; Tam, A.R.; Wu, T.C.; Lung, D.C.; Yip, C.C.Y.; Cai, J.P.; Chan, J.M.C.; Chik, T.S.H.; et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: An observational cohort study. Lancet. Infect. Dis. 2020, 20, 565–574. [Google Scholar] [CrossRef]
- Bennett, S.J.; Yalcin, D.; Privatt, S.R.; Ngalamika, O.; Lidenge, S.J.; West, J.T.; Wood, C. Antibody epitope profiling of the KSHV LANA protein using VirScan. PLoS Pathog. 2022, 18, e1011033. [Google Scholar] [CrossRef]

| Demographics | Baseline (n = 9) | 1st Shot (n = 9) | 2nd Shot (n = 9) | Booster (n = 9) |
|---|---|---|---|---|
| Age|Mean (SD) | 47 (13.23) | – | – | – |
| Sex|Male (Female) | 5 (4) | – | – | – |
| Race|n | W (8), H (1) | – | – | – |
| Sample Collection | May 20–January 21 | February 21 | February–March 21 | October–December 21 |
| Vaccination | – | January 21 | February 21 | October–December 21 |
| Antibody | ||||
| RBD/Spike Titer B****, C** | n (max %) | |||
| Below LOD | 7 (78%) | 1 | 1 | 0 |
| Low | 0 | 1 | 0 | 0 |
| Medium | 2 | 6 (67%) | 3 | 0 |
| High | 0 | 1 | 5 (56%) | 9 (100%) |
| NC Titer | n (max %) | |||
| Below LOD | 6 (67%) | 7 (78%) | 5 (56%) | 6 (67%) |
| Low | 3 | 2 | 3 | 2 |
| Medium | 0 | 0 | 1 | 1 |
| High | 0 | 0 | 0 | 0 |
| Delta Titer A***, B*** | n (max %) | |||
| Below LOD | 6 (67%) | 1 | 0 | 0 |
| Low | 1 | 1 | 0 | 0 |
| Medium | 2 | 4 (45%) | 1 | 0 |
| High | 0 | 3 | 8 (89%) | 9 (100%) |
| Total IgG A**, B*, D** | ||||
| Median (mg/mL) (IQR) | 3.178 (2.070) | 2.024 (1.740) | 2.013 (0.633) | 1.949 (2.492) |
| Neutralization A**, B*, C**** | ||||
| Serum NT50|Mean (SD) | 18.151 (25.302) | 141.159 (182.859) | 2199.893 (1785.858) | 3433.839 (2451.889) |
| Sequencing Data | ||||
| Total counts/Replicate | ||||
| Median (IQR) | 1,576,212 (201,681.25) | 1,640,594.5 (118,177.75) | 1,604,607 (179,009.5) | 1,569,583 (166,601) |
| Fraction of aligned reads (%) | >97% | >97% | >97% | >97% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yalcin, D.; Bennett, S.J.; Sheehan, J.; Trauth, A.J.; Tso, F.Y.; West, J.T.; Hagensee, M.E.; Ramsay, A.J.; Wood, C. Longitudinal Variations in Antibody Responses against SARS-CoV-2 Spike Epitopes upon Serial Vaccinations. Int. J. Mol. Sci. 2023, 24, 7292. https://doi.org/10.3390/ijms24087292
Yalcin D, Bennett SJ, Sheehan J, Trauth AJ, Tso FY, West JT, Hagensee ME, Ramsay AJ, Wood C. Longitudinal Variations in Antibody Responses against SARS-CoV-2 Spike Epitopes upon Serial Vaccinations. International Journal of Molecular Sciences. 2023; 24(8):7292. https://doi.org/10.3390/ijms24087292
Chicago/Turabian StyleYalcin, Dicle, Sydney J. Bennett, Jared Sheehan, Amber J. Trauth, For Yue Tso, John T. West, Michael E. Hagensee, Alistair J. Ramsay, and Charles Wood. 2023. "Longitudinal Variations in Antibody Responses against SARS-CoV-2 Spike Epitopes upon Serial Vaccinations" International Journal of Molecular Sciences 24, no. 8: 7292. https://doi.org/10.3390/ijms24087292
APA StyleYalcin, D., Bennett, S. J., Sheehan, J., Trauth, A. J., Tso, F. Y., West, J. T., Hagensee, M. E., Ramsay, A. J., & Wood, C. (2023). Longitudinal Variations in Antibody Responses against SARS-CoV-2 Spike Epitopes upon Serial Vaccinations. International Journal of Molecular Sciences, 24(8), 7292. https://doi.org/10.3390/ijms24087292






