Influence of Genetics on the Response to Omalizumab in Patients with Severe Uncontrolled Asthma with an Allergic Phenotype
Abstract
1. Introduction
2. Results
2.1. Characteristics of the Patients
2.2. Clinical Effectiveness of Omalizumab
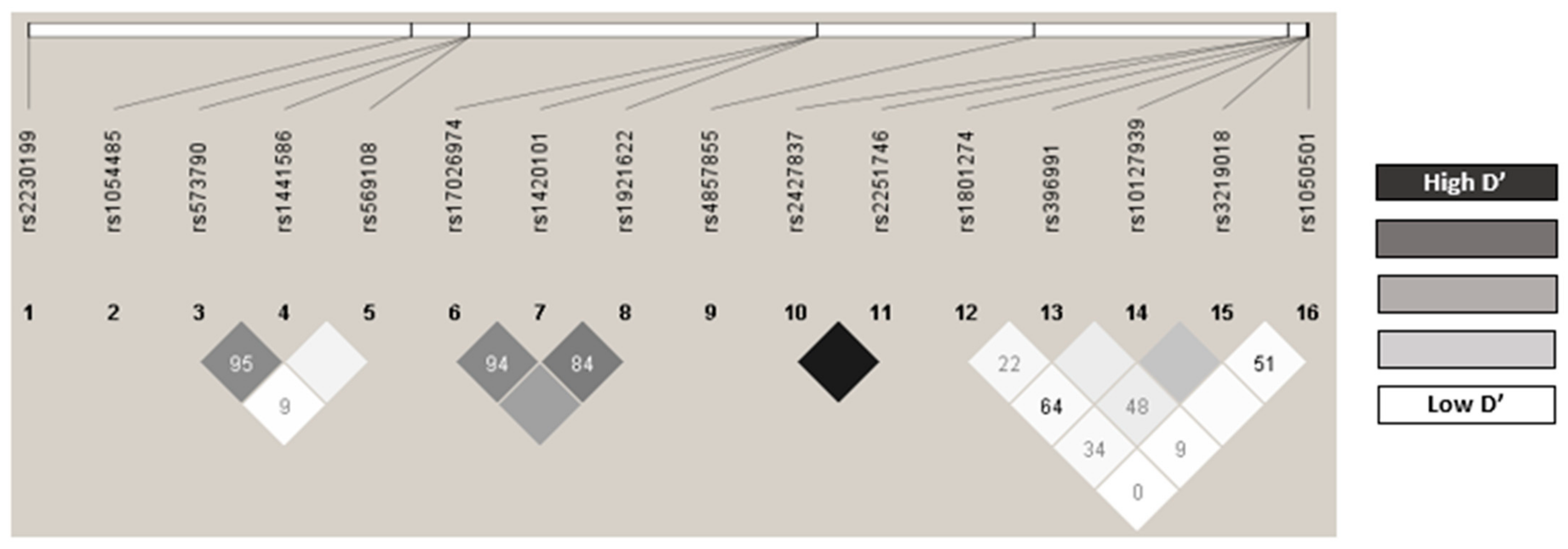
2.3. Distribution of the Genotypes Analyzed
2.4. Predictors of Omalizumab Response at 12 Months
2.4.1. Predictors of Response for Exacerbation Reduction
2.4.2. Predictors of Response for OCS Reduction
2.4.3. Predictors of Response for Lung Function Improvement
2.4.4. Predictors of Meeting at Least One Response Criterion
2.4.5. Predictors of Meeting at Least Two Response Criteria
2.4.6. Predictors of Meeting All Three Criteria
3. Discussion
4. Material and Methods
4.1. Study Design
4.2. Study Population
4.3. Ethics Statements
4.4. Socio-Demographic and Clinical Variables
4.5. Genetic Variables
4.5.1. DNA Isolation
4.5.2. Detection of Gene Polymorphisms and Quality Control
4.6. Response Variables
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sociedad Española de Neumología y Cirugía Torácica. GEMA 5.2. Guía Española para el Manejo del Asma; Luzán 5 Health Consulting, S.A.: Madrid, Spain, 2022. [Google Scholar]
- Abbafati, C.; Abbas, K.M.; Abbasi-Kangevari, M.; Abd-Allah, F.; Abdelalim, A.; Abdollahi, M.; Abdollahpour, I.; Abegaz, K.H.; Abolhassani, H.; Aboyans, V.; et al. Global Burden of 369 Diseases and Injuries in 204 Countries and Territories, 1990–2019: A Systematic Analysis for the Global Burden of Disease Study 2019. Lancet 2020, 396, 1204–1222. [Google Scholar] [CrossRef]
- Williams, S.A.; Wagner, S.; Kannan, H.; Bolge, S.C. The Association Between Asthma Control and Health Care Utilization, Work Productivity Loss and Health-Related Quality of Life. J. Occup. Environ. Med. 2009, 51, 780–785. [Google Scholar] [CrossRef] [PubMed]
- Bobolea, D.I. Fenotipos Del Asma Grave Del Adulto: Las Claves Para La Medicina Personalizada; Servicio de Neumología y Alergia Respiratoria. Hospital Clinic de Barcelona: Barcelona, Spain, 2017. [Google Scholar]
- Carretero Colomer, M. Omalizumab: Tratamiento Del Asma Alérgica Grave. Offarm 2007, 26, 120–122. [Google Scholar]
- Pelaia, C.; Calabrese, C.; Terracciano, R.; de Blasio, F.; Vatrella, A.; Pelaia, G. Omalizumab, the First Available Antibody for Biological Treatment of Severe Asthma: More than a Decade of Real-Life Effectiveness. Ther. Adv. Respir. Dis. 2018, 12, 1753466618810192. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.W.; Wu, P.C.; Hsu, C.L.; Hung, A.F. Anti-IgE Antibodies for the Treatment of IgE-Mediated Allergic Diseases. Adv. Immunol. 2007, 93, 63–119. [Google Scholar] [CrossRef]
- Presta, L.; Shields, R.; O’Connell, L.; Lahr, S.; Porter, J.; Gorman, C.; Jardieu, P. The Binding Site on Human Immunoglobulin E for Its High Affinity Receptor. J. Biol. Chem. 1994, 269, 26368–26373. [Google Scholar] [CrossRef]
- Lobo, E.D.; Hansen, R.J.; Balthasar, J.P. Antibody Pharmacokinetics and Pharmacodynamics. J. Pharm. Sci. 2004, 93, 2645–2668. [Google Scholar] [CrossRef]
- Nimmerjahn, F.; Ravetch, J.V. Fcγ Receptors: Old Friends and New Family Members. Immunity 2006, 24, 19–28. [Google Scholar] [CrossRef]
- Tracey, D.; Klareskog, L.; Sasso, E.H.; Salfeld, J.G.; Tak, P.P. Tumor Necrosis Factor Antagonist Mechanisms of Action: A Comprehensive Review. Pharmacol. Ther. 2008, 117, 244–279. [Google Scholar] [CrossRef]
- Inoue, H.; Ito, I.; Niimi, A.; Matsumoto, H.; Oguma, T.; Tajiri, T.; Iwata, T.; Nagasaki, T.; Kanemitsu, Y.; Morishima, T.; et al. Association of Interleukin 1 Receptor-like 1 Gene Polymorphisms with Eosinophilic Phenotype in Japanese Adults with Asthma. Respir. Investig. 2017, 55, 338–347. [Google Scholar] [CrossRef]
- Gudbjartsson, D.F.; Bjornsdottir, U.S.; Halapi, E.; Helgadottir, A.; Sulem, P.; Jonsdottir, G.M.; Thorleifsson, G.; Helgadottir, H.; Steinthorsdottir, V.; Stefansson, H.; et al. Sequence Variants Affecting Eosinophil Numbers Associate with Asthma and Myocardial Infarction. Nat. Genet. 2009, 41, 342–347. [Google Scholar] [CrossRef] [PubMed]
- Dijk, F.N.; Vijverberg, S.J.; Hernandez-Pacheco, N.; Repnik, K.; Karimi, L.; Mitratza, M.; Farzan, N.; Nawijn, M.C.; Burchard, E.G.; Engelkes, M.; et al. IL1RL1 Gene Variations Are Associated with Asthma Exacerbations in Children and Adolescents Using Inhaled Corticosteroids. Allergy 2020, 75, 984. [Google Scholar] [CrossRef] [PubMed]
- Ketelaar, M.E.; Westerlaken-van Ginkel, C.D.; Nawijn, M.C.; Dubois, A.E.J.; Koppelman, G.H. IL-1RL1a Serum Levels and IL1RL1 SNPs in the Prediction of Food Allergy. Clin. Exp. Allergy 2021, 51, 614. [Google Scholar] [CrossRef] [PubMed]
- Akhabir, L.; Sandford, A. Genetics of Interleukin 1 Receptor-Like 1 in Immune and Inflammatory Diseases. Curr. Genom. 2010, 11, 591. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, B.; Harmacek, L.; Long, Z.; Liang, J.; Lukin, K.; Leach, S.M.; O’Connor, B.; Gerber, A.N.; Hagman, J.; et al. The Transcription Factors GATA2 and MITF Regulate Gene Expression in Mast Cells and Are Required forIgE/Mast Cell-Mediated Anaphylaxis. J. Allergy Clin. Immunol. 2018, 142, 1173. [Google Scholar] [CrossRef]
- Ohmori, S.; Ishijima, Y.; Numata, S.; Takahashi, M.; Sekita, M.; Sato, T.; Chugun, K.; Yamamoto, M.; Ohneda, K. GATA2 and PU.1 Collaborate To Activate the Expression of the Mouse Ms4a2 Gene, Encoding FcεRIβ, through Distinct Mechanisms. Mol. Cell. Biol. 2019, 39, e00314-19. [Google Scholar] [CrossRef]
- Rees, H.A.; Liu, D.R. Base editing: Precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 2018, 19, 770–788, Erratum in Nat. Rev. Genet. 2018, 19, 801. [Google Scholar] [CrossRef]
- Auton, A.; Abecasis, G.R.; Altshuler, D.M.; Durbin, R.M.; Bentley, D.R.; Chakravarti, A.; Clark, A.G.; Donnelly, P.; Eichler, E.E.; Flicek, P.; et al. A Global Reference for Human Genetic Variation. Nature 2015, 526, 68. [Google Scholar] [CrossRef]
- Tabatabaian, F.; Ledford, D.K. Omalizumab for Severe Asthma: Toward Personalized Treatment Based on Biomarker Profile and Clinical History. J. Asthma Allergy 2018, 11, 53. [Google Scholar] [CrossRef]
- Bourdin, A.; Molinari, N.; Vachier, I.; Varrin, M.; Marin, G.; Gamez, A.S.; Paganin, F.; Chanez, P. Prognostic Value of Cluster Analysis of Severe Asthma Phenotypes. J. Allergy Clin. Immunol. 2014, 134, 1043–1050. [Google Scholar] [CrossRef]
- Chung, K.F. Managing Severe Asthma in Adults: Lessons from the ERS/ATS Guidelines. Curr. Opin. Pulm. Med. 2015, 21, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Fajt, M.L.; Wenzel, S.E. Development of New Therapies for Severe Asthma. Allergy. Asthma Immunol. Res. 2017, 9, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Hanania, N.A.; Alpan, O.; Hamilos, D.L.; Condemi, J.J.; Reyes-Rivera, I.; Zhu, J.; Rosen, K.E.; Eisner, M.D.; Wong, D.A.; Busse, W. Omalizumab in Severe Allergic Asthma Inadequately Controlled with Standard Therapy: A Randomized Trial. Ann. Intern. Med. 2011, 154, 573–582. [Google Scholar] [CrossRef] [PubMed]
- Solèr, M.; Matz, J.; Townley, R.; Buhl, R.; O’Brien, J.; Fox, H.; Thirlwell, J.; Gupta, N.; Della Cioppa, G. The Anti-IgE Antibody Omalizumab Reduces Exacerbations and Steroid Requirement in Allergic Asthmatics. Eur. Respir. J. 2001, 18, 254–261. [Google Scholar] [CrossRef] [PubMed]
- Busse, W.; Corren, J.; Lanier, B.Q.; McAlary, M.; Fowler-Taylor, A.; Cioppa, G.D.; Gupta, N. Omalizumab, Anti-IgE Recombinant Humanized Monoclonal Antibody, for the Treatment of Severe Allergic Asthma. J. Allergy Clin. Immunol. 2001, 108, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Normansell, R.; Walker, S.; Milan, S.J.; Walters, E.H.; Nair, P. Omalizumab for Asthma in Adults and Children. Cochrane Database Syst. Rev. 2014, 2014, CD003559. [Google Scholar] [CrossRef] [PubMed]
- Holguin, F.; Cardet, J.C.; Chung, K.F.; Diver, S.; Ferreira, D.S.; Fitzpatrick, A.; Gaga, M.; Kellermeyer, L.; Khurana, S.; Knight, S.; et al. Management of Severe Asthma: A European Respiratory Society/American Thoracic Society Guideline. Eur. Respir. J. 2020, 55, 1900588. [Google Scholar] [CrossRef]
- Adachi, M.; Kozawa, M.; Yoshisue, H.; Lee Milligan, K.; Nagasaki, M.; Sasajima, T.; Miyamoto, T.; Ohta, K. Real-World Safety and Efficacy of Omalizumab in Patients with Severe Allergic Asthma: A Long-Term Post-Marketing Study in Japan. Respir. Med. 2018, 141, 56–63. [Google Scholar] [CrossRef]
- Paganin, F.; Mangiapan, G.; Proust, A.; Prudhomme, A.; Attia, J.; Marchand-Adam, S.; Pellet, F.; Milhe, F.; Melloni, B.; Bernady, A.; et al. Lung Function Parameters in Omalizumab Responder Patients: An Interesting Tool? Allergy 2017, 72, 1953–1961. [Google Scholar] [CrossRef]
- Verma, P.; Randhawa, I.; Klaustermeyer, W.B. Clinical Efficacy of Omalizumab in an Elderly Veteran Population with Severe Asthma. Allergy Asthma Proc. 2011, 32, 346–350. [Google Scholar] [CrossRef]
- Humbert, M.; Beasley, R.; Ayres, J.; Slavin, R.; Hébert, J.; Bousquet, J.; Beeh, K.M.; Ramos, S.; Canonica, G.W.; Hedgecock, S.; et al. Benefits of Omalizumab as Add-on Therapy in Patients with Severe Persistent Asthma Who Are Inadequately Controlled despite Best Available Therapy (GINA 2002 Step 4 Treatment): INNOVATE. Allergy 2005, 60, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Ortega, H.; Katz, L.; Gunsoy, N.; Keene, O.; Yancey, S. Blood Eosinophil Counts Predict Treatment Response in Patients with Severe Eosinophilic Asthma. J. Allergy Clin. Immunol. 2015, 136, 825–826. [Google Scholar] [CrossRef] [PubMed]
- Humbert, M.; Taillé, C.; Mala, L.; Le Gros, V.; Just, J.; Molimard, M.; Agossou, M.; Appere De Vecchi, C.; Barbare, E.; Barbry, M.; et al. Omalizumab Effectiveness in Patients with Severe Allergic Asthma According to Blood Eosinophil Count: The STELLAIR Study. Eur. Respir. J. 2018, 51, 1702523. [Google Scholar] [CrossRef] [PubMed]
- Harrison, T.W.; Chanez, P.; Menzella, F.; Canonica, G.W.; Louis, R.; Cosio, B.G.; Lugogo, N.L.; Mohan, A.; Burden, A.; McDermott, L.; et al. Onset of Effect and Impact on Health-Related Quality of Life, Exacerbation Rate, Lung Function, and Nasal Polyposis Symptoms for Patients with Severe Eosinophilic Asthma Treated with Benralizumab (ANDHI): A Randomised, Controlled, Phase 3b Trial. Lancet. Respir. Med. 2021, 9, 260–274. [Google Scholar] [CrossRef]
- Rojo-Tolosa, S.; González-Gutiérrez, M.V.; Sánchez-Martínez, J.A.; Jiménez-Gálvez, G.; Pineda-Lancheros, L.E.; Gálvez-Navas, J.M.; Jiménez-Morales, A.; Pérez-Ramírez, C.; Morales-García, C. Impact of Omalizumab in Patients with Severe Uncontrolled Asthma and Possible Predictive Biomarkers of Response: A Real-Life Study. Pharmaceutics 2023, 15, 523. [Google Scholar] [CrossRef]
- Sheehan, W.J.; Krouse, R.Z.; Calatroni, A.; Gergen, P.J.; Gern, J.E.; Gill, M.A.; Gruchalla, R.S.; Khurana Hershey, G.K.; Kattan, M.; Kercsmar, C.M.; et al. Aeroallergen Sensitization, Serum IgE, and Eosinophilia as Predictors of Response to Omalizumab Therapy During the Fall Season Among Children with Persistent Asthma. J. Allergy Clin. Immunol. Pract. 2020, 8, 3021–3028.e2. [Google Scholar] [CrossRef]
- Bousquet, J.; Humbert, M.; Gibson, P.G.; Kostikas, K.; Jaumont, X.; Pfister, P.; Nissen, F. Real-World Effectiveness of Omalizumab in Severe Allergic Asthma: A Meta-Analysis of Observational Studies. J. Allergy Clin. Immunol. Pract. 2021, 9, 2702–2714. [Google Scholar] [CrossRef]
- Kallieri, M.; Papaioannou, A.I.; Papathanasiou, E.; Ntontsi, P.; Papiris, S.; Loukides, S. Predictors of Response to Therapy with Omalizumab in Patients with Severe Allergic Asthma—A Real Life Study. Postgrad. Med. 2017, 129, 598–604. [Google Scholar] [CrossRef]
- Caminati, M.; Vianello, A.; Chieco Bianchi, F.; Festi, G.; Guarnieri, G.; Marchi, M.R.; Micheletto, C.; Olivieri, M.; Tognella, S.; Guerriero, M.; et al. Relevance of TH2 Markers in the Assessment and Therapeutic Management of Severe Allergic Asthma: A Real-Life Perspective. J. Investig. Allergol. Clin. Immunol. 2020, 30, 35–41. [Google Scholar] [CrossRef]
- Ullemar, V.; Magnusson, P.K.E.; Lundholm, C.; Zettergren, A.; Melén, E.; Lichtenstein, P.; Almqvist, C. Heritability and Confirmation of Genetic Association Studies for Childhood Asthma in Twins. Allergy 2016, 71, 230–238. [Google Scholar] [CrossRef]
- Ober, C.; Yao, T.-C. The Genetics of Asthma and Allergic Disease: A 21st Century Perspective. Immunol. Rev. 2011, 242, 10–30. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, S.F.; Ulrik, C.S.; Kyvik, K.O.; Ferreira, M.A.R.; Backer, V. Multivariate Genetic Analysis of Atopy Phenotypes in a Selected Sample of Twins. Clin. Exp. Allergy 2006, 36, 1382–1390. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Gaitsch, H.; Poon, H.; Cox, N.J.; Rzhetsky, A. Classification of Common Human Diseases Derived from Shared Genetic and Environmental Determinants. Nat. Genet. 2017, 49, 1319–1325. [Google Scholar] [CrossRef] [PubMed]
- Witte, J.S.; Visscher, P.M.; Wray, N.R. The Contribution of Genetic Variants to Disease Depends on the Ruler. Nat. Rev. Genet. 2014, 15, 765–776. [Google Scholar] [CrossRef]
- Rojo-Tolosa, S.; Pineda-Lancheros, L.E.; Gálvez-Navas, J.M.; Sánchez-Martínez, J.A.; González-Gutiérrez, M.V.; Fernández-Alonso, A.; Morales-García, C.; Jiménez-Morales, A.; Pérez-Ramírez, C. Association between Single Nucleotide Polymorphisms Related to Vitamin D Metabolism and the Risk of Developing Asthma. Nutrients 2023, 15, 823. [Google Scholar] [CrossRef]
- Spear, M.L.; Hu, D.; Pino-Yanes, M.; Huntsman, S.; Eng, C.; Levin, A.M.; Ortega, V.E.; White, M.J.; McGarry, M.E.; Thakur, N.; et al. A Genome-Wide Association and Admixture Mapping Study of Bronchodilator Drug Response in African Americans with Asthma. Pharm. J. 2018, 19, 249–259. [Google Scholar] [CrossRef]
- Vijverberg, S.J. Realizing Personalized Medicine in Asthmatic Children Requires Large-Scale Collaboration. Pediatr. Ther. 2015, 5, 2. [Google Scholar] [CrossRef]
- Farzan, N.; Vijverberg, S.J.H.; Arets, H.G.; Raaijmakers, J.A.M.; Maitland-van der Zee, A.H. Pharmacogenomics of Inhaled Corticosteroids and Leukotriene Modifiers: A Systematic Review. Clin. Exp. Allergy 2017, 47, 271–293. [Google Scholar] [CrossRef]
- Vijverberg, S.J.; Raaijmakers, J.A.; Maitland-Van Der Zee, A.H. ADRB2 Arg16 and the Need for Collaboration in Childhood Asthma Pharmacogenomics. Pharmacogenomics 2013, 14, 1937–1939. [Google Scholar] [CrossRef]
- Casale, T.B.; Luskin, A.T.; Busse, W.; Zeiger, R.S.; Trzaskoma, B.; Yang, M.; Griffin, N.M.; Chipps, B.E. Omalizumab Effectiveness by Biomarker Status in Patients with Asthma: Evidence From PROSPERO, A Prospective Real-World Study. J. Allergy Clin. Immunol. Pract. 2019, 7, 156–164.e1. [Google Scholar] [CrossRef]
- Ledford, D.; Busse, W.; Trzaskoma, B.; Omachi, T.A.; Rosén, K.; Chipps, B.E.; Luskin, A.T.; Solari, P.G. A Randomized Multicenter Study Evaluating Xolair Persistence of Response after Long-Term Therapy. J. Allergy Clin. Immunol. 2017, 140, 162–169.e2. [Google Scholar] [CrossRef] [PubMed]
- Busse, W.; Spector, S.; Rosén, K.; Wang, Y.; Alpan, O. High Eosinophil Count: A Potential Biomarker for Assessing Successful Omalizumab Treatment Effects. J. Allergy Clin. Immunol. 2013, 132, 485–486.e11. [Google Scholar] [CrossRef]
- Casale, T.B.; Chipps, B.E.; Rosén, K.; Trzaskoma, B.; Haselkorn, T.; Omachi, T.A.; Greenberg, S.; Hanania, N.A. Response to Omalizumab Using Patient Enrichment Criteria from Trials of Novel Biologics in Asthma. Allergy 2018, 73, 490–497. [Google Scholar] [CrossRef]
- Marzano, A.V.; Genovese, G.; Casazza, G.; Fierro, M.T.; Dapavo, P.; Crimi, N.; Ferrucci, S.; Pepe, P.; Liberati, S.; Pigatto, P.D.; et al. Predictors of Response to Omalizumab and Relapse in Chronic Spontaneous Urticaria: A Study of 470 Patients. J. Eur. Acad. Dermatol. Venereol. 2019, 33, 918–924. [Google Scholar] [CrossRef]
- Liao, E.C.; Chang, C.Y.; Hsieh, C.W.; Yu, S.J.; Yin, S.C.; Tsai, J.J. An Exploratory Pilot Study of Genetic Marker for IgE-Mediated Allergic Diseases with Expressions of FcεR1α and Cε. Int. J. Mol. Sci. 2015, 16, 9504. [Google Scholar] [CrossRef]
- Amo, G.; García-Menaya, J.; Campo, P.; Cordobés, C.; Serón, M.C.P.; Ayuso, P.; Esguevillas, G.; Blanca, M.; Agúndez, J.A.G.; García-Martín, E. A Nonsynonymous FCER1B SNP Is Associated with Risk of Developing Allergic Rhinitis and with IgE Levels. Sci. Rep. 2016, 6, 19724. [Google Scholar] [CrossRef] [PubMed]
- Dar, S.A.; Rai, G.; Ansari, M.A.; Akhter, N.; Gupta, N.; Sharma, S.; Haque, S.; Ramachandran, V.G.; Wahid, M.; Rudramurthy, S.M.; et al. FcɛR1α Gene Polymorphism Shows Association with High IgE and Anti-FcɛR1α in Chronic Rhinosinusitis with Nasal Polyposis. J. Cell. Biochem. 2018, 119, 4142–4149. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Ghosh, B. Promoter Polymorphism in the MS4A2 Gene and Asthma in the Indian Population. Int. Arch. Allergy Immunol. 2009, 149, 208–218. [Google Scholar] [CrossRef] [PubMed]
- Green, S.L.; Gaillard, M.C.; Song, E.; Dewar, J.B.; Halkas, A. Polymorphisms of the Beta Chain of the High-Affinity Immunoglobulin E Receptor (Fcepsilon RI-Beta) in South African Black and White Asthmatic and Nonasthmatic Individuals. Am. J. Respir. Crit. Care Med. 1998, 158, 1487–1492. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Hopkin, J.M. Atopy Phenotype in Subjects with Variants of the Beta Subunit of the High Affinity IgE Receptor. Thorax 1997, 52, 654–655. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.K.; Park, H.W.; Yang, J.S.; Oh, S.Y.; Chang, Y.S.; Shin, E.S.; Lee, J.E.; Kim, S.; Gho, Y.S.; Cho, S.H.; et al. Association and Functional Relevance of E237G, a Polymorphism of the High-Affinity Immunoglobulin E-Receptor Beta Chain Gene, to Airway Hyper-Responsiveness. Clin. Exp. Allergy 2007, 37, 592–598. [Google Scholar] [CrossRef] [PubMed]
- Shirakawa, T.; Mao, X.; Sasaki, S.; Enomoto, T.; Kawai, M.; Morimoto, K.; Hopkin, J. Association between Atopic Asthma and a Coding Variant of FcεRIβ in a Japanese Population. Hum. Mol. Genet. 1996, 5, 1129–1130. [Google Scholar] [CrossRef] [PubMed]
- Pavón-Romero, G.F.; Pérez-Rubio, G.; Ramírez-Jiménez, F.; Ambrocio-Ortiz, E.; Bañuelos-Ortiz, E.; Alvarado-Franco, N.; Xochipa-Ruiz, K.E.; Hernández-Juárez, E.; Flores-García, B.A.; Camarena, Á.E.; et al. MS4A2-Rs573790 Is Associated with Aspirin-Exacerbated Respiratory Disease: Replicative Study Using a Candidate Gene Strategy. Front. Genet. 2018, 9, 363. [Google Scholar] [CrossRef]
- Drouin, S.M.; Corry, D.B.; Kildsgaard, J.; Wetsel, R.A. Cutting Edge: The Absence of C3 Demonstrates a Role for Complement in Th2 Effector Functions in a Murine Model of Pulmonary Allergy. J. Immunol. 2001, 167, 4141–4145. [Google Scholar] [CrossRef]
- Van Der Pol, W.L.; Van De Winkel, J.G.J. IgG Receptor Polymorphisms: Risk Factors for Disease. Immunogenetics 1998, 48, 222–232. [Google Scholar] [CrossRef]
- Chai, L.; Song, Y.Q.; Leung, W.K. Genetic Polymorphism Studies in Periodontitis and Fcγ Receptors. J. Periodontal Res. 2012, 47, 273–285. [Google Scholar] [CrossRef]
- Cañete, J.D.; Suárez, B.; Hernández, M.V.; Sanmartí, R.; Rego, I.; Celis, R.; Moll, C.; Pinto, J.A.; Blanco, F.J.; Lozano, F. Influence of Variants of Fcγ Receptors IIA and IIIA on the American College of Rheumatology and European League Against Rheumatism Responses to Anti-Tumour Necrosis Factor α Therapy in Rheumatoid Arthritis. Ann. Rheum. Dis. 2009, 68, 1547–1552. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Lin, R.; Huang, J.; Guan, W.; Oetting, W.S.; Sriramarao, P.; Blumenthal, M.N. Functional Fcgamma receptor polymorphisms are associated with human allergy. PLoS ONE 2014, 9, e89196. [Google Scholar] [CrossRef]
- Dharajiya, N.; Vaidya, S.V.; Murai, H.; Cardenas, V.; Kurosky, A.; Boldogh, I.; Sur, S.A. FcγRIIb Inhibits Allergic Lung Inflammation in a Murine Model of Allergic Asthma. PLoS ONE 2010, 5, 9337. [Google Scholar] [CrossRef]
- Jiang, Y.; Hirose, S.; Abe, M.; Sanokawa-Akakura, R.; Ohtsuji, M.; Mi, X.; Li, N.; Xiu, Y.; Zhang, D.; Shirai, J.; et al. Polymorphisms in IgG Fc Receptor IIB Regulatory Regions Associated with Autoimmune Susceptibility. Immunogenetics 2000, 51, 429–435. [Google Scholar] [CrossRef]
- Gordon, E.D.; Palandra, J.; Wesolowska-Andersen, A.; Ringel, L.; Rios, C.L.; Lachowicz-Scroggins, M.E.; Sharp, L.Z.; Everman, J.L.; MacLeod, H.J.; Lee, J.W.; et al. IL1RL1 Asthma Risk Variants Regulate Airway Type 2 Inflammation. JCI Insight 2016, 1, 87871. [Google Scholar] [CrossRef] [PubMed]
- Hirasawa, R.; Shimizu, R.; Takahashi, S.; Osawa, M.; Takayanagi, S.; Kato, Y.; Onodera, M.; Minegishi, N.; Yamamoto, M.; Fukao, K.; et al. Essential and Instructive Roles of GATA Factors in Eosinophil Development. J. Exp. Med. 2002, 195, 1379–1386. [Google Scholar] [CrossRef] [PubMed]
- Agencia Española del Medicamento y Productos Sanitarios Ficha Técnica XOLAIR 150 Mg Solucion Inyectable. Available online: https://cima.aemps.es/cima/dochtml/p/05319008/P_05319008.html (accessed on 12 February 2023).
- Asthma Control Test (ACT). Available online: https://www.thoracic.org/members/assemblies/assemblies/srn/questionaires/act.php (accessed on 4 May 2022).
- Alvarez-Gutiérrez, F.J.; Blanco-Aparicio, M.; Casas-Maldonado, F.; Plaza, V.; González-Barcala, F.J.; Carretero-Gracia, J.Á.; Castilla-Martínez, M.; Cisneros, C.; Diaz-Pérez, D.; Domingo-Ribas, C.; et al. Documento de consenso de asma grave en adultos. Actualización 2022. Open Respir. Arch. 2022, 4, 100192. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013. [Google Scholar]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and Visualization of LD and Haplotype Maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef]
- Solé, X.; Guinó, E.; Valls, J.; Iniesta, R.; Moreno, V. SNPStats: A Web Tool for the Analysis of Association Studies. Bioinformatics 2006, 22, 1928–1929. [Google Scholar] [CrossRef]
| N | % | Mean ± SD/ p50 (p25, p75) | |
|---|---|---|---|
| Sex | |||
| Women | 48 | 65.33 | |
| Men | 26 | 34.67 | |
| Age of starting BT (years) | 74 | 46.89 ± 16.74 | |
| Years with asthma | 74 | 8 (5, 14) | |
| BMI (kg/m2) | |||
| <25 | 17 | 23.29 | |
| >25 | 56 | 76.71 | |
| Previous respiratory disease | |||
| Yes | 19 | 25.33 | |
| No | 55 | 79.67 | |
| Smoking status | |||
| Non-smoker | 56 | 74.67 | |
| Active smoker | 3 | 4 | |
| Ex-smoker | 16 | 21.33 | |
| Polyposis | |||
| Yes | 18 | 24 | |
| No | 56 | 76 | |
| Allergies | |||
| Yes | 58 | 77.3 | |
| No | 16 | 22.67 | |
| GERD | |||
| Yes | 14 | 18.67 | |
| No | 61 | 81.33 | |
| SAHS | |||
| Yes | 24 | 32 | |
| No | 51 | 68 | |
| COPD | |||
| Yes | 20 | 22.67 | |
| No | 55 | 77.33 | |
| Age of diagnosis (years) | 74 | 43 (34, 54) | |
| <18 | 10 | 13.33 | |
| >18 | 65 | 88.67 | |
| ICS (µg/day) | 74 | 500 (250, 1000) | |
| OCS courses in previous year | |||
| Yes | 56 | 74.67 | |
| No | 19 | 25.33 | |
| Baseline %FEV1 | |||
| <80 | 42 | 59.15 | |
| >80 | 29 | 40.85 | |
| Exacerbations in previous year | |||
| Yes | 48 | 64 | |
| No | 27 | 36 | |
| Baseline blood eosinophils (cells/µL) | |||
| <300 | 37 | 54.41 | |
| >300 | 31 | 45.59 | |
| Baseline IgE (IU/mL) | 65 | 359 (151, 980) | |
| Years with omalizumab | |||
| <5 | 52 | 69.33 | |
| >5 | 23 | 30.67 | |
| Change of BT | |||
| Yes | 37 | 49.33 | |
| No | 38 | 50.67 |
| Response Variable | N | % |
|---|---|---|
| Responsive for 1 criterion | ||
| Yes | 71 | 95.95 |
| No | 3 | 4.05 |
| Responsive for 2 criteria | ||
| Yes | 63 | 85.15 |
| No | 11 | 14.86 |
| Responsive for 3 criteria | ||
| Yes | 31 | 45.59 |
| No | 37 | 54.41 |
| Reduction in OCS ≥ 50% | ||
| Yes | 49 | 66.22 |
| No | 25 | 33.78 |
| Reduction in exacerbations ≥ 50% | ||
| Yes | 64 | 86.49 |
| No | 10 | 13.51 |
| Increase in %FEV1 ≥ 10% or %FEV1 ≥ 80% | ||
| Yes | 52 | 76.47 |
| No | 16 | 23.53 |
| OR (95% CI) | p-Value | |
|---|---|---|
| Response in respect of exacerbation reduction | ||
| Polyposis (No) | 4.22 (0.95–19.63) | 0.050 |
| IL1RL1 rs17026974 (AG vs. AA) | 19.07 (1.27–547) | 0.040 |
| IL1RL1 rs17026974 (GG vs. AA) | 16.76 (1.22–438.76) | 0.041 |
| Response in respect of reduction of OCS | ||
| Eosinophils (>300 cll/µL) | 2.93 (1.01–9.29) | 0.055 |
| Age of starting omalizumab | 0.95 (0.91–0.99) | 0.032 |
| Response in respect of improved lung function | ||
| COPD (No) | 12.16 (2.45–79.49) | 0.004 |
| GATA2 rs4857855 (T vs. CC) | 15.98 (1.52–519.57) | 0.052 |
| FCGR2A rs1801274 (AG vs. AA) | 13.75 (2.14–142.68) | 0.012 |
| FCGR2A rs1801274 (GG vs. AA) | 7.46 (0.94–89.12) | 0.076 |
| FCGR2B rs3219018 (C vs. GG) | 8.6 (1.12–117.15) | 0.052 |
| Meeting at least 1 criterion | ||
| FCER1A rs2251746 (TT vs. CC) | 24 (0.77–804.57) | 0.045 |
| Meeting at least 2 criteria | ||
| Age of diagnosis | 0.93 (0.88–0.99) | 0.018 |
| Meeting all 3 criteria | ||
| BMI (<25) | 14.23 (3.31–100.77) | 0.001 |
| C3 rs2230199 (C vs. GG) | 3 (1.01–9.92) | 0.063 |
| Gene | SNP | dbSNP ID | Assay ID |
|---|---|---|---|
| FCER1A (1q23) | T > C | rs2251746 | C___1840470_20 |
| G > A | rs2427837 | C__16233438_20 | |
| FCER1B (11q12-13) | T > C | rs1441586 | C___1842226_10 |
| T > C | rs573790 | C____900105_20 | |
| T > G | rs1054485 | C___2932371_10 | |
| A > G | rs569108 | C____900116_10 | |
| C3 (19p13.3) | G > C | rs2230199 | C__26330755_10 |
| FCGR2A (1q23.3) | A > G | rs1801274 | C___9077561_20 |
| FCGR2B (1q23.3) | G > C | rs3219018 | ANPRZAZ |
| T > C | rs1050501 | ANRWUVX | |
| FCGR3A (1q23.3) | A > C | rs10127939 | C__57480226_10 |
| A > C | rs396991 | C__25815666_10 | |
| IL1RL1 (2q12) | C > T | rs1420101 | C___8906009_20 |
| G > A | rs17026974 | C__33551182_10 | |
| G > A | rs1921622 | C___1226146_10 | |
| GATA2 (3q21) | C > T | rs4857855 | C__11231076_10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rojo-Tolosa, S.; Sánchez-Martínez, J.A.; Pineda-Lancheros, L.E.; Gálvez-Navas, J.M.; González-Gutiérrez, M.V.; Jiménez-Gálvez, G.; Pérez-Ramírez, C.; Morales-García, C.; Jiménez-Morales, A. Influence of Genetics on the Response to Omalizumab in Patients with Severe Uncontrolled Asthma with an Allergic Phenotype. Int. J. Mol. Sci. 2023, 24, 7029. https://doi.org/10.3390/ijms24087029
Rojo-Tolosa S, Sánchez-Martínez JA, Pineda-Lancheros LE, Gálvez-Navas JM, González-Gutiérrez MV, Jiménez-Gálvez G, Pérez-Ramírez C, Morales-García C, Jiménez-Morales A. Influence of Genetics on the Response to Omalizumab in Patients with Severe Uncontrolled Asthma with an Allergic Phenotype. International Journal of Molecular Sciences. 2023; 24(8):7029. https://doi.org/10.3390/ijms24087029
Chicago/Turabian StyleRojo-Tolosa, Susana, José Antonio Sánchez-Martínez, Laura Elena Pineda-Lancheros, José María Gálvez-Navas, María Victoria González-Gutiérrez, Gonzalo Jiménez-Gálvez, Cristina Pérez-Ramírez, Concepción Morales-García, and Alberto Jiménez-Morales. 2023. "Influence of Genetics on the Response to Omalizumab in Patients with Severe Uncontrolled Asthma with an Allergic Phenotype" International Journal of Molecular Sciences 24, no. 8: 7029. https://doi.org/10.3390/ijms24087029
APA StyleRojo-Tolosa, S., Sánchez-Martínez, J. A., Pineda-Lancheros, L. E., Gálvez-Navas, J. M., González-Gutiérrez, M. V., Jiménez-Gálvez, G., Pérez-Ramírez, C., Morales-García, C., & Jiménez-Morales, A. (2023). Influence of Genetics on the Response to Omalizumab in Patients with Severe Uncontrolled Asthma with an Allergic Phenotype. International Journal of Molecular Sciences, 24(8), 7029. https://doi.org/10.3390/ijms24087029








