Nyamanini Virus Nucleoprotein and Phosphoprotein Organize Viral Inclusion Bodies That Associate with Host Biomolecular Condensates in the Nucleus
Abstract
1. Introduction
2. Results
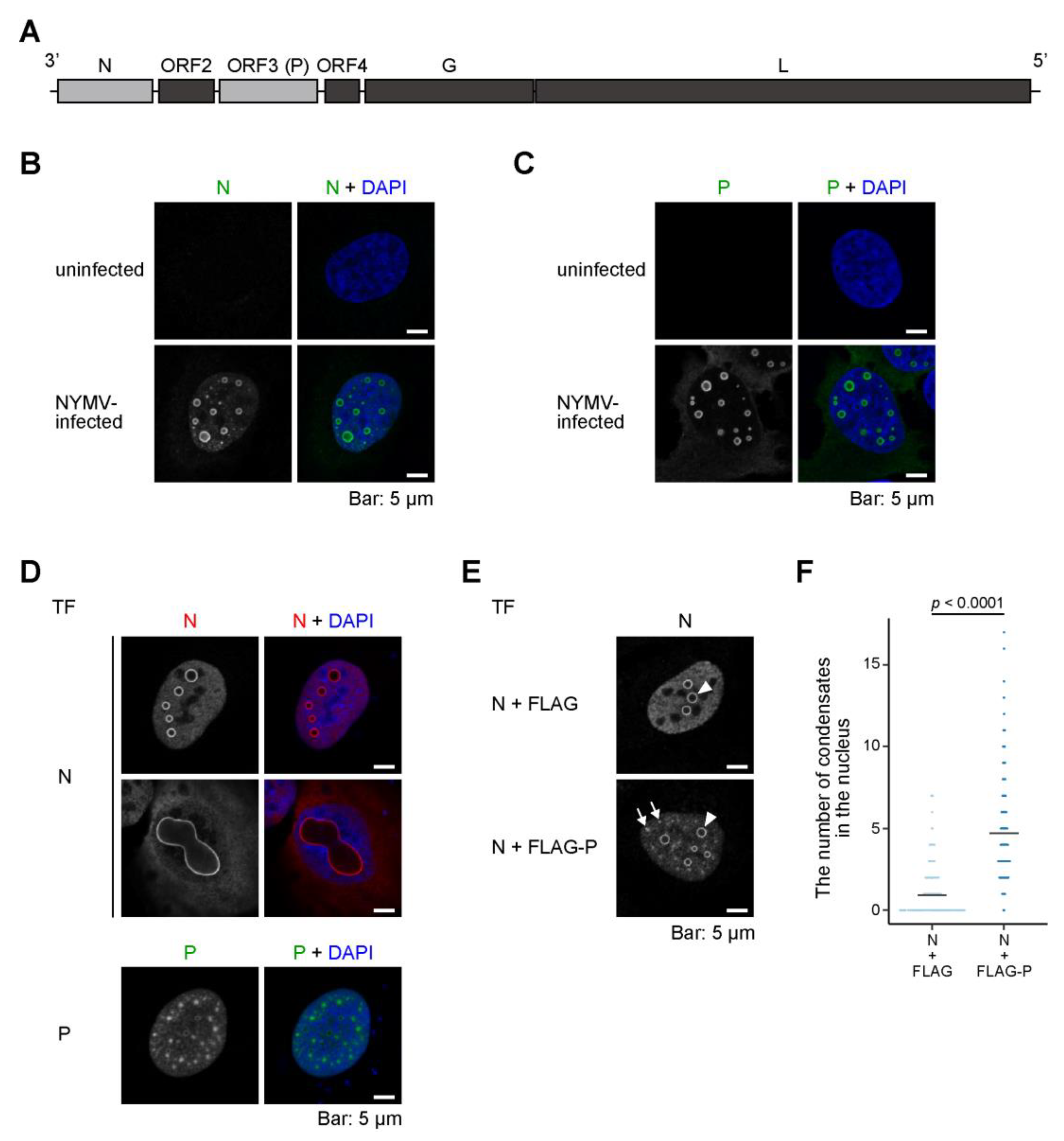
2.1. Both N and P Are Necessary for the Formation of an NYMV-Specific IB-Like Structure
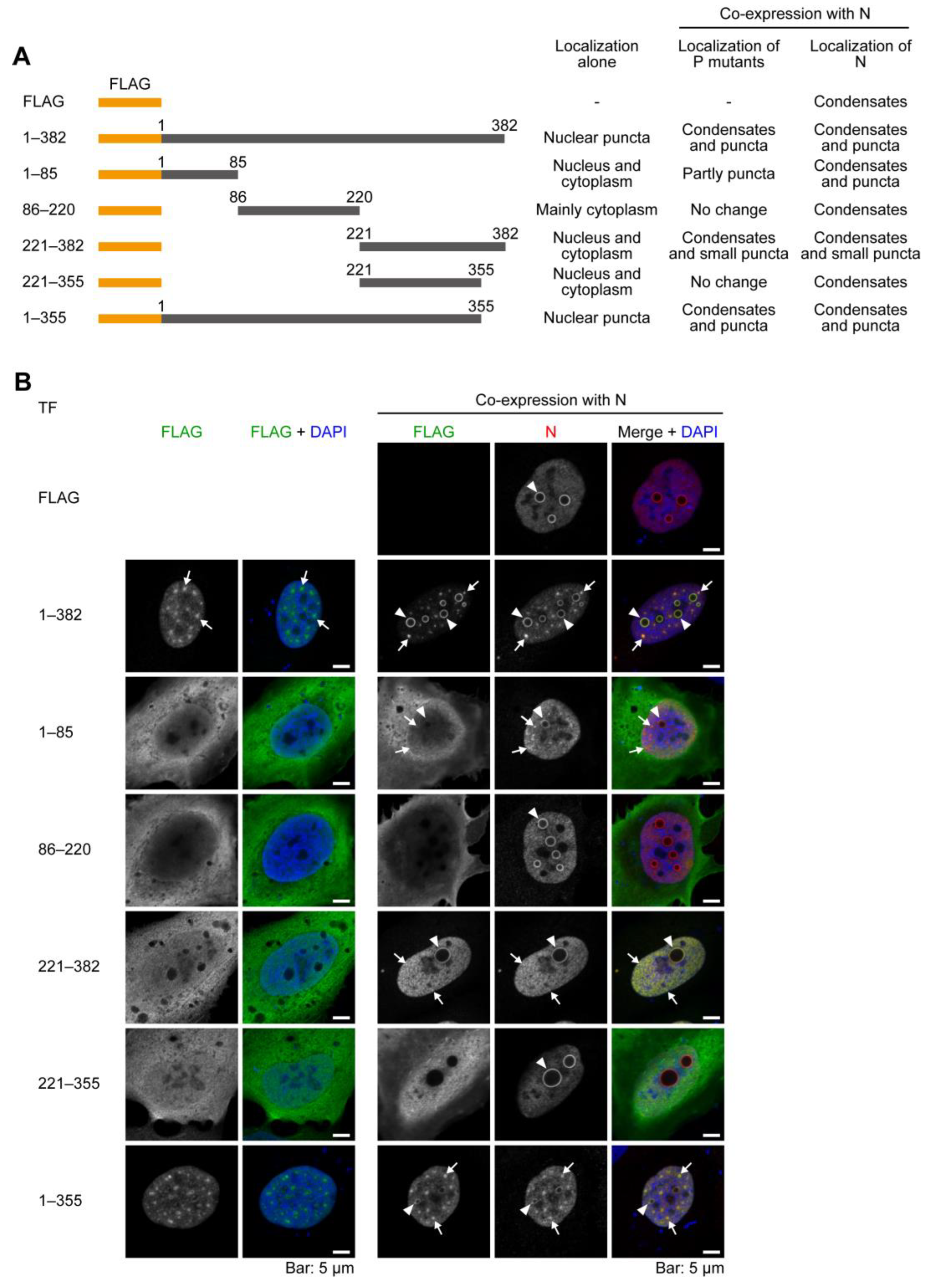
2.2. The C-Terminal 27 Amino Acid Residues of P Have the Ability to Be Recruited to the Condensates Formed by N
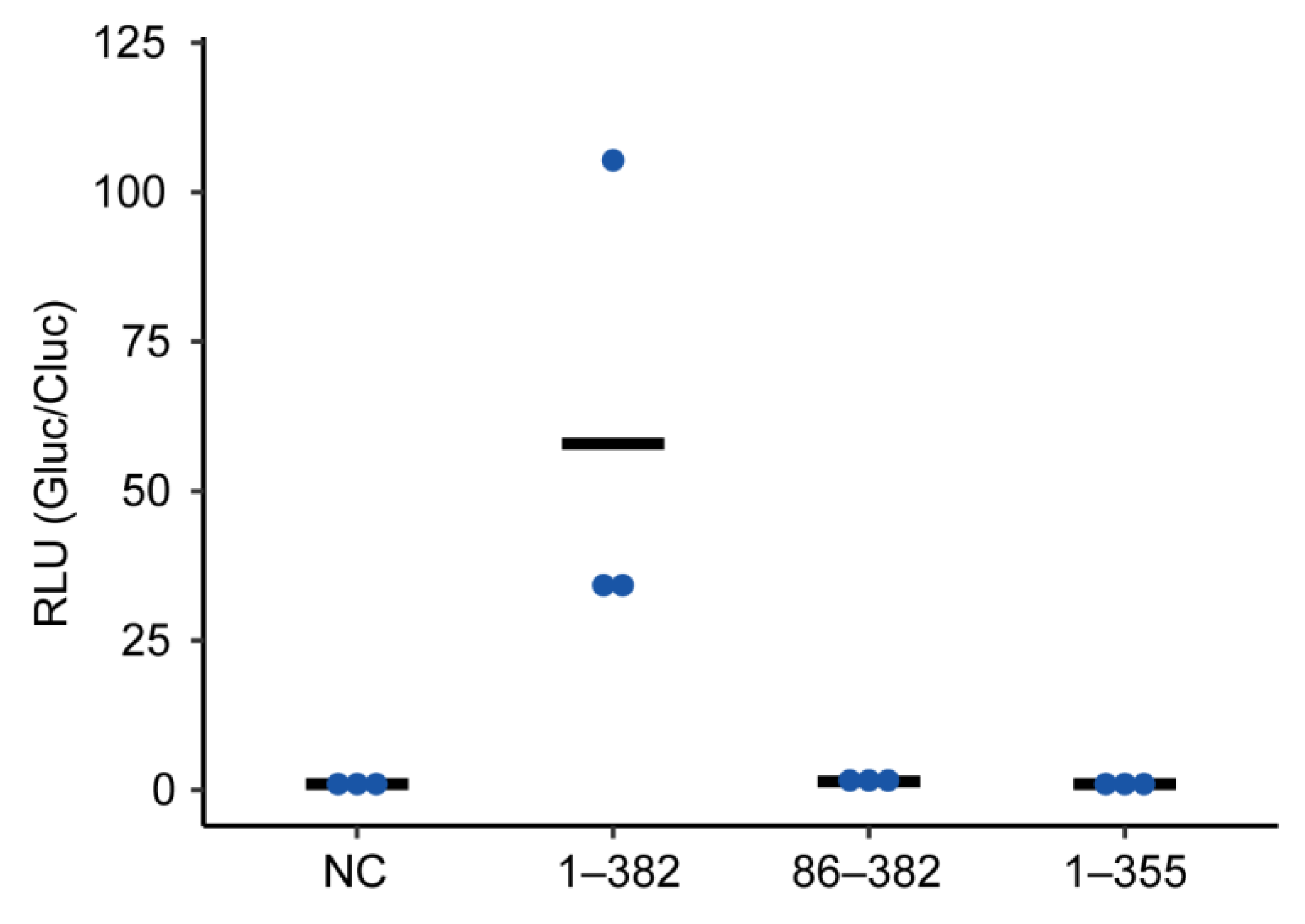
2.3. Truncated P Mutants Do Not Act as a Polymerase Co-Factor
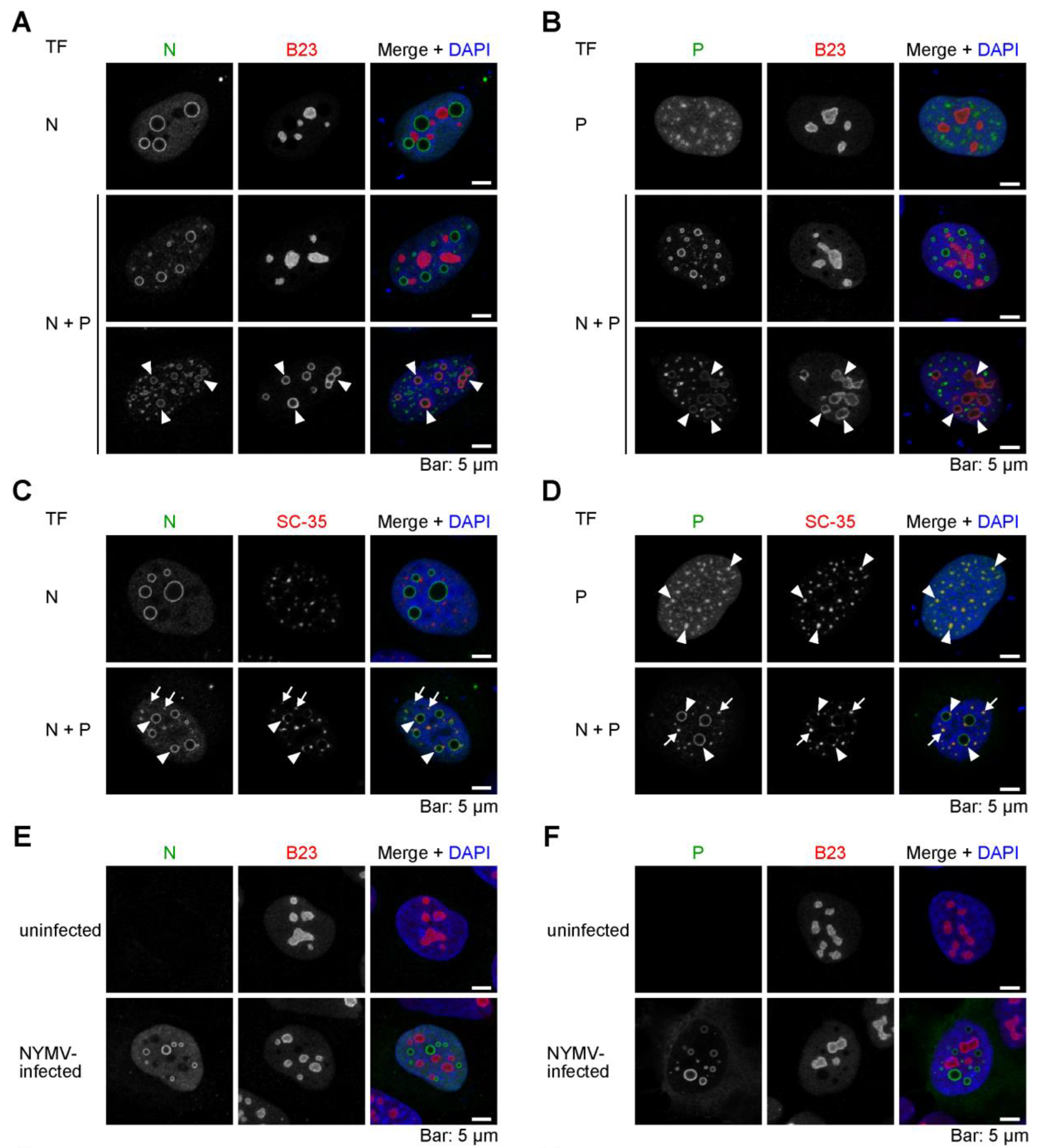
2.4. The Viral IB-like Structures of NYMV Colocalize with Cellular Biomolecular Condensates in the Nucleus
3. Discussion
4. Materials and Methods
4.1. Cells
4.2. Virus Preparation and Infection
4.3. DNA Constructs
4.4. Antibodies
4.5. Immunofluorescence Microscopy
4.6. Minireplicon Assay
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cifuentes-Muñoz, N.; Branttie, J.; Slaughter, K.B.; Dutch, R.E. Human Metapneumovirus Induces Formation of Inclusion Bodies for Efficient Genome Replication and Transcription. J. Virol. 2017, 91, e01282-17. [Google Scholar] [CrossRef]
- Lahaye, X.; Vidy, A.; Pomier, C.; Obiang, L.; Harper, F.; Gaudin, Y.; Blondel, D. Functional Characterization of Negri Bodies (NBs) in Rabies Virus-Infected Cells: Evidence That NBs Are Sites of Viral Transcription and Replication. J. Virol. 2009, 83, 7948–7958. [Google Scholar] [CrossRef]
- Heinrich, B.S.; Cureton, D.K.; Rahmeh, A.A.; Whelan, S.P.J. Protein Expression Redirects Vesicular Stomatitis Virus RNA Synthesis to Cytoplasmic Inclusions. PLoS Pathog. 2010, 6, e1000958. [Google Scholar] [CrossRef] [PubMed]
- Hoenen, T.; Shabman, R.S.; Groseth, A.; Herwig, A.; Weber, M.; Schudt, G.; Dolnik, O.; Basler, C.F.; Becker, S.; Feldmann, H. Inclusion Bodies Are a Site of Ebolavirus Replication. J. Virol. 2012, 86, 11779–11788. [Google Scholar] [CrossRef]
- Ringel, M.; Heiner, A.; Behner, L.; Halwe, S.; Sauerhering, L.; Becker, N.; Dietzel, E.; Sawatsky, B.; Kolesnikova, L.; Maisner, A. Nipah Virus Induces Two Inclusion Body Populations: Identification of Novel Inclusions at the Plasma Membrane. PLoS Pathog. 2019, 15, e1007733. [Google Scholar] [CrossRef]
- Lifland, A.W.; Jung, J.; Alonas, E.; Zurla, C.; Crowe, J.E.; Santangelo, P.J. Human Respiratory Syncytial Virus Nucleoprotein and Inclusion Bodies Antagonize the Innate Immune Response Mediated by MDA5 and MAVS. J. Virol. 2012, 86, 8245–8258. [Google Scholar] [CrossRef]
- Fricke, J.; Koo, L.Y.; Brown, C.R.; Collins, P.L. P38 and OGT Sequestration into Viral Inclusion Bodies in Cells Infected with Human Respiratory Syncytial Virus Suppresses MK2 Activities and Stress Granule Assembly. J. Virol. 2013, 87, 1333–1347. [Google Scholar] [CrossRef]
- Kaiser, M.N. Viruses in Ticks. I. Natural Infections of Argas (Persicargas) arboreus by Quaranfil and Nyamanini Viruses and Absence of Infections in A. (P.) persicus in Egypt. Am. J. Trop. Med. Hyg. 1966, 15, 964–975. [Google Scholar] [CrossRef]
- Kemp, G.E.; Lee, V.H.; Moore, D.L. Isolation of Nyamanini and Quaranfil Viruses from Argas (Persicargas) arboreus Ticks in Nigeria. J. Med. Entomol. 1975, 12, 535–537. [Google Scholar] [CrossRef]
- Taylor, R.M.; Hurlbut, H.S.; Work, T.H.; Kingston, J.R.; Hoogstraal, H. Arboviruses Isolated from ARGAS TICKS IN Egypt: Quaranfil, Chenuda, and Nyamanini. Am. J. Trop. Med. Hyg. 1966, 15, 76–86. [Google Scholar] [CrossRef] [PubMed]
- Mihindukulasuriya, K.A.; Nguyen, N.L.; Wu, G.; Huang, H.V.; da Rosa, A.P.A.T.; Popov, V.L.; Tesh, R.B.; Wang, D. Nyamanini and Midway Viruses Define a Novel Taxon of RNA Viruses in the Order Mononegavirales. J. Virol. 2009, 83, 5109–5116. [Google Scholar] [CrossRef]
- Dietzgen, R.G.; Firth, A.E.; Jiāng, D.; Junglen, S.; Kondo, H.; Kuhn, J.H.; Paraskevopoulou, S.; Vasilakis, N. ICTV Report Consortium ICTV Virus Taxonomy Profile: Nyamiviridae 2021. J. Gen. Virol. 2021, 102, 001681. [Google Scholar] [CrossRef]
- Herrel, M.; Hoefs, N.; Staeheli, P.; Schneider, U. Tick-Borne Nyamanini Virus Replicates in the Nucleus and Exhibits Unusual Genome and Matrix Protein Properties. J. Virol. 2012, 86, 10739–10747. [Google Scholar] [CrossRef]
- Herrel, M.; Haag, L.; Nilsson, J.; Staeheli, P.; Schneider, U. Reverse Genetics Identifies the Product of Open Reading Frame 4 as an Essential Particle Assembly Factor of Nyamanini Virus. J. Virol. 2013, 87, 8257–8260. [Google Scholar] [CrossRef]
- Cubitt, B.; de la Torre, J.C. Borna Disease Virus (BDV), a Nonsegmented RNA Virus, Replicates in the Nuclei of Infected Cells Where Infectious BDV Ribonucleoproteins Are Present. J. Virol. 1994, 68, 1371–1381. [Google Scholar] [CrossRef]
- Kuwata, R.; Isawa, H.; Hoshino, K.; Tsuda, Y.; Yanase, T.; Sasaki, T.; Kobayashi, M.; Sawabe, K. RNA Splicing in a New Rhabdovirus from Culex Mosquitoes. J. Virol. 2011, 85, 6185–6196. [Google Scholar] [CrossRef] [PubMed]
- Son, K.-N.; Liang, Z.; Lipton, H.L. Double-Stranded RNA Is Detected by Immunofluorescence Analysis in RNA and DNA Virus Infections, Including Those by Negative-Stranded RNA Viruses. J. Virol. 2015, 89, 9383–9392. [Google Scholar] [CrossRef] [PubMed]
- Galloux, M.; Tarus, B.; Blazevic, I.; Fix, J.; Duquerroy, S.; Eléouët, J.-F. Characterization of a Viral Phosphoprotein Binding Site on the Surface of the Respiratory Syncytial Nucleoprotein. J. Virol. 2012, 86, 8375–8387. [Google Scholar] [CrossRef]
- Zhang, S.; Chen, L.; Zhang, G.; Yan, Q.; Yang, X.; Ding, B.; Tang, Q.; Sun, S.; Hu, Z.; Chen, M. An Amino Acid of Human Parainfluenza Virus Type 3 Nucleoprotein Is Critical for Template Function and Cytoplasmic Inclusion Body Formation. J. Virol. 2013, 87, 12457–12470. [Google Scholar] [CrossRef]
- Hirai, Y.; Tomonaga, K.; Horie, M. Borna Disease Virus Phosphoprotein Triggers the Organization of Viral Inclusion Bodies by Liquid-Liquid Phase Separation. Int. J. Biol. Macromol. 2021, 192, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Schneider, J.; Dauber, B.; Melén, K.; Julkunen, I.; Wolff, T. Analysis of Influenza B Virus NS1 Protein Trafficking Reveals a Novel Interaction with Nuclear Speckle Domains. J. Virol. 2009, 83, 701–711. [Google Scholar] [CrossRef] [PubMed]
- Miyake, T.; Farley, C.M.; Neubauer, B.E.; Beddow, T.P.; Hoenen, T.; Engel, D.A. Ebola Virus Inclusion Body Formation and RNA Synthesis Are Controlled by a Novel Domain of Nucleoprotein Interacting with VP35. J. Virol. 2020, 94, e02100-19. [Google Scholar] [CrossRef] [PubMed]
- Becker, S.; Rinne, C.; Hofsäss, U.; Klenk, H.D.; Mühlberger, E. Interactions of Marburg Virus Nucleocapsid Proteins. Virology 1998, 249, 406–417. [Google Scholar] [CrossRef]
- Chenik, M.; Chebli, K.; Gaudin, Y.; Blondel, D. In Vivo Interaction of Rabies Virus Phosphoprotein (P) and Nucleoprotein (N): Existence of Two N-Binding Sites on P Protein. J. Gen. Virol. 1994, 75 Pt 11, 2889–2896. [Google Scholar] [CrossRef] [PubMed]
- Nikolic, J.; Le Bars, R.; Lama, Z.; Scrima, N.; Lagaudrière-Gesbert, C.; Gaudin, Y.; Blondel, D. Negri Bodies Are Viral Factories with Properties of Liquid Organelles. Nat. Commun. 2017, 8, 58. [Google Scholar] [CrossRef]
- Zhou, Y.; Su, J.M.; Samuel, C.E.; Ma, D. Measles Virus Forms Inclusion Bodies with Properties of Liquid Organelles. J. Virol. 2019, 93, e00948-19. [Google Scholar] [CrossRef] [PubMed]
- Galloux, M.; Risso-Ballester, J.; Richard, C.; Fix, J.; Rameix-Welti, M.-A.; Eléouët, J.-F. Minimal Elements Required for the Formation of Respiratory Syncytial Virus Cytoplasmic Inclusion Bodies In Vivo and In Vitro. MBio 2020, 11, e01202-20. [Google Scholar] [CrossRef]
- Derdowski, A.; Peters, T.R.; Glover, N.; Qian, R.; Utley, T.J.; Burnett, A.; Williams, J.V.; Spearman, P.; Crowe, J.E. Human Metapneumovirus Nucleoprotein and Phosphoprotein Interact and Provide the Minimal Requirements for Inclusion Body Formation. J. Gen. Virol. 2008, 89, 2698–2708. [Google Scholar] [CrossRef] [PubMed]
- Heinrich, B.S.; Maliga, Z.; Stein, D.A.; Hyman, A.A.; Whelan, S.P.J. Phase Transitions Drive the Formation of Vesicular Stomatitis Virus Replication Compartments. MBio 2018, 9, e02290-17. [Google Scholar] [CrossRef]
- Kobayashi, T.; Shoya, Y.; Koda, T.; Takashima, I.; Lai, P.K.; Ikuta, K.; Kakinuma, M.; Kishi, M. Nuclear Targeting Activity Associated with the Amino Terminal Region of the Borna Disease Virus Nucleoprotein. Virology 1998, 243, 188–197. [Google Scholar] [CrossRef] [PubMed]
- Shoya, Y.; Kobayashi, T.; Koda, T.; Ikuta, K.; Kakinuma, M.; Kishi, M. Two Proline-Rich Nuclear Localization Signals in the Amino- and Carboxyl-Terminal Regions of the Borna Disease Virus Phosphoprotein. J. Virol. 1998, 72, 9755–9762. [Google Scholar] [CrossRef] [PubMed]
- Motohashi, K. A Simple and Efficient Seamless DNA Cloning Method Using SLiCE from Escherichia coli Laboratory Strains and Its Application to SLiP Site-Directed Mutagenesis. BMC Biotechnol. 2015, 15, 47. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hirai, Y.; Horie, M. Nyamanini Virus Nucleoprotein and Phosphoprotein Organize Viral Inclusion Bodies That Associate with Host Biomolecular Condensates in the Nucleus. Int. J. Mol. Sci. 2023, 24, 6550. https://doi.org/10.3390/ijms24076550
Hirai Y, Horie M. Nyamanini Virus Nucleoprotein and Phosphoprotein Organize Viral Inclusion Bodies That Associate with Host Biomolecular Condensates in the Nucleus. International Journal of Molecular Sciences. 2023; 24(7):6550. https://doi.org/10.3390/ijms24076550
Chicago/Turabian StyleHirai, Yuya, and Masayuki Horie. 2023. "Nyamanini Virus Nucleoprotein and Phosphoprotein Organize Viral Inclusion Bodies That Associate with Host Biomolecular Condensates in the Nucleus" International Journal of Molecular Sciences 24, no. 7: 6550. https://doi.org/10.3390/ijms24076550
APA StyleHirai, Y., & Horie, M. (2023). Nyamanini Virus Nucleoprotein and Phosphoprotein Organize Viral Inclusion Bodies That Associate with Host Biomolecular Condensates in the Nucleus. International Journal of Molecular Sciences, 24(7), 6550. https://doi.org/10.3390/ijms24076550





