Genome-Wide Characterization and Expression Analysis of Transcription Factor Families in Desert Moss Syntrichia caninervis under Abiotic Stresses
Abstract
1. Introduction
2. Results
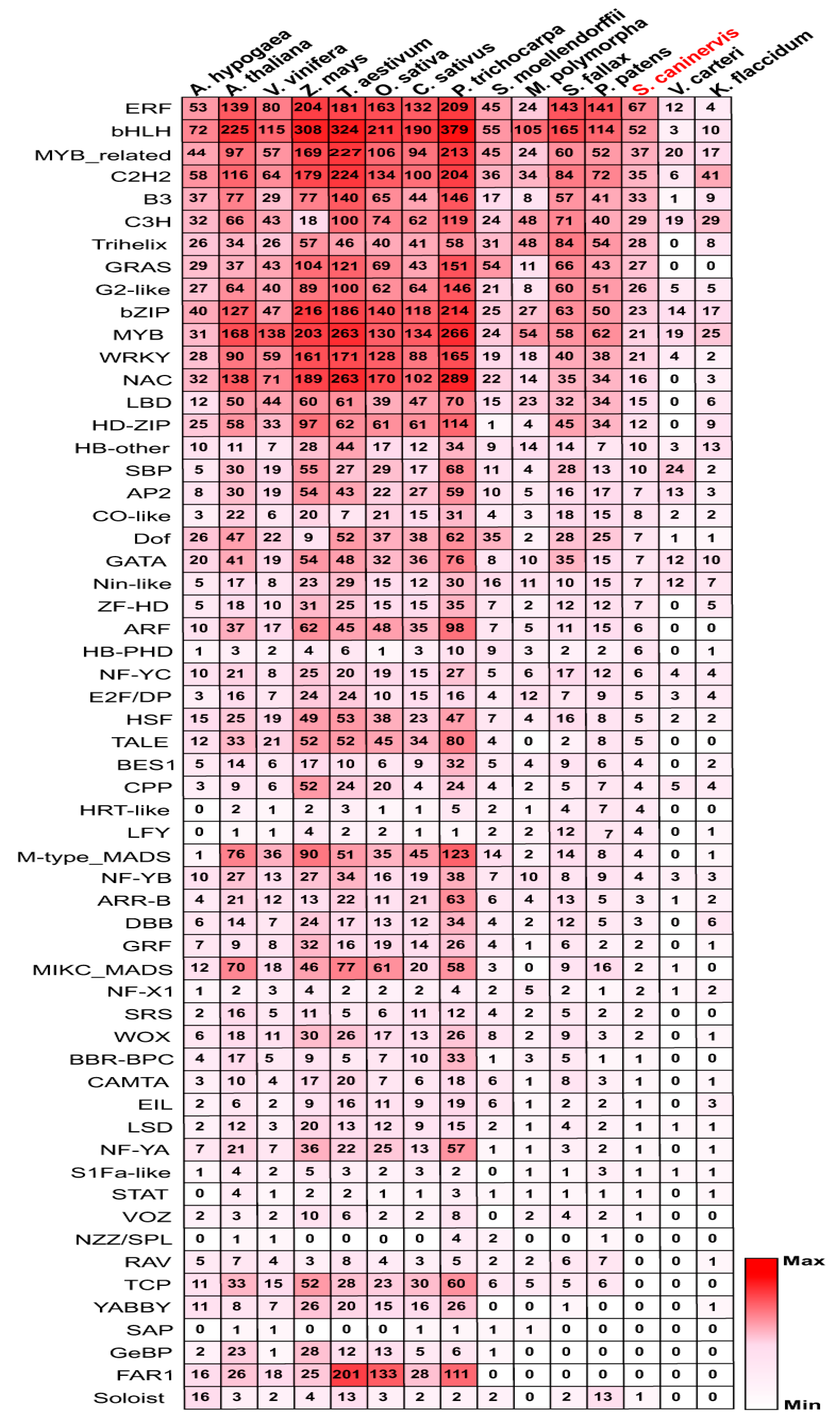
2.1. Identification of S. caninervis TFs
2.2. Chromosomal Mapping of TF Genes in S. caninervis
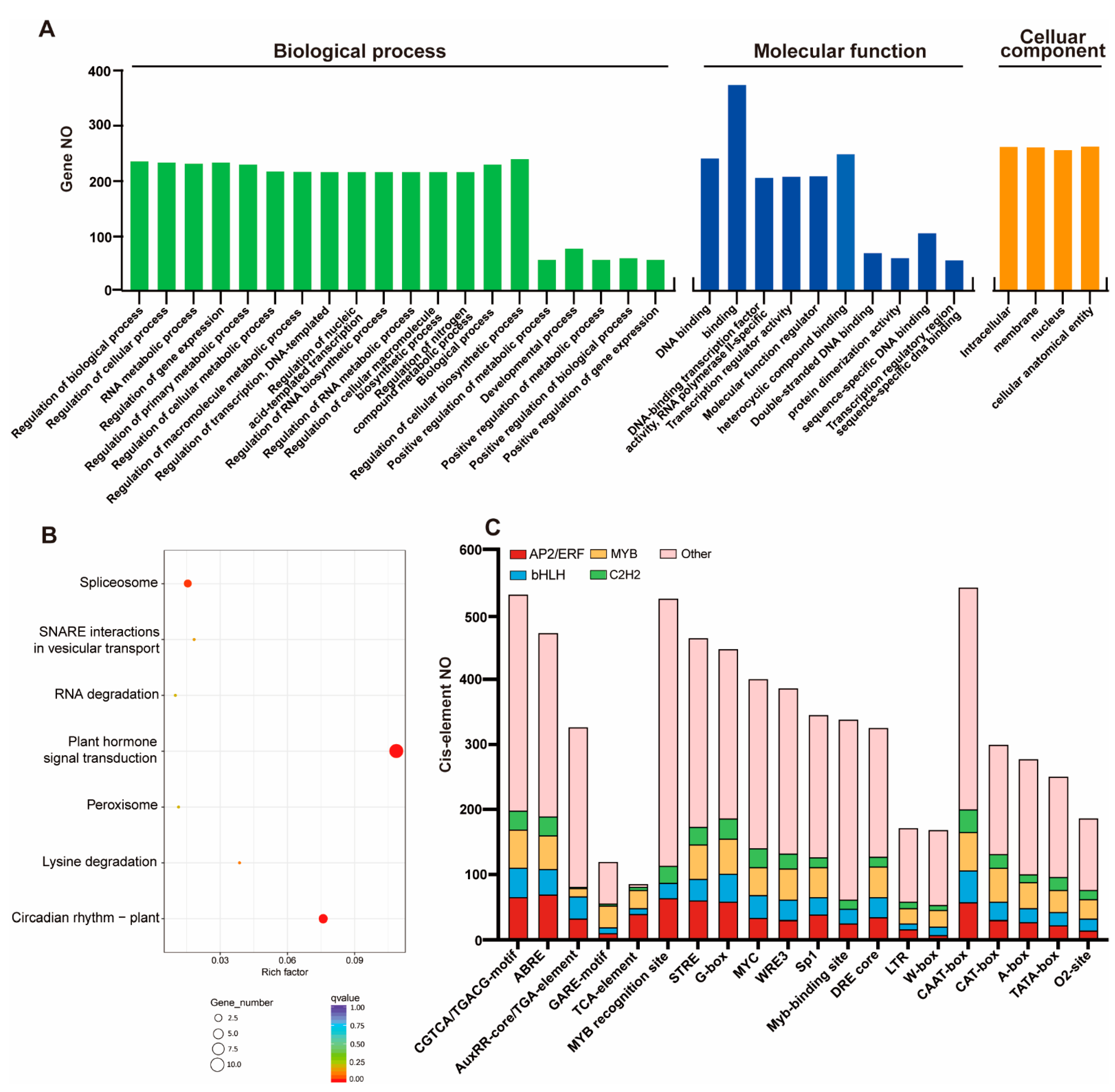
2.3. GO Term, KEGG Pathway, and Cis-Element Analysis of Tfs in S. caninervis
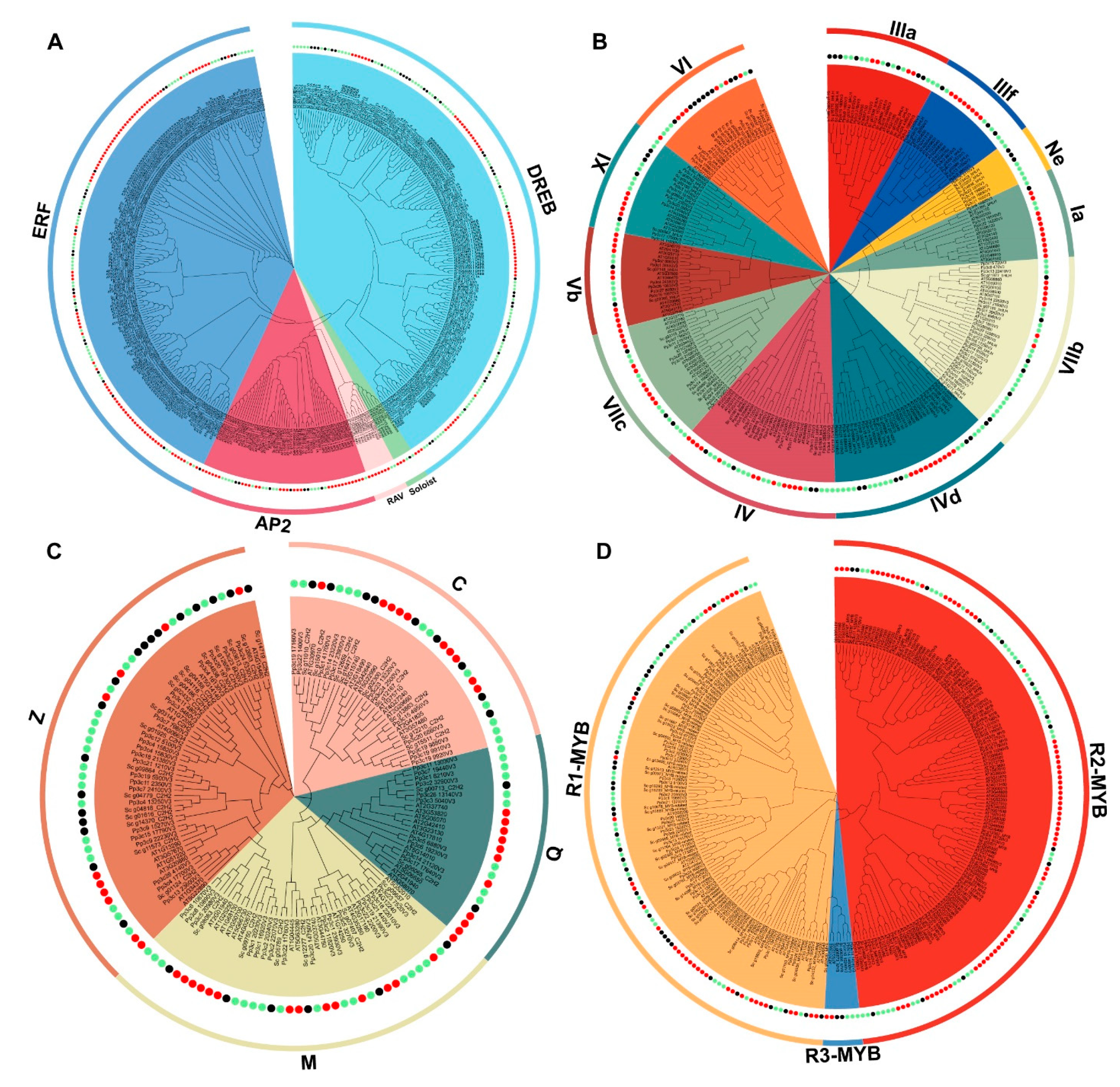
2.4. Evolutionary Conservation of the Top Four TF Families in S. caninervis
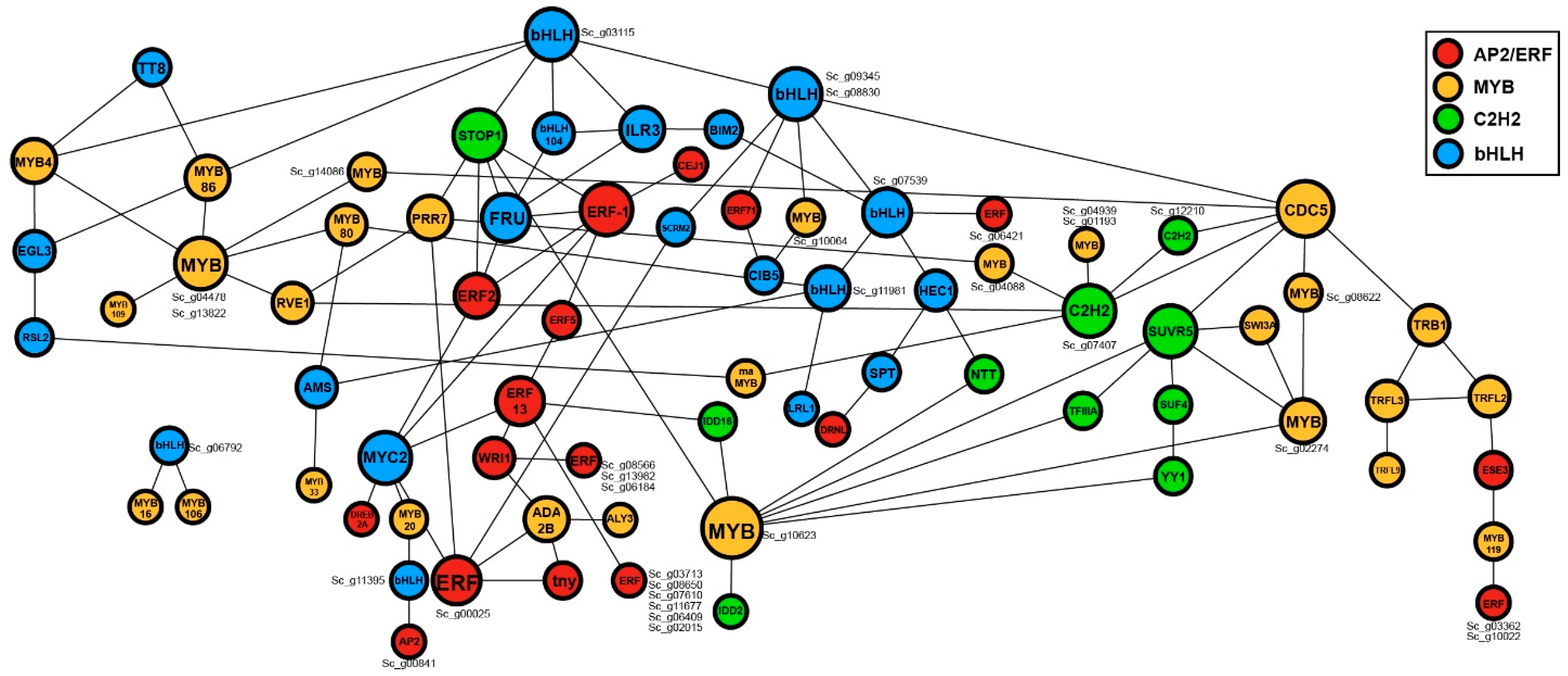
2.5. Protein-Protein Interactions Analysis of the Top Four TF Families
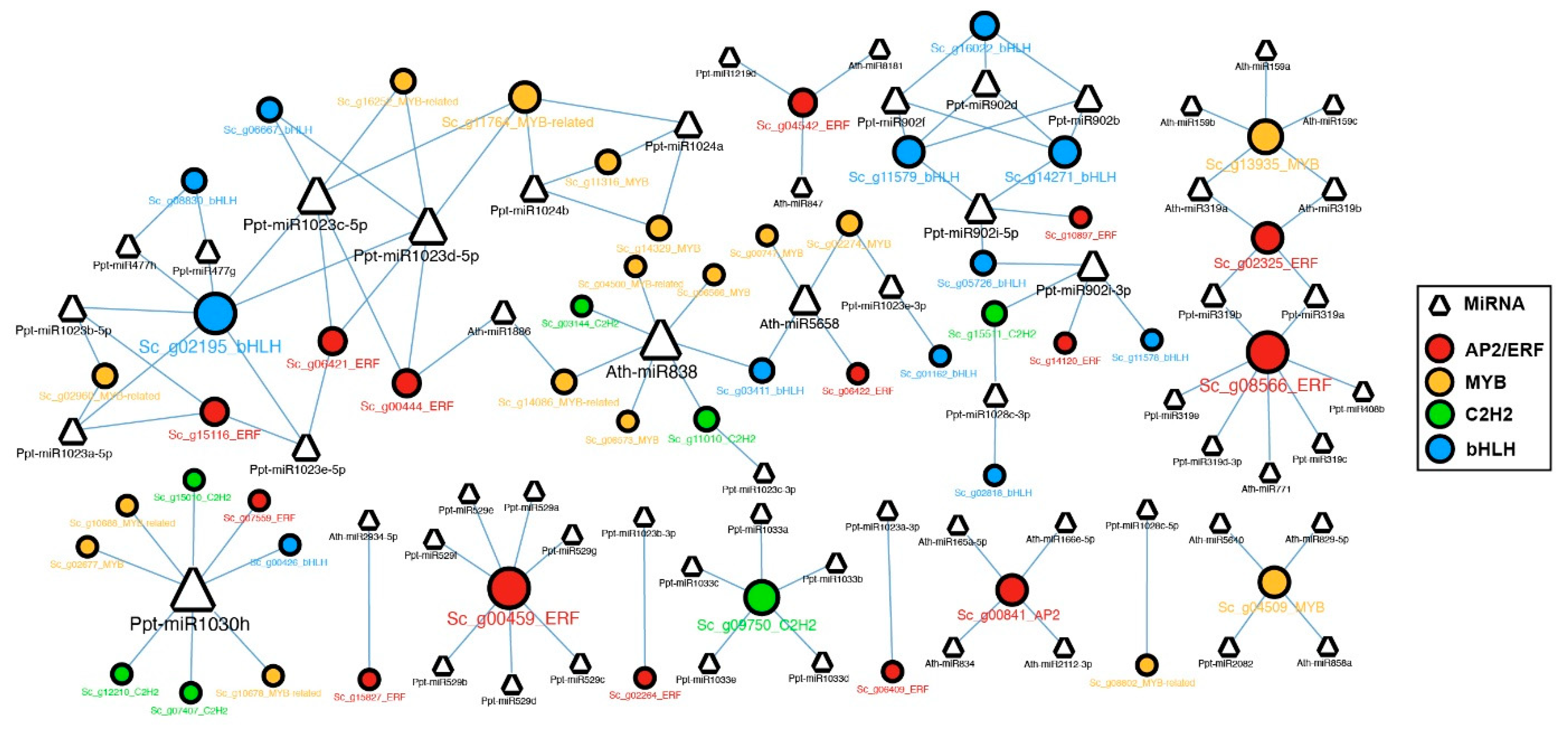
2.6. MicroRNAs and Their Target Transcription Factors
2.7. Expression Profile Analysis of Four Overrepresented TFs under Chilling and Freezing Stresses Based on Transcriptome Data
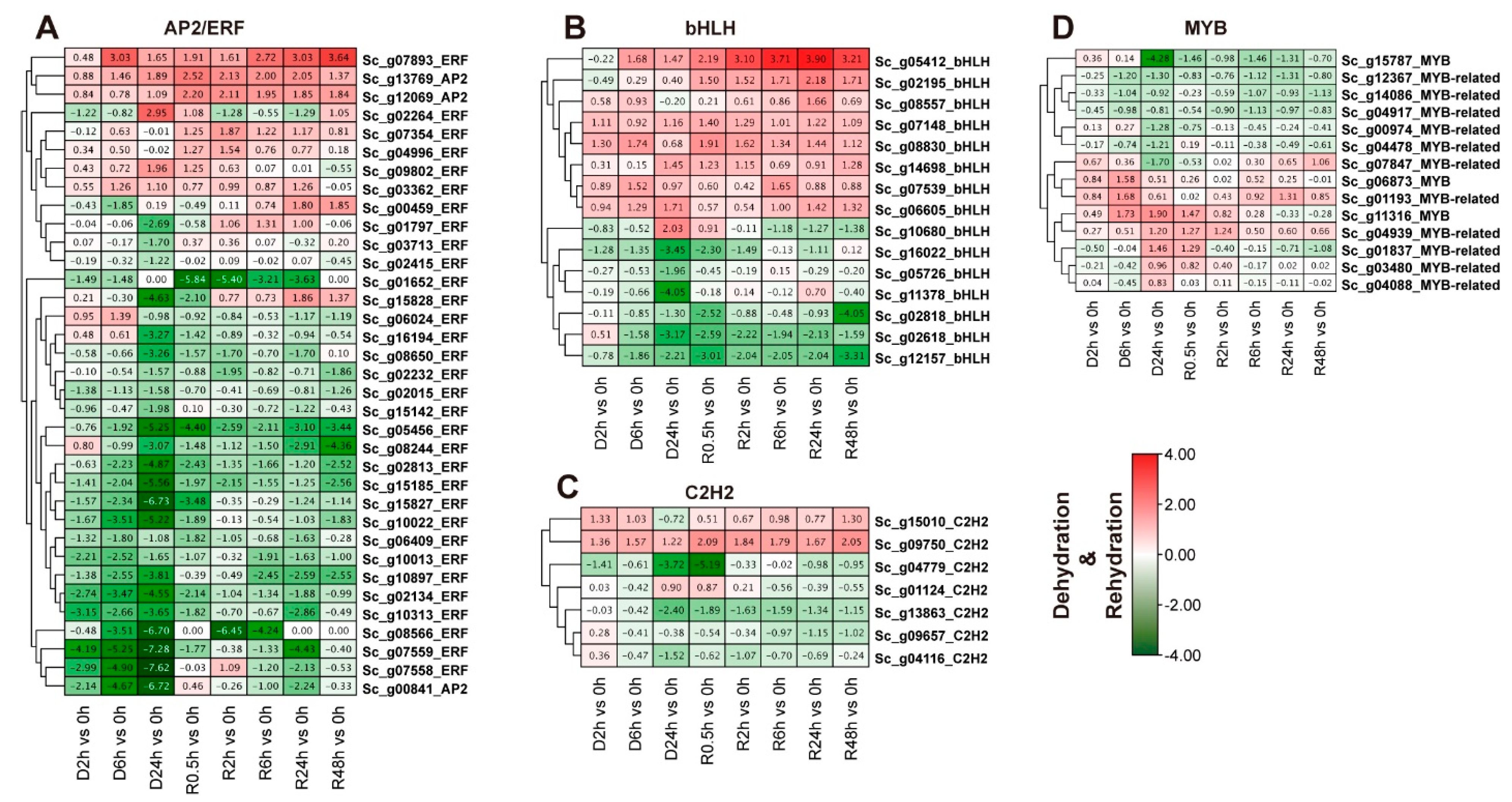
2.8. Expression Profile Analysis of Four Overrepresented TFs under Dehydration-Rehydration Stress Based on Transcriptome Data
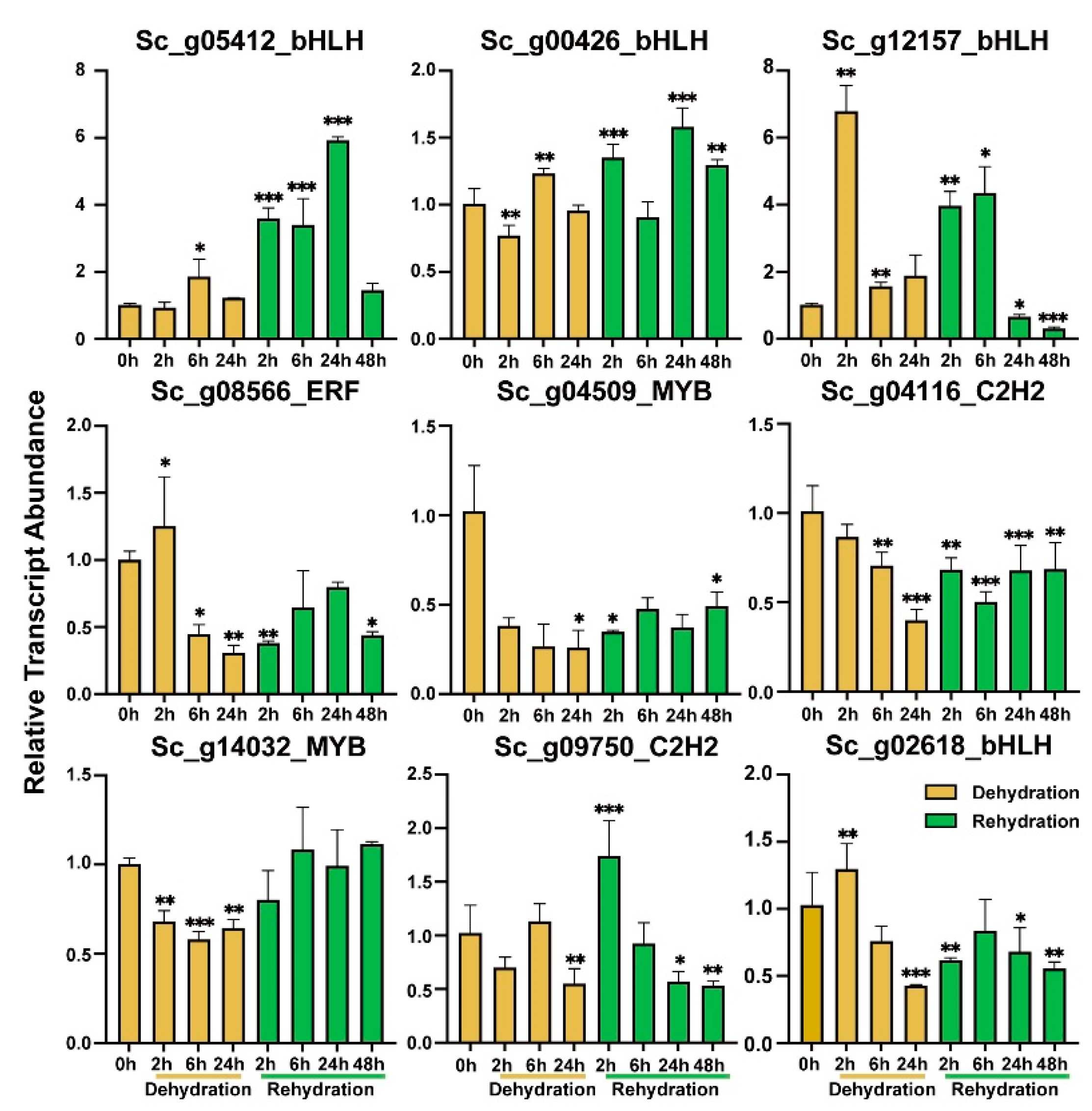
2.9. Validation of TFs Transcript Abundance by RT-qPCR
3. Discussion
3.1. Genome-Wide Identification and Comparative Analysis of TFs
3.2. The Functional Prediction of the Top Four TF Families by Comprehensive Analysis
3.3. Transcript Abundances of AP2/ERF, MYB, bHLH, and C2H2-Zinc Finger Genes Associated with Cold and Dehydration-Rehydration Stresses
4. Material and Methods
4.1. Identification and Genetic Mapping of S. caninervis TFs
4.2. Bioinformatics Analyses of S. Caninervis TF Families
4.3. Expression Profiling of TFs in Stressed S. caninervis by Abiotic Stress
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alejo-jacuinde, G.; Herrera-estrella, L. Exploring the High Variability of Vegetative Desiccation Tolerance in Pteridophytes. Plants 2022, 11, 1222. [Google Scholar] [CrossRef] [PubMed]
- Ben Rejeb, I.; Pastor, V.; Mauch-Mani, B. Plant Responses to Simultaneous Biotic and Abiotic Stress: Molecular Mechanisms. Plants 2014, 3, 458–475. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.V.; Davis, D.G. Abiotic Stress Alters Transcript Profiles and Activity of Glutathione S-Transferase, Glutathione Peroxidase, and Glutathione Reductase in Euphorbia esula. Physiol. Plant. 2004, 120, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Ito, Y.; Yamaguchi-Shinozaki, K. Transcriptional Regulatory Networks in Response to Abiotic Stresses in Arabidopsis and Grasses. Plant Physiol. 2009, 149, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.S.; Kayani, M.A.; Amjad, M. Transcription Factors as Tools to Engineer Enhanced Drought Stress Tolerance in Plants. Biotechnol. Prog. 2011, 27, 297–306. [Google Scholar] [CrossRef]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis Transcription Factors: Genome-Wide Comparative Analysis among Eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef]
- Ptashne, M. How eukaryotic transcriptional activators work. Nature 1988, 335, 683–689. [Google Scholar] [CrossRef]
- Xu, W.; Grain, D.; Bobet, S.; Le Gourrierec, J.; Thévenin, J.; Kelemen, Z.; Lepiniec, L.; Dubos, C. Complexity and Robustness of the Flavonoid Transcriptional Regulatory Network Revealed by Comprehensive Analyses of MYB-BHLH-WDR Complexes and Their Targets in Arabidopsis Seed. New Phytol. 2014, 202, 132–144. [Google Scholar] [CrossRef]
- Stracke, R.; Ishihara, H.; Huep, G.; Barsch, A.; Mehrtens, F.; Niehaus, K.; Weisshaar, B. Differential Regulation of Closely Related R2R3-MYB Transcription Factors Controls Flavonol Accumulation in Different Parts of the Arabidopsis thaliana Seedling. Plant J. 2007, 50, 660–677. [Google Scholar] [CrossRef]
- Franco-zorrilla, J.M.; López-vidriero, I.; Carrasco, J.L.; Godoy, M.; Vera, P.; Solano, R. DNA-Binding Specificities of Plant Transcription Factors and Their Potential to Define Target Genes. Proc. Natl. Acad. Sci. USA 2014, 111, 2367–2372. [Google Scholar] [CrossRef]
- Tian, F.; Yang, D.; Meng, Y.; Jin, J.; Gao, G. PlantRegMap: Charting Functional Regulatory Maps in Plants. Nucliec Acid Res. 2020, 48, 1104–1113. [Google Scholar] [CrossRef] [PubMed]
- Qu, L.J.; Zhu, Y.X. Transcription Factor Families in Arabidopsis: Major Progress and Outstanding Issues for Future Research. Curr. Opin. Plant Biol. 2006, 9, 544–549. [Google Scholar] [CrossRef] [PubMed]
- Chacón-Cerdas, R.; Barboza-Barquero, L.; Albertazzi, F.J.; Rivera-Méndez, W. Transcription Factors Controlling Biotic Stress Response in Potato Plants. Physiol. Mol. Plant Pathol. 2020, 112, 101527. [Google Scholar] [CrossRef]
- Seo, E.; Choi, D. Functional Studies of Transcription Factors Involved in Plant Defenses in the Genomics Era. Brief. Funct. Genom. 2015, 14, 260–267. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Fu, X.X.; Li, R.Q.; Zhao, X.; Liu, Y.; Li, M.H.; Zwaenepoel, A.; Ma, H.; Goffinet, B.; Guan, Y.L.; et al. The Hornwort Genome and Early Land Plant Evolution. Nat. Plants 2020, 6, 107–118. [Google Scholar] [CrossRef]
- Catarino, B.; Hetherington, A.J.; Emms, D.M.; Kelly, S.; Dolan, L. The Stepwise Increase in the Number of Transcription Factor Families in the Precambrian Predated the Diversification of Plants on Land. Mol. Biol. Evol. 2016, 33, 2815–2819. [Google Scholar] [CrossRef]
- Zhang, Y. The Microstructure and Formation of Biological Soil Crusts in Their Early Developmental Stage. Chin. Sci. Bull. 2005, 50, 117–121. [Google Scholar] [CrossRef]
- Wood, W.; Neal, D.T. A New Look at Habits and the Habit-Goal Interface. Psychol. Rev. 2007, 114, 843–863. [Google Scholar] [CrossRef]
- Yang, H.; Zhang, D.; Wang, J.; Wood, A.J.; Zhang, Y. Molecular Cloning of a Stress-Responsive Aldehyde Dehydrogenase Gene ScALDH21 from the Desiccation-Tolerant Moss Syntrichia caninervis and Its Responses to Different Stresses. Mol. Biol. Rep. 2012, 39, 2645–2652. [Google Scholar] [CrossRef]
- Li, X.; Zhang, D.; Gao, B.; Liang, Y.; Yang, H.; Wang, Y.; Wood, A.J. Transcriptome-Wide Identification, Classification, and Characterization of AP2/ERF Family Genes in the Desert Moss Syntrichia caninervis. Front. Plant Sci. 2017, 8, 262. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, Y.; Yang, H.; Liang, Y.; Li, X.; Oliver, M.J.; Zhang, D. Functional Aspects of Early Light-induced Protein (Elip) Genes from the Desiccation-tolerant Moss Syntrichia caninervis. Int. J. Mol. Sci. 2020, 21, 1411. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, D.; Li, H.; Gao, B.; Yang, H.; Zhang, Y.; Wood, A.J. Characterization of Reference Genes for RT-QPCR in the Desert Moss Syntrichia caninervis in Response to Abiotic Stress and Desiccation/Rehydration. Front. Plant Sci. 2015, 6, 38. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Zhang, D.; Li, X.; Yang, H.; Wood, A.J. De Novo Assembly and Characterization of the Transcriptome in the Desiccation-Tolerant Moss Syntrichia caninervis. BMC Res. Notes 2014, 7, 490. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, X.; Zhang, K.; Zhang, D.; Guan, K. An ABSCISIC ACID INSENSITIVE3-like Gene from the Desert Moss Syntrichia caninervis Confers Abiotic Stress Tolerance and Reduces ABA Sensitivity. Plant Cell Tissue Organ Cult. 2018, 133, 417–435. [Google Scholar] [CrossRef]
- Liu, J.; Yang, R.; Liang, Y.; Wang, Y.; Li, X. The DREB A-5 Transcription Factor ScDREB5 From Syntrichia caninervis Enhanced Salt Tolerance by Regulating Jasmonic Acid Biosynthesis in Transgenic Arabidopsis. Front. Plant Sci. 2022, 13, 857396. [Google Scholar] [CrossRef]
- Li, H.; Zhang, D.; Li, X.; Guan, K.; Yang, H. Novel DREB A-5 Subgroup Transcription Factors from Desert Moss (Syntrichia caninervis) Confers Multiple Abiotic Stress Tolerance to Yeast. J. Plant Physiol. 2016, 194, 45–53. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, C.; Huang, M.; Zhang, Y. Functional Analysis of ScABI3 from Syntrichia caninervis Mitt. in Medicago sativa L. Agronomy 2022, 12, 2238. [Google Scholar] [CrossRef]
- Eckardt, N.A. DREB Duo Defines Distinct Drought and Cold Response Pathways. Plant Cell 2019, 31, 1196–1197. [Google Scholar] [CrossRef]
- Silva, A.T.; Gao, B.; Fisher, K.M.; Mishler, B.D.; Ekwealor, J.T.B.; Stark, L.R.; Li, X.; Zhang, D.; Bowker, M.A.; Brinda, J.C.; et al. To Dry Perchance to Live: Insights from the Genome of the Desiccation-Tolerant Biocrust Moss Syntrichia caninervis. Plant J. 2021, 105, 1339–1356. [Google Scholar] [CrossRef]
- Zhang, T.; Cui, Z.; Li, Y.; Kang, Y.; Song, X.; Wang, J. Genome-Wide Identification and Expression Analysis of MYB Transcription Factor Superfamily in Dendrobium catenatum. Front. Genet. 2021, 12, 714696. [Google Scholar] [CrossRef]
- Keren, H.; Lev-Maor, G.; Ast, G. Alternative Splicing and Evolution: Diversification, Exon Definition and Function. Nat. Rev. Genet. 2010, 11, 345–355. [Google Scholar] [CrossRef] [PubMed]
- Stockinger, E.J.; Gilmour, S.J.; Thomashow, M.F. Arabidopsis thaliana CBF1 Encodes an AP2 Domain-Containing Transcriptional Activator That Binds to the C-Repeat/DRE, a Cis-Acting DNA Regulatory Element That Stimulates Transcription in Response to Low Temperature and Water Deficit. Proc. Natl. Acad. Sci. USA 1997, 94, 1035–1040. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, K.; Todaka, D.; Mizoi, J.; Yoshida, T.; Kidokoro, S.; Matsukura, S.; Takasaki, H.; Sakurai, T.; Yamamoto, Y.Y.; Yoshiwara, K.; et al. Identification of Cis-Acting Promoter Elements in Cold-and Dehydration-Induced Transcriptional Pathways in Arabidopsis, Rice, and Soybean. DNA Res. 2012, 19, 37–49. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Ni, D.; Shen, J.; Deng, S.; Xuan, H. Genome-Wide Identification of the AP2/ERF Gene Family and Functional Analysis of GmAP2/ERF144 for Drought Tolerance in Soybean. Front. Plant Sci. 2022, 13, 848766. [Google Scholar] [CrossRef]
- Li, Q.; Zhang, L.; Chen, P.; Wu, C.; Zhang, H. Genome-Wide Identification of APETALA2/ ETHYLENE RESPONSIVE FACTOR Transcription Factors in Cucurbita moschata and Their Involvement in Ethylene Response. Front. Plant Sci. 2022, 13, 847754. [Google Scholar] [CrossRef]
- Pires, N.; Dolan, L. Origin and Diversification of Basic-Helix-Loop-Helix Proteins in Plants. Mol. Biol. Evol. 2010, 27, 862–874. [Google Scholar] [CrossRef]
- Song, Y.; Zhang, X.; Li, M.; Yang, H.; Fu, D.; Lv, J.; Ding, Y.; Gong, Z.; Shi, Y.; Yang, S. The Direct Targets of CBFs: In Cold Stress Response and Beyond. J. Integr. Plant Biol. 2021, 63, 1874–1887. [Google Scholar] [CrossRef]
- Zhou, H.; He, Y.; Zhu, Y.; Li, M.; Song, S.; Bo, W.; Li, Y. Comparative Transcriptome Profiling Reveals Cold Stress Responsiveness in Two Contrasting Chinese Jujube Cultivars. BMC Plant Biol. 2020, 20, 240. [Google Scholar] [CrossRef]
- Agarwal, P.K.; Agarwal, P.; Reddy, M.K.; Sopory, S.K. Role of DREB Transcription Factors in Abiotic and Biotic Stress Tolerance in Plants. Plant Cell Rep. 2006, 25, 1263–1274. [Google Scholar] [CrossRef]
- Dai, X.; Zhao, P.X. PsRNATarget: A Plant Small RNA Target Analysis Server. Nucleic Acids Res. 2011, 46, 49–54. [Google Scholar] [CrossRef]
- Guo, Z.; Kuang, Z.; Wang, Y.; Zhao, Y.; Tao, Y.; Cheng, C.; Yang, J.; Lu, X.; Hao, C.; Wang, T.; et al. PmiREN: A Comprehensive Encyclopedia of Plant MiRNAs. Nucleic Acids Res. 2020, 48, 1114–1121. [Google Scholar] [CrossRef]
- He, K.; Guo, A.; Gao, G.; Zhu, Q.; Liu, X.; Zhang, H.; Chen, X.; Gu, X.; Luo, J. Computational Identification of Plant Transcription Factors and the Construction of the PlantTFDB Database. Methods Mol. Biol. 2010, 674, 351–368. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Tian, F.; Yang, D.; Meng, Y.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a Central Hub for Transcription Factors and Regulatory Interactions in Plants. Nucliec Acid Res. 2017, 45, 1040–1045. [Google Scholar] [CrossRef]
- Feller, A.; MacHemer, K.; Braun, E.L.; Grotewold, E. Evolutionary and Comparative Analysis of MYB and BHLH Plant Transcription Factors. Plant J. 2011, 66, 94–116. [Google Scholar] [CrossRef]
- Perroud, P.F.; Haas, F.B.; Hiss, M.; Ullrich, K.K.; Alboresi, A.; Amirebrahimi, M.; Barry, K.; Bassi, R.; Bonhomme, S.; Chen, H.; et al. The Physcomitrella patens Gene Atlas Project: Large-Scale RNA-Seq Based Expression Data. Plant J. 2018, 95, 168–182. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Gao, B.; Zhang, D.; Liang, Y.; Liu, X.; Zhao, J.; Zhang, J.; Wood, A.J. Identification, Classification, and Functional Analysis of AP2/ERF Family Genes in the Desert Moss Bryum argenteum. Int. J. Mol. Sci. 2018, 19, 3637. [Google Scholar] [CrossRef] [PubMed]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB Transcription Factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Reboledo, G.; Agorio, A.; León, I.P. De Moss Transcription Factors Regulating Development and Defense Responses to Stress. Exp. Bot. 2022, 73, 4546–4561. [Google Scholar] [CrossRef]
- Englbrecht, C.C.; Schoof, H.; Böhm, S. Conservation, Diversification and Expansion of C2H2 Zinc Finger Proteins in the Arabidopsis thaliana Genome. BMC Genom. 2004, 5, 39. [Google Scholar] [CrossRef]
- Ritonga, F.N.; Chen, S. Physiological and Molecular Mechanism Involved in Cold Stress Tolerance in Plants. Plants 2020, 9, 560. [Google Scholar] [CrossRef]
- Müller, M.; Munné-bosch, S. Ethylene Response Factors: A Key Regulatory Hub in Hormone and Stress Signaling. Plant Physiol. 2015, 169, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Liang, Z.; Dai, X.; Zhang, Y.; Mahboub, S.; Ngu, D.W. Predicting Transcriptional Responses to Cold Stress across Plant Species. Proc. Natl. Acad. Sci. USA 2021, 118, e2026330118. [Google Scholar] [CrossRef] [PubMed]
- Knight, H.; Zarka, D.G.; Okamoto, H.; Thomashow, M.F.; Knight, M.R. Abscisic Acid Induces CBF Gene Transcription and Subsequent Induction of Cold-Regulated Genes via the CRT Promoter Element. Plant Physiol. 2004, 135, 1710–1717. [Google Scholar] [CrossRef]
- Göös, H.; Kinnunen, M.; Salokas, K.; Tan, Z.; Liu, X.; Zhang, Q.; Wei, G.; Varjosalo, M.; Yadav, L. Human trancriprion factor proten interaction Networks. Nat. Commun. 2022, 13, 766. [Google Scholar] [CrossRef] [PubMed]
- An, J.P.; Li, R.; Qu, F.J.; You, C.X.; Wang, X.F.; Hao, Y.J. R2R3-MYB Transcription Factor MdMYB23 Is Involved in the Cold Tolerance and Proanthocyanidin Accumulation in Apple. Plant J. 2018, 96, 562–577. [Google Scholar] [CrossRef]
- Nyikó, T.; Auber, A.; Szabadkai, L.; Benkovics, A.; Auth, M.; Mérai, Z.; Kerényi, Z.; Dinnyés, A.; Nagy, F.; Silhavy, D. Expression of the ERF1 Translation Termination Factor Is Controlled by an Autoregulatory Circuit Involving Readthrough and Nonsense-Mediated Decay in Plants. Nucleic Acids Res. 2017, 45, 4174–4188. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhang, L.; Zhang, H.; Chen, L.; Yu, D. ERF1 Delays Flowering through Direct Inhibition of FLOWERING LOCUS T Expression in Arabidopsis. J. Integr. Plant Biol. 2021, 63, 1712–1723. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo, O.; Piqueras, R.; Sánchez-Serrano, J.J.; Solano, R. ETHYLENE RESPONSE FACTOR1 Integrates Signals from Ethylene and Jasmonate Pathways in Plant Defense. Plant Cell 2003, 15, 165–178. [Google Scholar] [CrossRef]
- Oh, E.; Kang, H.; Yamaguchi, S.; Park, J.; Lee, D.; Kamiya, Y.; Choi, G. Genome-Wide Analysis of Genes Targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during Seed Germination in Arabidopsis. Plant Cell 2009, 21, 403–419. [Google Scholar] [CrossRef]
- Song, C.; Cao, Y.; Dai, J.; Li, G.; Manzoor, M.A.; Chen, C.; Deng, H. The Multifaceted Roles of MYC2 in Plants: Toward Transcriptional Reprogramming and Stress Tolerance by Jasmonate Signaling. Front. Plant Sci. 2022, 13, 868874. [Google Scholar] [CrossRef]
- Qin, G.; Mallik, S.; Mitra, R.; Li, A.; Jia, P.; Eischen, C.M.; Zhao, Z. MicroRNA and Transcription Factor Co-Regulatory Networks and Subtype Classification of Seminoma and Non-Seminoma in Testicular Germ Cell Tumors. Sci. Rep. 2020, 10, 852. [Google Scholar] [CrossRef]
- Singh, P.; Dutta, P.; Chakrabarty, D. MiRNAs Play Critical Roles in Response to Abiotic Stress by Modulating Cross-Talk of Phytohormone Signaling. Plant Cell Rep. 2021, 40, 1617–1630. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Zhang, Q.; Zhao, Y.-Q.; Yang, J.; He, H.-B.; Jia, G.-X. The Lre-MiR159a-LrGAMYB Pathway Mediates Resistance to Grey Mould Infection in Lilium regale. Mol. Plant Pathol. 2020, 21, 749–760. [Google Scholar] [CrossRef] [PubMed]
- Biniaz, Y.; Tahmasebi, A.; Afsharifar, A.; Tahmasebi, A.; Poczai, P. Meta-Analysis of Common and Differential Transcriptomic Responses to Biotic and Abiotic Stresses in Arabidopsis thaliana. Plants 2022, 11, 502. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Badola, P.K.; Gautam, H.; Trivedi, P.K. Heterologous Expression of Arabidopsis MiR858 Modulates Biosynthesis of Secondary Metabolites and Affects Drought Tolerance in Tobacco. Plant Cell Tissue Organ Cult. 2022, 152, 287–298. [Google Scholar] [CrossRef]
- Snigdha, M.; Prasath, D. Transcriptomic Analysis to Reveal the Differentially Expressed MiRNA Targets and Their MiRNAs in Response to Ralstonia solanacearum in Ginger Species. BMC Plant Biol. 2021, 21, 355. [Google Scholar] [CrossRef]
- Li, X.; Liang, Y.; Gao, B.; Mijiti, M.; Bozorov, T.A.; Yang, H.; Zhang, D.; Wood, A.J. ScDREB10, an A-5c Type of DREB Gene of the Desert Moss Syntrichia caninervis, Confers Osmotic and Salt Tolerances to Arabidopsis. Genes 2019, 10, 146. [Google Scholar] [CrossRef]
- Liang, Y.; Li, X.; Zhang, D.; Gao, B.; Yang, H.; Wang, Y.; Guan, K.; Wood, A.J. ScDREB8, a Novel A-5 Type of DREB Gene in the Desert Moss Syntrichia caninervis, Confers Salt Tolerance to Arabidopsis. Plant Physiol. Biochem. 2017, 120, 242–251. [Google Scholar] [CrossRef]
- Feng, K.; Hou, X.L.; Xing, G.M.; Liu, J.X.; Duan, A.Q.; Xu, Z.S.; Li, M.Y.; Zhuang, J.; Xiong, A.S. Advances in AP2/ERF Super-Family Transcription Factors in Plant. Crit. Rev. Biotechnol. 2020, 40, 750–776. [Google Scholar] [CrossRef] [PubMed]
- Hiss, M.; Laule, O.; Meskauskiene, R.M.; Arif, M.A.; Decker, E.L.; Erxleben, A.; Frank, W.; Hanke, S.T.; Lang, D.; Martin, A.; et al. Large-Scale Gene Expression Profiling Data for the Model Moss Physcomitrella Patens Aid Understanding of Developmental Progression, Culture and Stress Conditions. Plant J. 2014, 79, 530–539. [Google Scholar] [CrossRef]
- Beike, A.K.; Lang, D.; Zimmer, A.D.; Florian, W.; Trautmann, D.; Wiedemann, G.; Beyer, P.; Decker, E.L.; Reski, R. Insights from the Cold Transcriptome of Physcomitrella patens: Global Specialization Pattern of Conserved Transcriptional Regulators and Identification of Orphan Genes Involved in Cold Acclimation. Plant J. 2015, 2, 869–881. [Google Scholar] [CrossRef]
- Oh, E.; Yamaguchi, S.; Hu, J.; Yusuke, J.; Jung, B.; Paik, I.; Lee, H.S.; Sun, T.P.; Kamiya, Y.; Choi, G. PIL5, a Phytochrome-Interacting BHLH Protein, Regulates Gibberellin Responsiveness by Binding Directly to the GAI and RGA Promoters in Arabidopsis Seeds. Plant Cell 2007, 4, 1192–1208. [Google Scholar] [CrossRef]
- Jin, R.; Kim, H.S.; Yu, T.; Zhang, A.; Yang, Y.; Liu, M.; Yu, W.; Zhao, P.; Zhang, Q.; Cao, Q.; et al. Identification and Function Analysis of BHLH Genes in Response to Cold Stress in Sweet potato. Plant Physiol. Biochem. 2021, 169, 224–235. [Google Scholar] [CrossRef]
- Le Hir, R.; Castelain, M.; Chakraborti, D.; Moritz, T.; Dinant, S.; Bellini, C. AtbHLH68 Transcription Factor Contributes to the Regulation of ABA Homeostasis and Drought Stress Tolerance in Arabidopsis thaliana. Physiol. Plant. 2017, 160, 312–327. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Yang, B.; Deyholos, M.K. Functional Characterization of the Arabidopsis BHLH92 Transcription Factor in Abiotic Stress. Mol. Genet. Genom. 2009, 282, 503–516. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Cao, X.; Shi, S.; Zhao, N.; Li, D.; Fang, P.; Chen, X.; Qi, W.; Zhang, Z. Comparative RNA-Seq Analysis Reveals a Critical Role for Brassinosteroids in Rose (Rosa hybrida) Petal Defense against Botrytis cinerea Infection. BMC Genet. 2018, 19, 62. [Google Scholar] [CrossRef]
- Segreto, R.; Hassel, K.; Bardal, R.; Stenøien, H.K. Desiccation Tolerance and Natural Cold Acclimation Allow Cryopreservation of Bryophytes without Pretreatment or Use of Cryoprotectants. Bryologist 2010, 113, 760–769. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The Protein Families Database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. Protein-Protein Interaction Networks, Integrated over the Tree of Life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef]
- Oliver, M.J.; Hudgeons, J.; Dowd, S.E.; Payton, P.R. A Combined Subtractive Suppression Hybridization and Expression Profiling Strategy to Identify Novel Desiccation Response Transcripts from Tortula ruralis Gametophytes. Physiol. Plant. 2009, 136, 437–460. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript Assembly and Quantification by RNA-Seq Reveals Unannotated Transcripts and Isoform Switching during Cell Differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential Expression Analysis for Sequence Count Data. Genome Biol. 2010, 11, 106. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Li, X.; Yang, R.; Liang, Y.; Gao, B.; Li, S.; Bai, W.; Oliver, M.J. The ScAPD1-like Gene from the Desert Moss Syntrichia caninervis Enhances Resistance to Verticillium dahliae via Phenylpropanoid Gene Regulation. Plant J. 2022, 113, 75–91. [Google Scholar] [CrossRef]
- Li, Q.Q.; Skinner, J.; Bennett, J.E. Evaluation of Reference Genes for Real-Time Quantitative PCR Studies in Candida Glabrata Following Azole Treatment. BMC Mol. Biol. 2012, 13, 22. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salih, H.; Bai, W.; Zhao, M.; Liang, Y.; Yang, R.; Zhang, D.; Li, X. Genome-Wide Characterization and Expression Analysis of Transcription Factor Families in Desert Moss Syntrichia caninervis under Abiotic Stresses. Int. J. Mol. Sci. 2023, 24, 6137. https://doi.org/10.3390/ijms24076137
Salih H, Bai W, Zhao M, Liang Y, Yang R, Zhang D, Li X. Genome-Wide Characterization and Expression Analysis of Transcription Factor Families in Desert Moss Syntrichia caninervis under Abiotic Stresses. International Journal of Molecular Sciences. 2023; 24(7):6137. https://doi.org/10.3390/ijms24076137
Chicago/Turabian StyleSalih, Haron, Wenwan Bai, Mingqi Zhao, Yuqing Liang, Ruirui Yang, Daoyuan Zhang, and Xiaoshuang Li. 2023. "Genome-Wide Characterization and Expression Analysis of Transcription Factor Families in Desert Moss Syntrichia caninervis under Abiotic Stresses" International Journal of Molecular Sciences 24, no. 7: 6137. https://doi.org/10.3390/ijms24076137
APA StyleSalih, H., Bai, W., Zhao, M., Liang, Y., Yang, R., Zhang, D., & Li, X. (2023). Genome-Wide Characterization and Expression Analysis of Transcription Factor Families in Desert Moss Syntrichia caninervis under Abiotic Stresses. International Journal of Molecular Sciences, 24(7), 6137. https://doi.org/10.3390/ijms24076137






