ADAM10 Gene Variants in AD Patients and Their Relationship to CSF Protein Levels
Abstract
1. Introduction
2. Results
2.1. Sequencing of ADAM10
2.2. ADAM10 CSF Level Determinations by ELISA
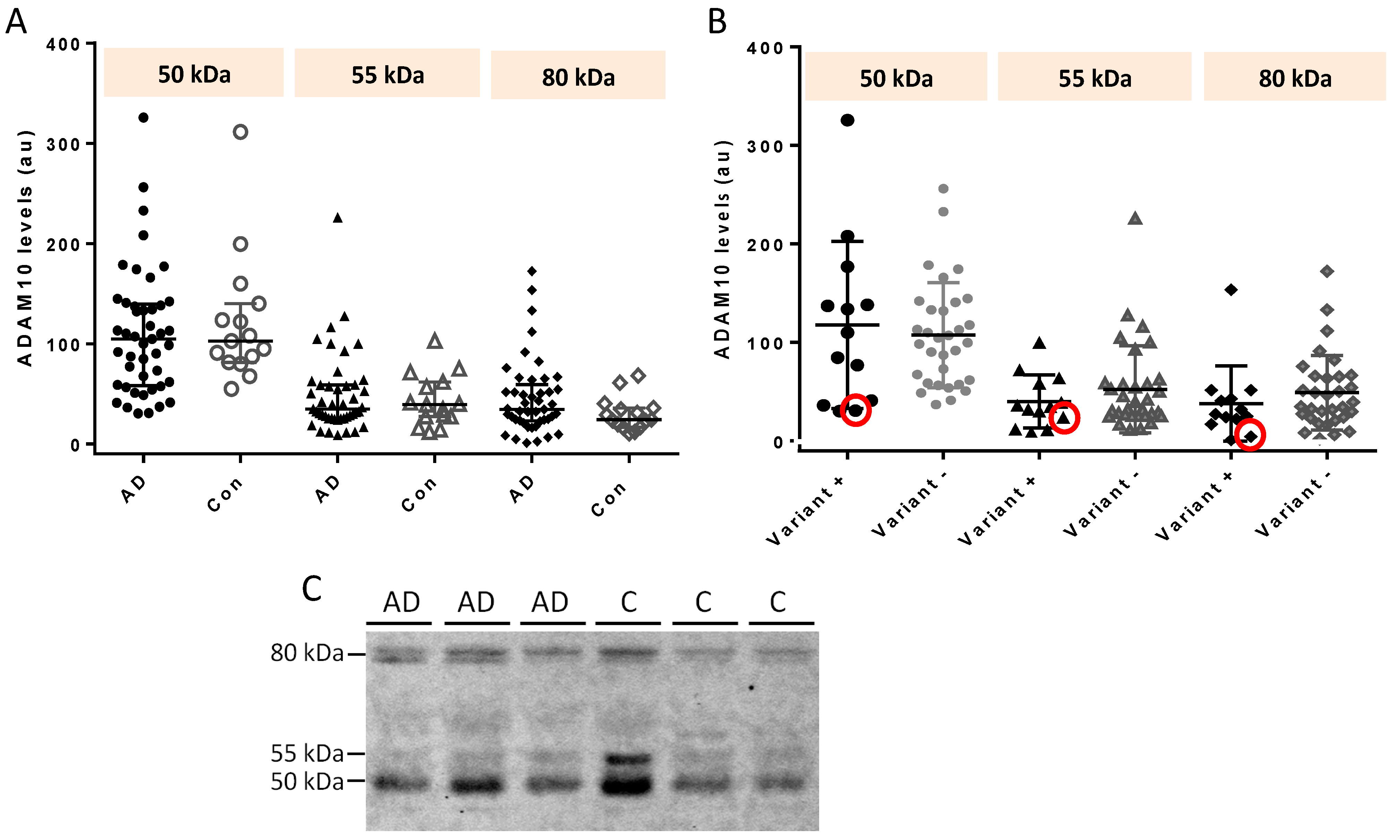
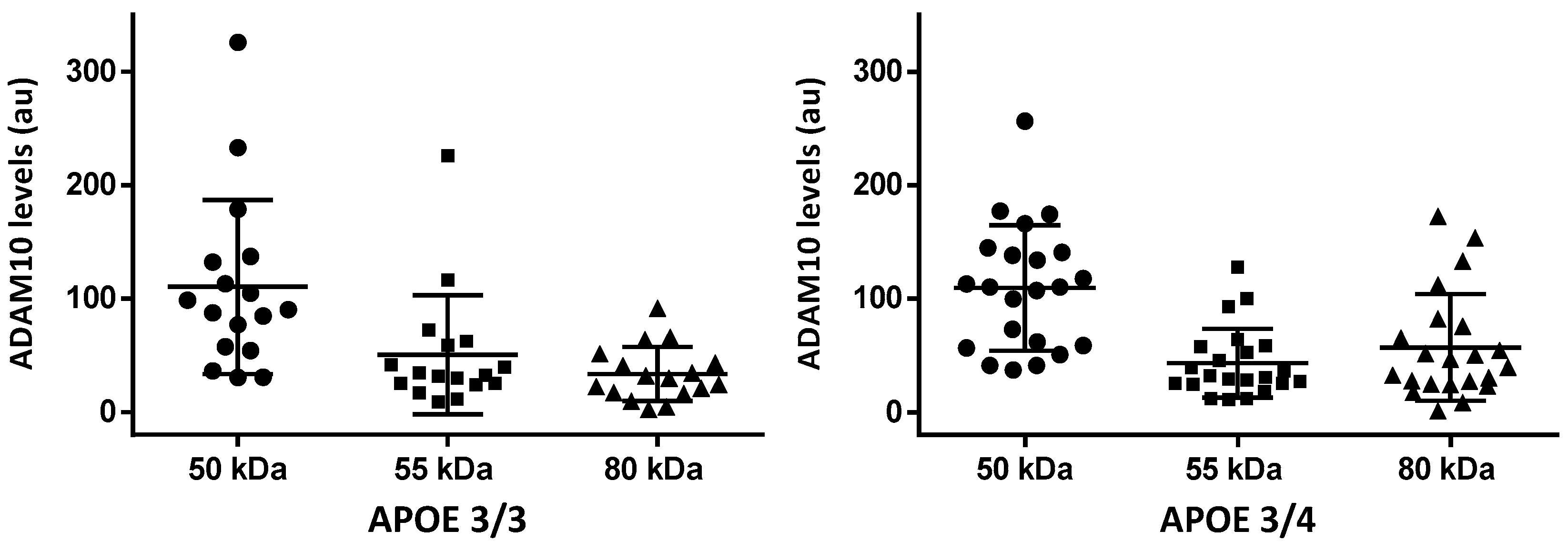
2.3. ADAM10 CSF Species by Western Blot
3. Discussion
4. Methods
4.1. Participants
4.2. Genetic Study
4.3. CSF Analyses
4.4. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
References
- Selkoe, D.J.; Hardy, J. The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol. Med. 2016, 8, 595–608. [Google Scholar] [CrossRef] [PubMed]
- Haass, C.; Kaether, C.; Thinakaran, G.; Sisodia, S. Trafficking and Proteolytic Processing of APP. Cold Spring Harb. Perspect. Med. 2012, 2, a006270. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, P.-H.; Wang, H.; Dislich, B.; Colombo, A.; Zeitschel, U.; Ellwart, J.W.; Kremmer, E.; Roßner, S.; Lichtenthaler, S.F. ADAM10 is the physiologically relevant, constitutive α-secretase of the amyloid precursor protein in primary neurons. EMBO J. 2010, 29, 3020–3032. [Google Scholar] [CrossRef] [PubMed]
- Hitschler, L.; Lang, T. The transmembrane domain of the amyloid precursor protein is required for antiamyloidogenic processing by α-secretase ADAM10. J. Biol. Chem. 2022, 298, 101911. [Google Scholar] [CrossRef] [PubMed]
- Peron, R.; Vatanabe, I.P.; Manzine, P.R.; Camins, A.; Cominetti, M.R. Alpha-Secretase ADAM10 Regulation: Insights into Alzheimer’s Disease Treatment. Pharmaceuticals 2018, 11, 12. [Google Scholar] [CrossRef]
- Musardo, S.; Marcello, E. Synaptic dysfunction in Alzheimer’s disease: From the role of amyloid β-peptide to the α-secretase ADAM10. Eur. J. Pharmacol. 2017, 817, 30–37. [Google Scholar] [CrossRef]
- Manzine, P.R.; Ettcheto, M.; Cano, A.; Busquets, O.; Marcello, E.; Pelucchi, S.; Di Luca, M.; Endres, K.; Olloquequi, J.; Camins, A.; et al. ADAM10 in Alzheimer’s disease: Pharmacological modulation by natural compounds and its role as a peripheral marker. Biomed. Pharmacother. 2019, 113, 108661. [Google Scholar] [CrossRef]
- Postina, R.; Schroeder, A.; Dewachter, I.; Bohl, J.; Schmitt, U.; Kojro, E.; Prinzen, C.; Endres, K.; Hiemke, C.; Blessing, M.; et al. A disintegrin-metalloproteinase prevents amyloid plaque formation and hippocampal defects in an Alzheimer disease mouse model. J. Clin. Investig. 2004, 113, 1456–1464. [Google Scholar] [CrossRef]
- Esteve, P.; Rueda-Carrasco, J.; Mateo, M.I.; Martin-Bermejo, M.J.; Draffin, J.; Pereyra, G.; Sandonís, Á.; Crespo, I.; Moreno, I.; Aso, E.; et al. Elevated levels of Secreted-Frizzled-Related-Protein 1 contribute to Alzheimer’s disease pathogenesis. Nat. Neurosci. 2019, 22, 1258–1268. [Google Scholar] [CrossRef]
- Epis, R.; Marcello, E.; Gardoni, F.; Vastagh, C.; Malinverno, M.; Balducci, C.; Colombo, A.; Borroni, B.; Vara, H.; Dell’Agli, M.; et al. Blocking ADAM10 synaptic trafficking generates a model of sporadic Alzheimer’s disease. Brain 2010, 133, 3323–3335. [Google Scholar] [CrossRef]
- Elsworthy, R.J.; Hill, E.J.; Dunleavy, C.; Aldred, S. The role of ADAM10 in astrocytes: Implications for Alzheimer’s disease. Front. Aging Neurosci. 2022, 14, 1056507. [Google Scholar] [CrossRef] [PubMed]
- Musardo, S.; Therin, S.; Pelucchi, S.; D’Andrea, L.; Stringhi, R.; Ribeiro, A.; Manca, A.; Balducci, C.; Pagano, J.; Sala, C.; et al. The development of ADAM10 endocytosis inhibitors for the treatment of Alzheimer’s disease. Mol. Ther. 2022, 30, 2474–2490. [Google Scholar] [CrossRef] [PubMed]
- Colciaghi, F.; Borroni, B.; Pastorino, L.; Marcello, E.; Zimmermann, M.; Cattabeni, F.; Padovani, A.; Di Luca, M. α-Secretase ADAM10 as well as αAPPs is reduced in platelets and CSF of Alzheimer Disease patients. Mol. Med. 2002, 8, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Manzine, P.R.; Marcello, E.; Borroni, B.; Kamphuis, W.; Hol, E.; Padovani, A.; Nascimento, C.C.; Bueno, P.D.G.; Vale, F.A.C.; Pavarini, S.C.I.; et al. ADAM10 gene expression in the blood cells of Alzheimer’s disease patients and mild cognitive impairment subjects. Biomarkers 2015, 20, 196–201. [Google Scholar] [CrossRef] [PubMed]
- Esteve, A.S.; García-Ayllón, M.-S.; Gobom, J.; Alom, J.; Zetterberg, H.; Blennow, K.; Sáez-Valero, J. Levels of ADAM10 are reduced in Alzheimer’s disease CSF. J. Neuroinflammation 2018, 15, 213. [Google Scholar] [CrossRef] [PubMed]
- Bram, J.M.D.F.; Talib, L.L.; Joaquim, H.P.G.; Sarno, T.A.; Gattaz, W.F.; Forlenza, O.V. Protein levels of ADAM10, BACE1, and PSEN1 in platelets and leukocytes of Alzheimer’s disease patients. Eur. Arch. Psychiatry Clin. Neurosci. 2018, 269, 963–972. [Google Scholar] [CrossRef]
- Cai, H.; Pang, Y.; Wang, Q.; Qin, W.; Wei, C.; Li, Y.; Li, T.; Li, F.; Wang, Q.; Li, Y.; et al. Proteomic profiling of circulating plasma exosomes reveals novel biomarkers of Alzheimer’s disease. Alzheimer’s Res. Ther. 2022, 14, 181. [Google Scholar] [CrossRef]
- Schuck, F.; Wolf, D.; Fellgiebel, A.; Endres, K. Increase of α-Secretase ADAM10 in Platelets Along Cognitively Healthy Aging. J. Alzheimer’s Dis. 2016, 50, 817–826. [Google Scholar] [CrossRef]
- Vatanabe, I.P.; Peron, R.; Grigoli, M.M.; Pelucchi, S.; De Cesare, G.; Magalhães, T.; Manzine, P.; Balthazar, M.F.; Di Luca, M.; Marcello, E.; et al. ADAM10 Plasma and CSF Levels Are Increased in Mild Alzheimer’s Disease. Int. J. Mol. Sci. 2021, 22, 2416. [Google Scholar] [CrossRef]
- Seifert, A.; Düsterhöft, S.; Wozniak, J.; Koo, C.Z.; Tomlinson, M.G.; Nuti, E.; Rossello, A.; Cuffaro, D.; Yildiz, D.; Ludwig, A. The metalloproteinase ADAM10 requires its activity to sustain surface expression. Cell. Mol. Life Sci. 2020, 78, 715–732. [Google Scholar] [CrossRef]
- Kunkle, B.W.; Grenier-Boley, B.; Sims, R.; Bis, J.C.; Damotte, V.; Naj, A.C.; Boland, A.; Vronskaya, M.; Van Der Lee, S.J.; Amlie-Wolf, A.; et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet. 2019, 51, 414–430. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Suh, J.; Romano, D.; Truong, M.H.; Mullin, K.; Hooli, B.; Norton, D.; Tesco, G.; Elliott, K.; Wagner, S.L.; et al. Potential late-onset Alzheimer’s disease-associated mutations in the ADAM10 gene attenuate α-secretase activity. Hum. Mol. Genet. 2009, 18, 3987–3996. [Google Scholar] [CrossRef] [PubMed]
- Suh, J.; Choi, S.H.; Romano, D.M.; Gannon, M.A.; Lesinski, A.N.; Kim, D.Y.; Tanzi, R.E. ADAM10 Missense Mutations Potentiate β-Amyloid Accumulation by Impairing Prodomain Chaperone Function. Neuron 2013, 80, 385–401. [Google Scholar] [CrossRef]
- Agüero, P.; Sainz, M.J.; García-Ayllón, M.-S.; Sáez-Valero, J.; Téllez, R.; Guerrero-López, R.; Pérez-Pérez, J.; Jiménez-Escrig, A.; Gómez-Tortosa, E. α-Secretase nonsense mutation (ADAM10 Tyr167*) in familial Alzheimer’s disease. Alzheimer’s Res. Ther. 2020, 12, 139. [Google Scholar] [CrossRef] [PubMed]
- Song, J.-H.; Yu, J.-T.; Liu, M.; Yan, C.-Z.; Tan, L. Genetic association between ADAM10 gene polymorphism and Alzheimer’s disease in a Northern Han Chinese population. Brain Res. 2011, 1421, 78–81. [Google Scholar] [CrossRef]
- Zeng, F.; Shen, C.; Liu, Y.-H.; Li, J.; Zhu, J.; Wang, Y.-R.; Yan, J.-C.; Gao, C.-Y.; Zhou, H.-D.; Deng, J.; et al. Genetic Association Between APP, ADAM10 Gene Polymorphism, and Sporadic Alzheimer’s Disease in the Chinese Population. Neurotox. Res. 2015, 27, 284–291. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.-H.; Chen, W.; Jiang, L.-Y.; Yang, Y.-X.; Yao, L.-F.; Li, K.-S. Influence of ADAM10 Polymorphisms on Plasma Level of Soluble Receptor for Advanced Glycation End Products and The Association with Alzheimer’s Disease Risk. Front. Genet. 2018, 9, 540. [Google Scholar] [CrossRef] [PubMed]
- Bekris, L.M.; Lutz, F.; Li, G.; Galasko, D.R.; Farlow, M.R.; Quinn, J.F.; Kaye, J.A.; Leverenz, J.B.; Tsuang, D.W.; Montine, T.J.; et al. ADAM10 expression and promoter haplotype in Alzheimer’s disease. Neurobiol. Aging 2012, 33, 2229.e1–2229.e9. [Google Scholar] [CrossRef]
- Cai, G.; Atzmon, G.; Naj, A.C.; Beecham, G.W.; Barzilai, N.; Haines, J.L.; Sano, M.; Pericak-Vance, M.; Buxbaum, J.D. Evidence against a role for rare ADAM10 mutations in sporadic Alzheimer Disease. Neurobiol. Aging 2012, 33, 416–417. [Google Scholar] [CrossRef]
- Laws, S.M.; Eckart, K.; Friedrich, P.; Kurz, A.; Förstl, H.; Riemenschneider, M. Lack of evidence to support the association of polymorphisms within the alpha- and beta-secretase genes (ADAM10/BACE1) with Alzheimer’s disease. Neurobiol. Aging 2011, 32, 541–543. [Google Scholar] [CrossRef]
- Jansen, I.E.; Savage, J.E.; Watanabe, K.; Bryois, J.; Williams, D.; Steinberg, S.; Sealock, J.; Karlsson, I.; Hägg, S.; Athanasiu, L.; et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat. Genet. 2019, 51, 404–413. [Google Scholar] [CrossRef]
- Holstege, H.; Hulsman, M.; Charbonnier, C.; Grenier-Boley, B.; Quenez, O.; Grozeva, D.; van Rooij, J.; Sims, R.; Ahmad, S.; Amin, N.; et al. Exome sequencing identifies rare damaging variants in ATP8B4 and ABCA1 as novel risk factors for Alzheimer’s Disease. Nat. Genet. 2022, 54, 1786–1794. [Google Scholar] [CrossRef] [PubMed]
- Oliveira Monteiro, M.P.; Salheb Oliveira, D.S.M.; Manzine, P.; Monteiro, M.P.O.; Oliveira, D.S.; Manzine, P.; Nascimento, C.C.; Orlandi, A.D.S.; de Oliveira Gomes, G.; Orlandi, F.D.S.; et al. ADAM10 plasma levels predict worsening in cognition of older adults: A 3-year follow-up study. Alzheimer Res. Ther. 2021, 13, 18. [Google Scholar] [CrossRef] [PubMed]
- Goldman, J.S.; Farmer, J.M.; Wood, E.M.; Johnson, J.K.; Boxer, A.; Neuhaus, J.; Lomen-Hoerth, C.; Wilhelmsen, K.C.; Lee, V.M.-Y.; Grossman, M.; et al. Comparison of family histories in FTLD subtypes and related tauopathies. Neurology 2005, 65, 1817–1819. [Google Scholar] [CrossRef] [PubMed]
- Kircher, M.; Witten, D.M.; Jain, P.; O‘Roak, B.J.; Cooper, G.M.; Shendure, J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014, 46, 310–315. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Anesthesia Analg. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- Jack, C.R., Jr.; Bennett, D.A.; Blennow, K.; Carrillo, M.C.; Dunn, B.; Haeberlein, S.B.; Holtzman, D.M.; Jagust, W.; Jessen, F.; Karlawish, J.; et al. NIA-AA Research Framework: Toward a biological definition of Alzheimer’s disease. Alzheimer Dement. 2018, 14, 535–562. [Google Scholar] [CrossRef]
| ADAM10 Analysis | AD Cases | Controls |
|---|---|---|
| Sequencing | n = 103 | n = 96 |
| Gender M/F | 45/58 | 34/62 |
| Age (yrs) | 68 ± 6 (57–80) | 93.2 ± 3 (90–105) |
| Age at onset (AO) | 66 ± 6 (52–80) | -- |
| MMSE | 22.4 ± 3.6 | 28.7 ± 1.2 |
| APOE | 3/3 (42%); 3/4 (45%); 4/4 (12%) | 2/3 (11%); 3/3 (83%); 3/4 (6%) |
| Goldman # | 1 (9%), 2 (39%), 3 (9%), 3.5 (25%), 4 (18%) | -- |
| CSF ELISA | n = 83 | n = 37 * |
| AD biomarkers | β42/β40 < 0.068, Ttau > 410, p-tau > 59 | β42/β40 >0.068, Ttau <410, p-tau <40 |
| 1st kit | n = 27. AO: 66 ± 5; M/F: 11/16 | n = 14. Age: 66 ± 5; M/F: 5/9 |
| 2nd kit | n = 27. AO: 63 ± 6; M/F: 13/14 | n = 11. Age: 61 ± 10; M/F: 6/5 |
| 3rd kit | n = 29. AO: 68 ± 6; M/F: 15/14 | n = 12. Age: 51 ± 16; M/F: 6/6 |
| CSF Western blot | n = 45 | n = 15 |
| AO: 66 ± 5 (52–75); M/F: 20/25 | Age: 62 ± 12 (32–76); M/F: 8/7 |
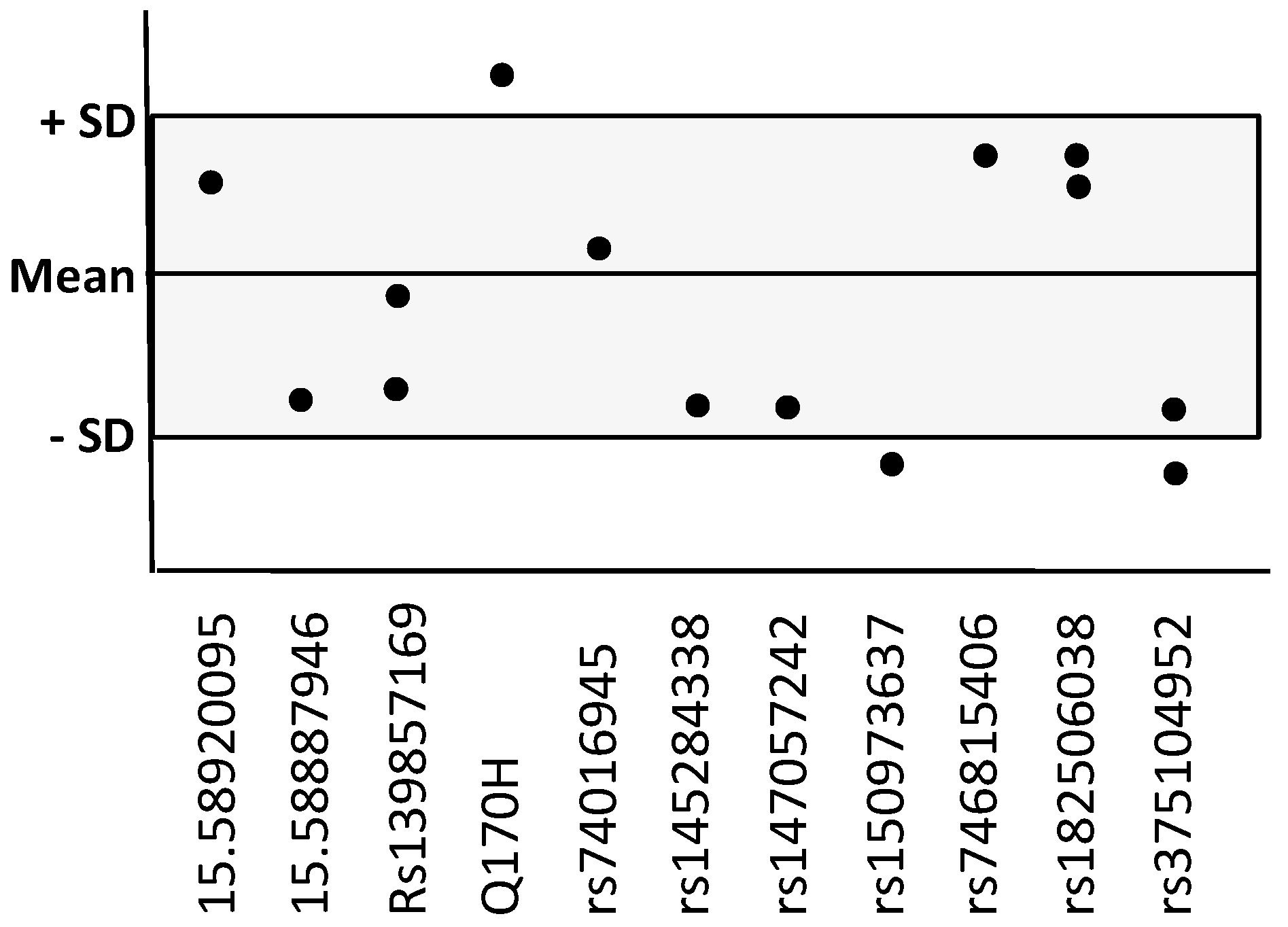
| ADAM10 Variants dbSNP ID | MAF gnomAD (v3.1.1) | Present in n AD Cases/Frequency | Present in n Nonagenarians/Frequency |
|---|---|---|---|
| Non-described (chr pos) | |||
| 15.58920095 (A-G) | -- | n = 1 0.01 | -- |
| 15.58887946 (3′UTR C-T) | -- | n = 1 0.01 | -- |
| Rare variants | |||
| rs139857169 | 0.007 | n = 2 0.02 | n = 2 0.02 |
| rs61751103 (Q170H) | 0.0014 | n = 3 0.03 | n = 1 0.01 |
| rs74016945 (3′UTR) | 0.0089 | n = 1 0.01 | n = 6 0.06 * |
| rs192779774 (G-T) | 0.0042 | -- | n = 2 0.02 |
| rs201948093 | 0.0018 | -- | n = 1 0.01 |
| rs145284338 | 0.0068 | n = 1 0.01 | -- |
| rs147057242(3′UTR) | 0.0052 | n = 1 0.01 | -- |
| rs150973637 (A-C) | 0.00013 | n = 2 0.02 | -- |
| rs746815406 | 0.00003 | n = 1 0.01 | -- |
| rs182506038(3′UTR) | 0.00054 | n = 2 0.02 | -- |
| rs375104952 (T-C) | 0.00010 | n = 2 0.02 | -- |
| ADAM10 (ng/mL) | AD Cases n = 83 | Controls n = 37 | p (Student Test) |
|---|---|---|---|
| 1 kit | Total n = 27 2.1 ± 0.3 Early-onset (n = 9) 2.3 ± 0.2 Late-onset (n = 18) 2.0 ± 0.3 Variant + (n = 4) 2.0 ± 0.3 | 2.2 ± 0.1 (n = 14) | 0.59 Early vs late-onset: 0.012 Variant + vs-: ns |
| 2 kit | Total n = 27 6.4 ± 0.4 Early-onset (n = 15) 6.4 ± 0.4 Late-onset (n =12) 6.3 ± 0.4 Variant + (n = 8) 6.4 ± 0.4 | 5.8 ± 1.1 (n = 11) | 0.16 Early vs late-onset: 0.86 Variant + vs-: ns |
| 3 kit | Total n = 29 7.5 ± 0.6 Early-onset (n = 8) 7.4 ± 0.6 Late-onset (n = 21) 7.5 ± 0.6 Variant + (n = 2) 7.7 ± 0.1 | 6.7 ± 0.9 (n = 12) | 0.006 Early vs late-onset: 0.79 Variant + vs-: ns |
| 50 kDa | 55 kDa | 80 KDa | |
|---|---|---|---|
| AD cases (n = 45) | 105 (58.8, 138) | 34.7 (25.4, 58.7) | 34.4 (23.4, 54.4) |
| Controls (n = 15) | 103 (84.3, 132) | 39.3 (27.5, 59.2) | 24.4 (18.1, 35.1) |
| p = 0.567 | p = 0.986 | p = 0.140 | |
| Early-onset AD (n = 16) | 113 (64.8, 139) | 38.0 (26.2, 58.8) | 27.4 (16.8, 57.7) |
| Late-onset AD (n = 29) | 98.7 (58.8, 137) | 32.2 (25.4, 57.2) | 39.6 (25.0, 54.4) |
| p = 0.749 | p = 0.953 | p = 0.245 | |
| AD variant carriers (n = 13) | 110 (41.1, 138) | 34.7 (24.0, 58.7) | 27.3 (22.5, 43.0) |
| AD non-carriers (n = 32) | 102 (61.1, 135) | 35.9 (25.5, 58.1) | 37.3 (24.1, 65.5) |
| p = 0.940 | p = 0.582 | p = 0.225 | |
| Q170H carrier (n = 1) | 30.8 | 24.0 | 4.6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Agüero-Rabes, P.; Pérez-Pérez, J.; Cremades-Jimeno, L.; García-Ayllón, M.-S.; Gea-González, A.; Sainz, M.J.; Mahillo-Fernández, I.; Téllez, R.; Cárdaba, B.; Sáez-Valero, J.; et al. ADAM10 Gene Variants in AD Patients and Their Relationship to CSF Protein Levels. Int. J. Mol. Sci. 2023, 24, 6113. https://doi.org/10.3390/ijms24076113
Agüero-Rabes P, Pérez-Pérez J, Cremades-Jimeno L, García-Ayllón M-S, Gea-González A, Sainz MJ, Mahillo-Fernández I, Téllez R, Cárdaba B, Sáez-Valero J, et al. ADAM10 Gene Variants in AD Patients and Their Relationship to CSF Protein Levels. International Journal of Molecular Sciences. 2023; 24(7):6113. https://doi.org/10.3390/ijms24076113
Chicago/Turabian StyleAgüero-Rabes, Pablo, Julián Pérez-Pérez, Lucía Cremades-Jimeno, María-Salud García-Ayllón, Adriana Gea-González, María José Sainz, Ignacio Mahillo-Fernández, Raquel Téllez, Blanca Cárdaba, Javier Sáez-Valero, and et al. 2023. "ADAM10 Gene Variants in AD Patients and Their Relationship to CSF Protein Levels" International Journal of Molecular Sciences 24, no. 7: 6113. https://doi.org/10.3390/ijms24076113
APA StyleAgüero-Rabes, P., Pérez-Pérez, J., Cremades-Jimeno, L., García-Ayllón, M.-S., Gea-González, A., Sainz, M. J., Mahillo-Fernández, I., Téllez, R., Cárdaba, B., Sáez-Valero, J., & Gómez-Tortosa, E. (2023). ADAM10 Gene Variants in AD Patients and Their Relationship to CSF Protein Levels. International Journal of Molecular Sciences, 24(7), 6113. https://doi.org/10.3390/ijms24076113






