Emerging Personalized Opportunities for Enhancing Translational Readthrough in Rare Genetic Diseases and Beyond
Abstract
1. Introduction
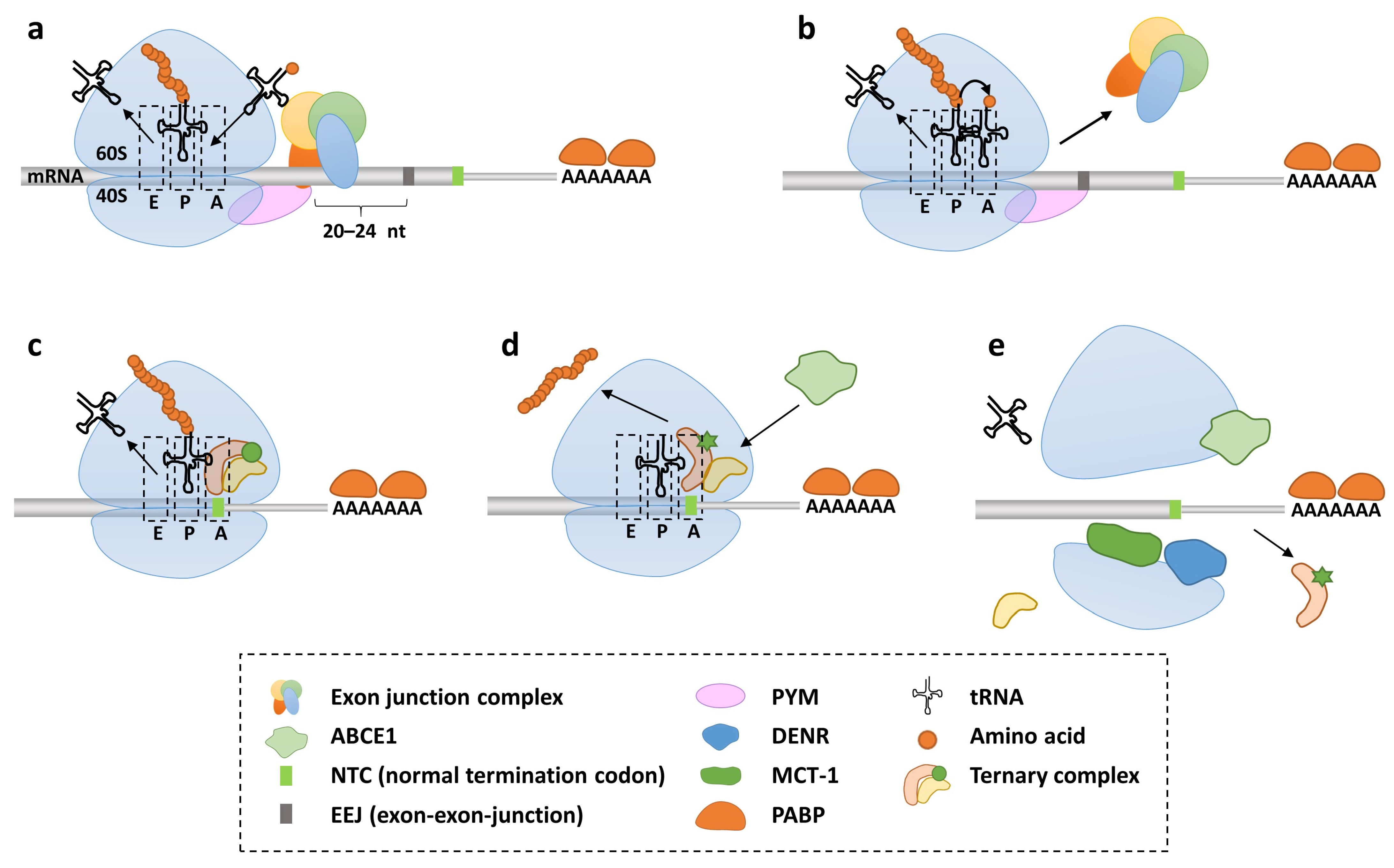
2. Molecular Mechanisms of Translation Termination and Nonsense Suppression
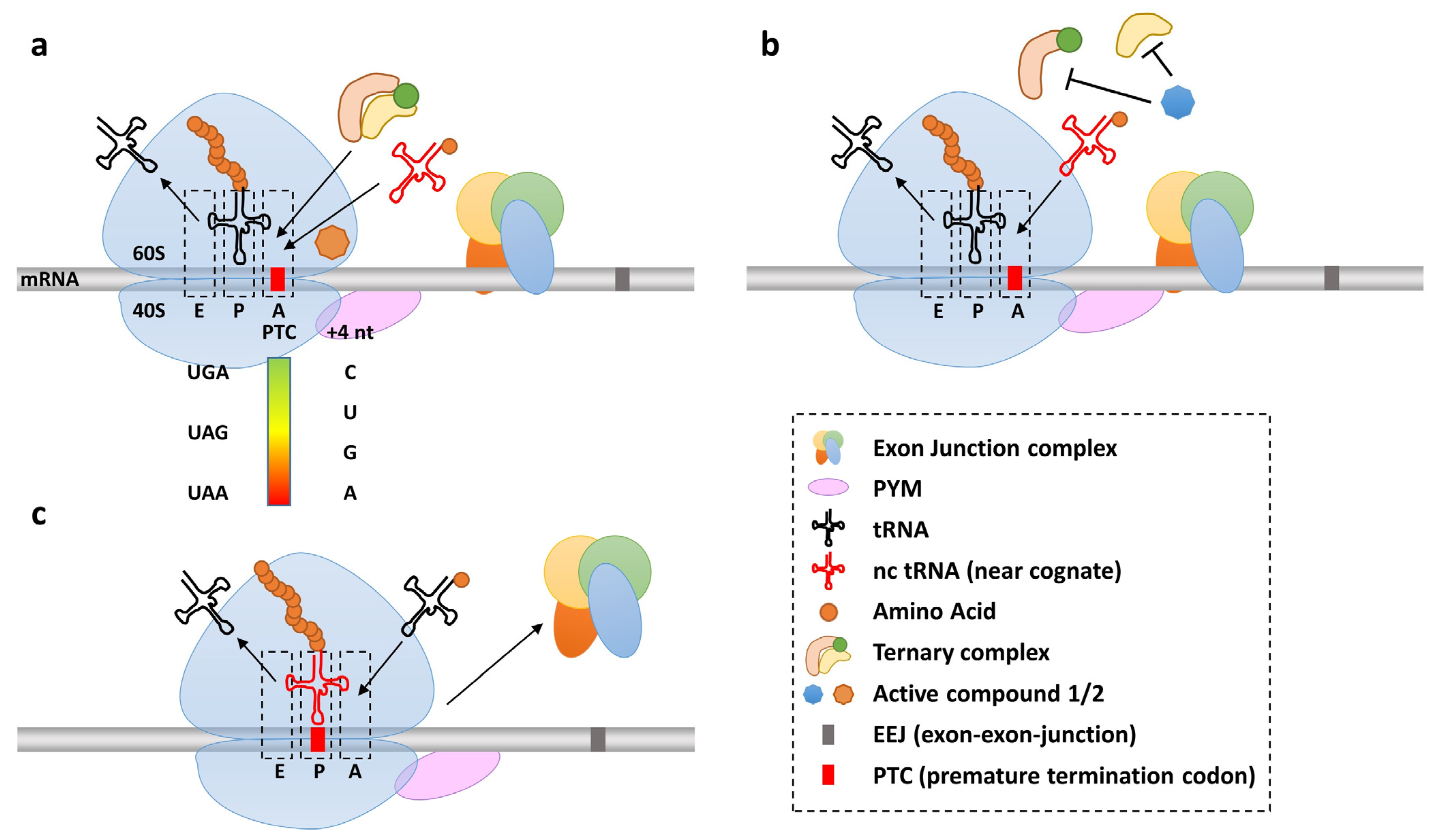
2.1. Natural and Induced Stop Codon Readthrough
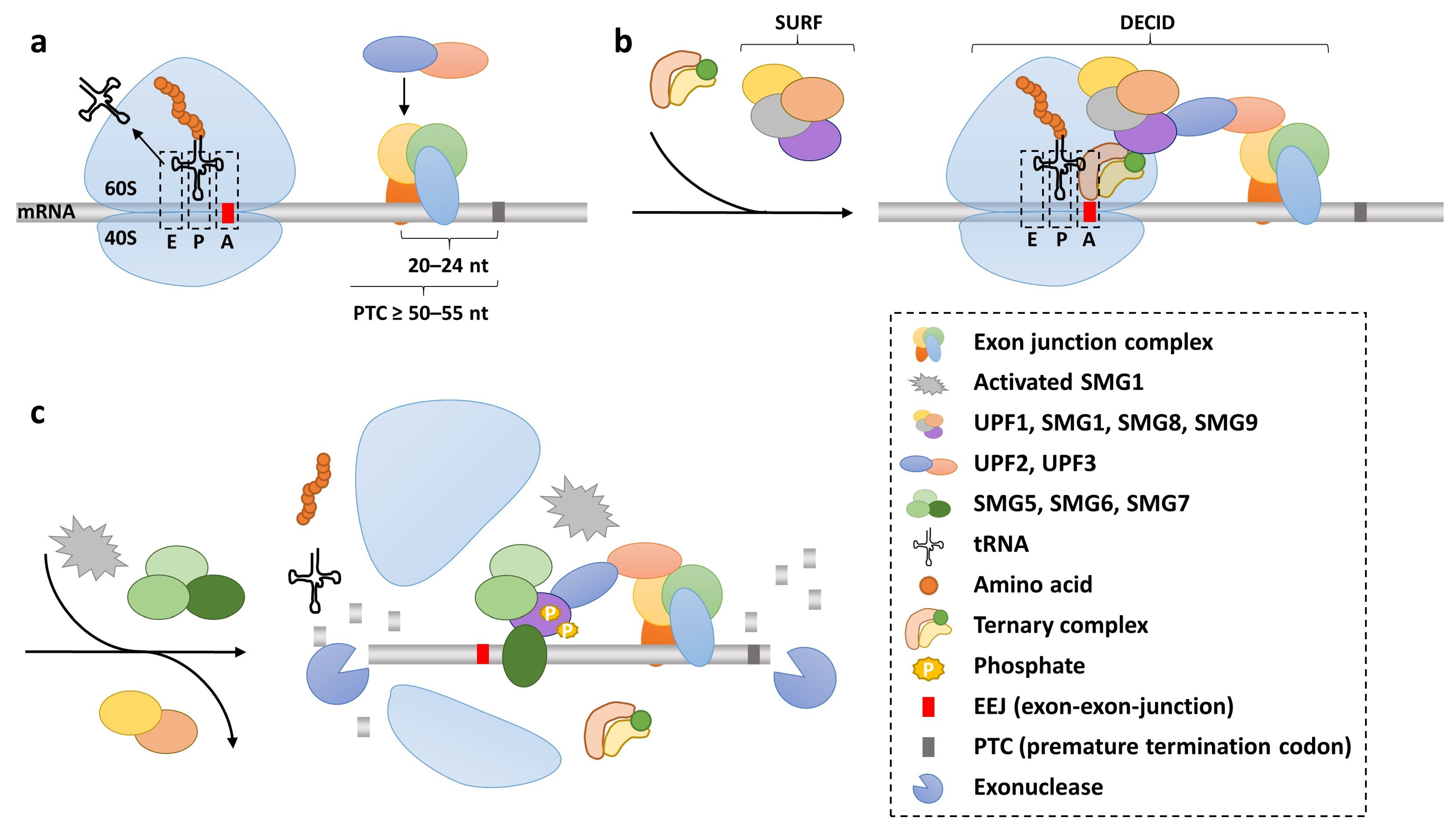
2.2. Translation Termination and Nonsense-Mediated Decay
2.3. Reporter Assays and Detection of Readthrough at Genomic Scale
3. Induction of Nonsense Suppression
3.1. Aminoglycoside Antibiotics
3.2. Non-Aminoglycosides
3.3. Novel Classes of Nonsense Suppressors
3.4. Interference with Release Factors and Translation Termination
3.5. Ribosome Editing to Boost Production Levels of Full Length Protein from PTC mRNAs
4. Nonsense Suppression in the Context of Rare Genetic Diseases and Cancer
4.1. Cystic Fibrosis
4.2. Retinitis Pigmentosa
4.3. Shwachman–Diamond Syndrome
4.4. Alport Syndrome
4.5. Breast Cancer
4.6. Epidermolysis Bullosa
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AG | aminoglycoside |
| CBC | cap binding complex |
| CF | cystic fibrosis |
| CFTR | cystic fibrosis transmembrane conductance regulator |
| DECID | decay-inducing complex |
| DMD | Duchenne muscular dystrophy |
| EB | epidermolysis bullosa |
| EJC | exon junction complex |
| eRF1 | eukaryotic release factor 1 |
| JEB | junctional epidermolysis bullosa |
| NMD | nonsense mediated mRNA decay |
| NTC | normal termination codon |
| PTC | premature termination codon |
| RFC | release factor complex |
| RPFs | ribosome-protected fragments |
| SCR | stop codon readthrough |
| TRIDs | translational-readthrough-inducing drugs |
References
- Mort, M.; Ivanov, D.; Cooper, D.N.; Chuzhanova, N.A. A meta-analysis of nonsense mutations causing human genetic disease. Hum. Mutat. 2008, 29, 1037–1047. [Google Scholar] [CrossRef] [PubMed]
- Keeling, K.M.; Xue, X.; Gunn, G.; Bedwell, D.M. Therapeutics based on stop codon readthrough. Annu. Rev. Genom. Hum. Genet. 2014, 15, 371–394. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.L.; Dougherty, J.P. Pharmaceutical therapies to recode nonsense mutations in inherited diseases. Pharmacol. Ther. 2012, 136, 227–266. [Google Scholar] [CrossRef]
- Morais, P.; Adachi, H.; Yu, Y.T. Suppression of Nonsense Mutations by New Emerging Technologies. Int. J. Mol. Sci. 2020, 21, 4394. [Google Scholar] [CrossRef]
- Martins-Dias, P.; Romão, L. Nonsense suppression therapies in human genetic diseases. Cell. Mol. Life Sci. 2021, 78, 4677–4701. [Google Scholar] [CrossRef]
- Manuvakhova, M.; Keeling, K.; Bedwell, D.M. Aminoglycoside antibiotics mediate context-dependent suppression of termination codons in a mammalian translation system. RNA 2000, 6, 1044–1055. [Google Scholar] [CrossRef]
- Harrell, L.; Melcher, U.; Atkins, J.F. Predominance of six different hexanucleotide recoding signals 3’ of read-through stop codons. Nucleic Acids Res. 2002, 30, 2011–2017. [Google Scholar] [CrossRef]
- Floquet, C.; Hatin, I.; Rousset, J.P.; Bidou, L. Statistical analysis of readthrough levels for nonsense mutations in mammalian cells reveals a major determinant of response to gentamicin. PLoS Genet. 2012, 8, e1002608. [Google Scholar] [CrossRef]
- Dabrowski, M.; Bukowy-Bieryllo, Z.; Zietkiewicz, E. Translational readthrough potential of natural termination codons in eucaryotes--The impact of RNA sequence. RNA Biol. 2015, 12, 950–958. [Google Scholar] [CrossRef]
- Lombardi, S.; Testa, M.F.; Pinotti, M.; Branchini, A. Molecular Insights into Determinants of Translational Readthrough and Implications for Nonsense Suppression Approaches. Int. J. Mol. Sci. 2020, 21, 9449. [Google Scholar] [CrossRef] [PubMed]
- Wangen, J.R.; Green, R. Stop codon context influences genome-wide stimulation of termination codon readthrough by aminoglycosides. eLife 2020, 9, e52611. [Google Scholar] [CrossRef] [PubMed]
- Schilff, M.; Sargsyan, Y.; Hofhuis, J.; Thoms, S. Stop Codon Context-Specific Induction of Translational Readthrough. Biomolecules 2021, 11, 1006. [Google Scholar] [CrossRef]
- Biziaev, N.; Sokolova, E.; Yanvarev, D.V.; Toropygin, I.Y.; Shuvalov, A.; Egorova, T.; Alkalaeva, E. Recognition of 3’ nucleotide context and stop codon readthrough are determined during mRNA translation elongation. J. Biol. Chem. 2022, 298, 102133. [Google Scholar] [CrossRef]
- Pranke, I.M.; Varilh, J.; Hatton, A.; Faucon, C.; Girodon, E.; Dreano, E.; Chevalier, B.; Karri, S.; Reix, P.; Durieu, I.; et al. The U UGA C sequence provides a favorable context to ELX-02 induced CFTR readthrough. J. Cyst. Fibros. 2022. [Google Scholar] [CrossRef] [PubMed]
- Nudelman, I.; Rebibo-Sabbah, A.; Cherniavsky, M.; Belakhov, V.; Hainrichson, M.; Chen, F.; Schacht, J.; Pilch, D.S.; Ben-Yosef, T.; Baasov, T. Development of novel aminoglycoside (NB54) with reduced toxicity and enhanced suppression of disease-causing premature stop mutations. J. Med. Chem. 2009, 52, 2836–2845. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.; Shao, S.; Murray, J.; Hegde, R.S.; Ramakrishnan, V. Structural basis for stop codon recognition in eukaryotes. Nature 2015, 524, 493–496. [Google Scholar] [CrossRef]
- Ou, X.; Cao, J.; Cheng, A.; Peppelenbosch, M.P.; Pan, Q. Errors in translational decoding: tRNA wobbling or misincorporation? PLoS Genet. 2019, 15, e1008017. [Google Scholar] [CrossRef]
- Firth, A.E.; Wills, N.M.; Gesteland, R.F.; Atkins, J.F. Stimulation of stop codon readthrough: Frequent presence of an extended 3’ RNA structural element. Nucleic Acids Res. 2011, 39, 6679–6691. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Hayashi, A.; Campagnoni, C.W.; Kimura, A.; Inuzuka, T.; Baba, H. L-MPZ, a novel isoform of myelin P0, is produced by stop codon readthrough. J. Biol. Chem. 2012, 287, 17765–17776. [Google Scholar] [CrossRef]
- Dunn, J.G.; Foo, C.K.; Belletier, N.G.; Gavis, E.R.; Weissman, J.S. Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. eLife 2013, 2, e01179. [Google Scholar] [CrossRef]
- Loughran, G.; Chou, M.Y.; Ivanov, I.P.; Jungreis, I.; Kellis, M.; Kiran, A.M.; Baranov, P.V.; Atkins, J.F. Evidence of efficient stop codon readthrough in four mammalian genes. Nucleic Acids Res. 2014, 42, 8928–8938. [Google Scholar] [CrossRef] [PubMed]
- Mangkalaphiban, K.; He, F.; Ganesan, R.; Wu, C.; Baker, R.; Jacobson, A. Transcriptome-wide investigation of stop codon readthrough in Saccharomyces cerevisiae. PLoS Genet. 2021, 17, e1009538. [Google Scholar] [CrossRef] [PubMed]
- Chkuaseli, T.; White, K.A. Complex and simple translational readthrough signals in pea enation mosaic virus 1 and potato leafroll virus, respectively. PLoS Pathog. 2022, 18, e1010888. [Google Scholar] [CrossRef] [PubMed]
- Karijolich, J.; Yu, Y.T. Converting nonsense codons into sense codons by targeted pseudouridylation. Nature 2011, 474, 395–398. [Google Scholar] [CrossRef]
- Nir, R.; Hoernes, T.P.; Muramatsu, H.; Faserl, K.; Karikó, K.; Erlacher, M.D.; Sas-Chen, A.; Schwartz, S. A systematic dissection of determinants and consequences of snoRNA-guided pseudouridylation of human mRNA. Nucleic Acids Res. 2022, 50, 4900–4916. [Google Scholar] [CrossRef]
- Kleppe, A.S.; Bornberg-Bauer, E. Robustness by intrinsically disordered C-termini and translational readthrough. Nucleic Acids Res. 2018, 46, 10184–10194. [Google Scholar] [CrossRef]
- Eswarappa, S.M.; Potdar, A.A.; Koch, W.J.; Fan, Y.; Vasu, K.; Lindner, D.; Willard, B.; Graham, L.M.; DiCorleto, P.E.; Fox, P.L. Programmed translational readthrough generates antiangiogenic VEGF-Ax. Cell 2014, 157, 1605–1618. [Google Scholar] [CrossRef] [PubMed]
- Loughran, G.; Jungreis, I.; Tzani, I.; Power, M.; Dmitriev, R.I.; Ivanov, I.P.; Kellis, M.; Atkins, J.F. Stop codon readthrough generates a C-terminally extended variant of the human vitamin D receptor with reduced calcitriol response. J. Biol. Chem. 2018, 293, 4434–4444. [Google Scholar] [CrossRef]
- Sahoo, S.; Singh, D.; Singh, A.; Pandit, M.; Vasu, K.; Som, S.; Pullagurla, N.J.; Laha, D.; Eswarappa, S.M. Identification and functional characterization of mRNAs that exhibit stop codon readthrough in Arabidopsis thaliana. J. Biol. Chem. 2022, 298, 102173. [Google Scholar] [CrossRef]
- Schueren, F.; Lingner, T.; George, R.; Hofhuis, J.; Dickel, C.; Gartner, J.; Thoms, S. Peroxisomal lactate dehydrogenase is generated by translational readthrough in mammals. eLife 2014, 3, e03640. [Google Scholar] [CrossRef]
- Karki, P.; Carney, T.D.; Maracci, C.; Yatsenko, A.S.; Shcherbata, H.R.; Rodnina, M.V. Tissue-specific regulation of translational readthrough tunes functions of the traffic jam transcription factor. Nucleic Acids Res. 2022, 50, 6001–6019. [Google Scholar] [CrossRef] [PubMed]
- De Bellis, M.; Pisani, F.; Mola, M.G.; Rosito, S.; Simone, L.; Buccoliero, C.; Trojano, M.; Nicchia, G.P.; Svelto, M.; Frigeri, A. Translational readthrough generates new astrocyte AQP4 isoforms that modulate supramolecular clustering, glial endfeet localization, and water transport. Glia 2017, 65, 790–803. [Google Scholar] [CrossRef] [PubMed]
- Sapkota, D.; Florian, C.; Doherty, B.M.; White, K.M.; Reardon, K.M.; Ge, X.; Garbow, J.R.; Yuede, C.M.; Cirrito, J.R.; Dougherty, J.D. Aqp4 stop codon readthrough facilitates amyloid-β clearance from the brain. Brain 2022, 145, 2982–2990. [Google Scholar] [CrossRef]
- Roy, B.; Friesen, W.J.; Tomizawa, Y.; Leszyk, J.D.; Zhuo, J.; Johnson, B.; Dakka, J.; Trotta, C.R.; Xue, X.; Mutyam, V.; et al. Ataluren stimulates ribosomal selection of near-cognate tRNAs to promote nonsense suppression. Proc. Natl. Acad. Sci. USA 2016, 113, 12508–12513. [Google Scholar] [CrossRef] [PubMed]
- Palmer, E.; Wilhelm, J.M.; Sherman, F. Phenotypic suppression of nonsense mutants in yeast by aminoglycoside antibiotics. Nature 1979, 277, 148–150. [Google Scholar] [CrossRef]
- Howard, M.; Frizzell, R.A.; Bedwell, D.M. Aminoglycoside antibiotics restore CFTR function by overcoming premature stop mutations. Nat. Med. 1996, 2, 467–469. [Google Scholar] [CrossRef]
- Barton-Davis, E.R.; Cordier, L.; Shoturma, D.I.; Leland, S.E.; Sweeney, H.L. Aminoglycoside antibiotics restore dystrophin function to skeletal muscles of mdx mice. J. Clin. Investig. 1999, 104, 375–381. [Google Scholar] [CrossRef] [PubMed]
- Kellermayer, R.; Szigeti, R.; Keeling, K.M.; Bedekovics, T.; Bedwell, D.M. Aminoglycosides as potential pharmacogenetic agents in the treatment of Hailey-Hailey disease. J. Investig. Dermatol. 2006, 126, 229–231. [Google Scholar] [CrossRef]
- Prokhorova, I.; Altman, R.B.; Djumagulov, M.; Shrestha, J.P.; Urzhumtsev, A.; Ferguson, A.; Chang, C.T.; Yusupov, M.; Blanchard, S.C.; Yusupova, G. Aminoglycoside interactions and impacts on the eukaryotic ribosome. Proc. Natl. Acad. Sci. USA 2017, 114, E10899–E10908. [Google Scholar] [CrossRef]
- Nagel-Wolfrum, K.; Moller, F.; Penner, I.; Baasov, T.; Wolfrum, U. Targeting Nonsense Mutations in Diseases with Translational Read-Through-Inducing Drugs (TRIDs). BioDrugs 2016, 30, 49–74. [Google Scholar] [CrossRef]
- Kramarski, L.; Arbely, E. Translational read-through promotes aggregation and shapes stop codon identity. Nucleic Acids Res. 2020, 48, 3747–3760. [Google Scholar] [CrossRef]
- Wohlgemuth, I.; Garofalo, R.; Samatova, E.; Gunenc, A.N.; Lenz, C.; Urlaub, H.; Rodnina, M.V. Translation error clusters induced by aminoglycoside antibiotics. Nat. Commun. 2021, 12, 1830. [Google Scholar] [CrossRef]
- Fu, X.; Wan, P.; Li, P.; Wang, J.; Guo, S.; Zhang, Y.; An, Y.; Ye, C.; Liu, Z.; Gao, J.; et al. Mechanism and Prevention of Ototoxicity Induced by Aminoglycosides. Front. Cell. Neurosci. 2021, 15, 692762. [Google Scholar] [CrossRef]
- Baradaran-Heravi, A.; Balgi, A.D.; Zimmerman, C.; Choi, K.; Shidmoossavee, F.S.; Tan, J.S.; Bergeaud, C.; Krause, A.; Flibotte, S.; Shimizu, Y.; et al. Novel small molecules potentiate premature termination codon readthrough by aminoglycosides. Nucleic Acids Res. 2016, 44, 6583–6598. [Google Scholar] [CrossRef] [PubMed]
- Rabea, S.M.; Baradaran-Heravi, A.; Balgi, A.D.; Krause, A.; Hosseini Farahabadi, S.; Roberge, M.; Grierson, D.S. 2-Aminothiazole-4-carboxamides Enhance Readthrough of Premature Termination Codons by Aminoglycosides. ACS Med. Chem. Lett. 2019, 10, 726–731. [Google Scholar] [CrossRef]
- Popadynec, M.; Baradaran-Heravi, A.; Alford, B.; Cameron, S.A.; Clinch, K.; Mason, J.M.; Rendle, P.M.; Zubkova, O.V.; Gan, Z.; Liu, H.; et al. Reducing the Toxicity of Designer Aminoglycosides as Nonsense Mutation Readthrough Agents for Therapeutic Targets. ACS Med. Chem. Lett. 2021, 12, 1486–1492. [Google Scholar] [CrossRef] [PubMed]
- Wolstencroft, E.C.; Mattis, V.; Bajer, A.A.; Young, P.J.; Lorson, C.L. A non-sequence-specific requirement for SMN protein activity: The role of aminoglycosides in inducing elevated SMN protein levels. Hum. Mol. Genet. 2005, 14, 1199–1210. [Google Scholar] [CrossRef] [PubMed]
- Mattis, V.B.; Rai, R.; Wang, J.; Chang, C.W.; Coady, T.; Lorson, C.L. Novel aminoglycosides increase SMN levels in spinal muscular atrophy fibroblasts. Hum. Genet. 2006, 120, 589–601. [Google Scholar] [CrossRef]
- Mattis, V.B.; Ebert, A.D.; Fosso, M.Y.; Chang, C.W.; Lorson, C.L. Delivery of a read-through inducing compound, TC007, lessens the severity of a spinal muscular atrophy animal model. Hum. Mol. Genet. 2009, 18, 3906–3913. [Google Scholar] [CrossRef]
- Heier, C.R.; DiDonato, C.J. Translational readthrough by the aminoglycoside geneticin (G418) modulates SMN stability in vitro and improves motor function in SMA mice in vivo. Hum. Mol. Genet. 2009, 18, 1310–1322. [Google Scholar] [CrossRef]
- Crawford, D.K.; Alroy, I.; Sharpe, N.; Goddeeris, M.M.; Williams, G. ELX-02 Generates Protein via Premature Stop Codon Read-Through without Inducing Native Stop Codon Read-Through Proteins. J. Pharmacol. Exp. Ther. 2020, 374, 264–272. [Google Scholar] [CrossRef]
- Le Hir, H.; Gatfield, D.; Izaurralde, E.; Moore, M.J. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 2001, 20, 4987–4997. [Google Scholar] [CrossRef]
- Gehring, N.H.; Lamprinaki, S.; Kulozik, A.E.; Hentze, M.W. Disassembly of exon junction complexes by PYM. Cell 2009, 137, 536–548. [Google Scholar] [CrossRef] [PubMed]
- Isken, O.; Maquat, L.E. The multiple lives of NMD factors: Balancing roles in gene and genome regulation. Nat. Rev. Genet. 2008, 9, 699–712. [Google Scholar] [CrossRef] [PubMed]
- Kurosaki, T.; Maquat, L.E. Nonsense-mediated mRNA decay in humans at a glance. J. Cell Sci. 2016, 129, 461–467. [Google Scholar] [CrossRef] [PubMed]
- Tan, K.; Stupack, D.G.; Wilkinson, M.F. Nonsense-mediated RNA decay: An emerging modulator of malignancy. Nat. Rev. Cancer 2022, 22, 437–451. [Google Scholar] [CrossRef]
- Khajavi, M.; Inoue, K.; Lupski, J.R. Nonsense-mediated mRNA decay modulates clinical outcome of genetic disease. Eur. J. Hum. Genet. 2006, 14, 1074–1081. [Google Scholar] [CrossRef]
- Ghosh, A.; Lima, C.D. Enzymology of RNA cap synthesis. Wiley Interdiscip. Rev. RNA 2010, 1, 152–172. [Google Scholar] [CrossRef]
- Hocine, S.; Singer, R.H.; Grunwald, D. RNA processing and export. Cold Spring Harb. Perspect. Biol. 2010, 2, a000752. [Google Scholar] [CrossRef]
- Wahle, E.; Kuhn, U. The mechanism of 3’ cleavage and polyadenylation of eukaryotic pre-mRNA. Prog. Nucleic Acid. Res. Mol. Biol. 1997, 57, 41–71. [Google Scholar]
- Le Hir, H.; Izaurralde, E.; Maquat, L.E.; Moore, M.J. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 2000, 19, 6860–6869. [Google Scholar] [CrossRef] [PubMed]
- Kataoka, N.; Diem, M.D.; Kim, V.N.; Yong, J.; Dreyfuss, G. Magoh, a human homolog of Drosophila mago nashi protein, is a component of the splicing-dependent exon-exon junction complex. EMBO J. 2001, 20, 6424–6433. [Google Scholar] [CrossRef]
- Chan, C.C.; Dostie, J.; Diem, M.D.; Feng, W.; Mann, M.; Rappsilber, J.; Dreyfuss, G. eIF4A3 is a novel component of the exon junction complex. RNA 2004, 10, 200–209. [Google Scholar] [CrossRef] [PubMed]
- Ishigaki, Y.; Li, X.; Serin, G.; Maquat, L.E. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell 2001, 106, 607–617. [Google Scholar] [CrossRef] [PubMed]
- Maquat, L.E.; Hwang, J.; Sato, H.; Tang, Y. CBP80-promoted mRNP rearrangements during the pioneer round of translation, nonsense-mediated mRNA decay, and thereafter. Cold Spring Harb. Symp. Quant. Biol. 2010, 75, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Choe, J.; Oh, N.; Park, S.; Lee, Y.K.; Song, O.K.; Locker, N.; Chi, S.G.; Kim, Y.K. Translation initiation on mRNAs bound by nuclear cap-binding protein complex CBP80/20 requires interaction between CBP80/20-dependent translation initiation factor and eukaryotic translation initiation factor 3g. J. Biol. Chem. 2012, 287, 18500–18509. [Google Scholar] [CrossRef]
- Diem, M.D.; Chan, C.C.; Younis, I.; Dreyfuss, G. PYM binds the cytoplasmic exon-junction complex and ribosomes to enhance translation of spliced mRNAs. Nat. Struct. Mol. Biol. 2007, 14, 1173–1179. [Google Scholar] [CrossRef]
- Bonetti, B.; Fu, L.; Moon, J.; Bedwell, D.M. The efficiency of translation termination is determined by a synergistic interplay between upstream and downstream sequences in Saccharomyces cerevisiae. J. Mol. Biol. 1995, 251, 334–345. [Google Scholar] [CrossRef]
- Zhouravleva, G.; Frolova, L.; Le Goff, X.; Le Guellec, R.; Inge-Vechtomov, S.; Kisselev, L.; Philippe, M. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 1995, 14, 4065–4072. [Google Scholar] [CrossRef]
- Inge-Vechtomov, S.; Zhouravleva, G.; Philippe, M. Eukaryotic release factors (eRFs) history. Biol. Cell 2003, 95, 195–209. [Google Scholar] [CrossRef]
- Salas-Marco, J.; Bedwell, D.M. GTP hydrolysis by eRF3 facilitates stop codon decoding during eukaryotic translation termination. Mol. Cell. Biol. 2004, 24, 7769–7778. [Google Scholar] [CrossRef]
- Bertram, G.; Bell, H.A.; Ritchie, D.W.; Fullerton, G.; Stansfield, I. Terminating eukaryote translation: Domain 1 of release factor eRF1 functions in stop codon recognition. RNA 2000, 6, 1236–1247. [Google Scholar] [CrossRef] [PubMed]
- Frolova, L.Y.; Tsivkovskii, R.Y.; Sivolobova, G.F.; Oparina, N.Y.; Serpinsky, O.I.; Blinov, V.M.; Tatkov, S.I.; Kisselev, L.L. Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA 1999, 5, 1014–1020. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Mugnier, P.; Das, A.K.; Webb, H.M.; Evans, D.R.; Tuite, M.F.; Hemmings, B.A.; Barford, D. The crystal structure of human eukaryotic release factor eRF1--mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell 2000, 100, 311–321. [Google Scholar] [CrossRef] [PubMed]
- Alkalaeva, E.Z.; Pisarev, A.V.; Frolova, L.Y.; Kisselev, L.L.; Pestova, T.V. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell 2006, 125, 1125–1136. [Google Scholar] [CrossRef]
- Ivanov, A.; Mikhailova, T.; Eliseev, B.; Yeramala, L.; Sokolova, E.; Susorov, D.; Shuvalov, A.; Schaffitzel, C.; Alkalaeva, E. PABP enhances release factor recruitment and stop codon recognition during translation termination. Nucleic Acids Res. 2016, 44, 7766–7776. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, P.; Peltz, S.W.; Ross, J. The poly(A)-poly(A)-binding protein complex is a major determinant of mRNA stability in vitro. Mol. Cell. Biol. 1989, 9, 659–670. [Google Scholar]
- McCaughan, K.K.; Brown, C.M.; Dalphin, M.E.; Berry, M.J.; Tate, W.P. Translational termination efficiency in mammals is influenced by the base following the stop codon. Proc. Natl. Acad. Sci. USA 1995, 92, 5431–5435. [Google Scholar] [CrossRef] [PubMed]
- Pisarev, A.V.; Skabkin, M.A.; Pisareva, V.P.; Skabkina, O.V.; Rakotondrafara, A.M.; Hentze, M.W.; Hellen, C.U.; Pestova, T.V. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol. Cell 2010, 37, 196–210. [Google Scholar] [CrossRef]
- Pisarev, A.V.; Hellen, C.U.; Pestova, T.V. Recycling of eukaryotic posttermination ribosomal complexes. Cell 2007, 131, 286–299. [Google Scholar] [CrossRef]
- Skabkin, M.A.; Skabkina, O.V.; Dhote, V.; Komar, A.A.; Hellen, C.U.; Pestova, T.V. Activities of Ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes. Dev. 2010, 24, 1787–1801. [Google Scholar] [CrossRef] [PubMed]
- Lejeune, F.; Ishigaki, Y.; Li, X.; Maquat, L.E. The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: Dynamics of mRNP remodeling. EMBO J. 2002, 21, 3536–3545. [Google Scholar] [CrossRef] [PubMed]
- Amrani, N.; Dong, S.; He, F.; Ganesan, R.; Ghosh, S.; Kervestin, S.; Li, C.; Mangus, D.A.; Spatrick, P.; Jacobson, A. Aberrant termination triggers nonsense-mediated mRNA decay. Biochem. Soc. Trans. 2006, 34 Pt 1, 39–42. [Google Scholar] [CrossRef]
- Leeds, P.; Peltz, S.W.; Jacobson, A.; Culbertson, M.R. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes. Dev. 1991, 5, 2303–2314. [Google Scholar] [CrossRef] [PubMed]
- Lykke-Andersen, J.; Shu, M.D.; Steitz, J.A. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell 2000, 103, 1121–1131. [Google Scholar] [CrossRef]
- Zund, D.; Muhlemann, O. Recent transcriptome-wide mapping of UPF1 binding sites reveals evidence for its recruitment to mRNA before translation. Translation 2013, 1, e26977. [Google Scholar] [CrossRef]
- Linde, L.; Boelz, S.; Neu-Yilik, G.; Kulozik, A.E.; Kerem, B. The efficiency of nonsense-mediated mRNA decay is an inherent character and varies among different cells. Eur. J. Hum. Genet. 2007, 15, 1156–1162. [Google Scholar] [CrossRef]
- Lejeune, F.; Maquat, L.E. Mechanistic links between nonsense-mediated mRNA decay and pre-mRNA splicing in mammalian cells. Curr. Opin. Cell. Biol. 2005, 17, 309–315. [Google Scholar] [CrossRef] [PubMed]
- Kashima, I.; Yamashita, A.; Izumi, N.; Kataoka, N.; Morishita, R.; Hoshino, S.; Ohno, M.; Dreyfuss, G.; Ohno, S. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes. Dev. 2006, 20, 355–367. [Google Scholar] [CrossRef]
- Yamashita, A.; Izumi, N.; Kashima, I.; Ohnishi, T.; Saari, B.; Katsuhata, Y.; Muramatsu, R.; Morita, T.; Iwamatsu, A.; Hachiya, T.; et al. SMG-8 and SMG-9, two novel subunits of the SMG-1 complex, regulate remodeling of the mRNA surveillance complex during nonsense-mediated mRNA decay. Genes. Dev. 2009, 23, 1091–1105. [Google Scholar] [CrossRef]
- Arias-Palomo, E.; Yamashita, A.; Fernandez, I.S.; Nunez-Ramirez, R.; Bamba, Y.; Izumi, N.; Ohno, S.; Llorca, O. The nonsense-mediated mRNA decay SMG-1 kinase is regulated by large-scale conformational changes controlled by SMG-8. Genes. Dev. 2011, 25, 153–164. [Google Scholar] [CrossRef]
- Okada-Katsuhata, Y.; Yamashita, A.; Kutsuzawa, K.; Izumi, N.; Hirahara, F.; Ohno, S. N- and C-terminal Upf1 phosphorylations create binding platforms for SMG-6 and SMG-5:SMG-7 during NMD. Nucleic Acids Res. 2012, 40, 1251–1266. [Google Scholar] [CrossRef]
- Lopez-Perrote, A.; Castano, R.; Melero, R.; Zamarro, T.; Kurosawa, H.; Ohnishi, T.; Uchiyama, A.; Aoyagi, K.; Buchwald, G.; Kataoka, N.; et al. Human nonsense-mediated mRNA decay factor UPF2 interacts directly with eRF3 and the SURF complex. Nucleic Acids Res. 2016, 44, 1909–1923. [Google Scholar] [CrossRef]
- Xue, X.; Mutyam, V.; Thakerar, A.; Mobley, J.; Bridges, R.J.; Rowe, S.M.; Keeling, K.M.; Bedwell, D.M. Identification of the amino acids inserted during suppression of CFTR nonsense mutations and determination of their functional consequences. Hum. Mol. Genet. 2017, 26, 3116–3129. [Google Scholar] [CrossRef]
- Ingolia, N.T.; Ghaemmaghami, S.; Newman, J.R.; Weissman, J.S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 2009, 324, 218–223. [Google Scholar] [CrossRef]
- McGlincy, N.J.; Ingolia, N.T. Transcriptome-wide measurement of translation by ribosome profiling. Methods 2017, 126, 112–129. [Google Scholar] [CrossRef] [PubMed]
- Kiniry, S.J.; Michel, A.M.; Baranov, P.V. Computational methods for ribosome profiling data analysis. Wiley Interdiscip. Rev. RNA 2019, 11, e1577. [Google Scholar] [CrossRef]
- Mudge, J.M.; Ruiz-Orera, J.; Prensner, J.R.; Brunet, M.A.; Calvet, F.; Jungreis, I.; Gonzalez, J.M.; Magrane, M.; Martinez, T.F.; Schulz, J.F.; et al. Standardized annotation of translated open reading frames. Nat. Biotechnol. 2022, 40, 994–999. [Google Scholar] [CrossRef]
- Pellegrino, S.; Terrosu, S.; Yusupova, G.; Yusupov, M. Inhibition of the Eukaryotic 80S Ribosome as a Potential Anticancer Therapy: A Structural Perspective. Cancers 2021, 13, 4392. [Google Scholar] [CrossRef] [PubMed]
- Venturini, A.; Borrelli, A.; Musante, I.; Scudieri, P.; Capurro, V.; Renda, M.; Pedemonte, N.; Galietta, L.J.V. Comprehensive Analysis of Combinatorial Pharmacological Treatments to Correct Nonsense Mutations in the CFTR Gene. Int. J. Mol. Sci. 2021, 22, 11972. [Google Scholar] [CrossRef] [PubMed]
- de Poel, E.; Spelier, S.; Suen, S.W.F.; Kruisselbrink, E.; Graeber, S.Y.; Mall, M.A.; Weersink, E.J.M.; van der Eerden, M.M.; Koppelman, G.H.; van der Ent, C.K.; et al. Functional Restoration of CFTR Nonsense Mutations in Intestinal Organoids. J. Cyst. Fibros. 2022, 21, 246–253. [Google Scholar] [CrossRef]
- Atanasova, V.S.; Jiang, Q.; Prisco, M.; Gruber, C.; Pinon Hofbauer, J.; Chen, M.; Has, C.; Bruckner-Tuderman, L.; McGrath, J.A.; Uitto, J.; et al. Amlexanox Enhances Premature Termination Codon Read-Through in COL7A1 and Expression of Full Length Type VII Collagen: Potential Therapy for Recessive Dystrophic Epidermolysis Bullosa. J. Investig. Dermatol. 2017, 137, 1842–1849. [Google Scholar] [CrossRef]
- Dabrowski, M.; Bukowy-Bieryllo, Z.; Jackson, C.L.; Zietkiewicz, E. Properties of Non-Aminoglycoside Compounds Used to Stimulate Translational Readthrough of PTC Mutations in Primary Ciliary Dyskinesia. Int. J. Mol. Sci. 2021, 22, 4923. [Google Scholar] [CrossRef]
- Welch, E.M.; Barton, E.R.; Zhuo, J.; Tomizawa, Y.; Friesen, W.J.; Trifillis, P.; Paushkin, S.; Patel, M.; Trotta, C.R.; Hwang, S.; et al. PTC124 targets genetic disorders caused by nonsense mutations. Nature 2007, 447, 87–91. [Google Scholar] [CrossRef] [PubMed]
- Michorowska, S. Ataluren-Promising Therapeutic Premature Termination Codon Readthrough Frontrunner. Pharmaceuticals 2021, 14, 785. [Google Scholar] [CrossRef]
- Peltz, S.W.; Welch, E.M.; Jacobson, A.; Trotta, C.R.; Naryshkin, N.; Sweeney, H.L.; Bedwell, D.M. Nonsense suppression activity of PTC124 (ataluren). Proc. Natl. Acad. Sci. USA 2009, 106, E64, author reply E65. [Google Scholar] [CrossRef]
- Wilschanski, M.; Miller, L.L.; Shoseyov, D.; Blau, H.; Rivlin, J.; Aviram, M.; Cohen, M.; Armoni, S.; Yaakov, Y.; Pugatsch, T.; et al. Chronic ataluren (PTC124) treatment of nonsense mutation cystic fibrosis. Eur. Respir. J. 2011, 38, 59–69. [Google Scholar] [CrossRef] [PubMed]
- McElroy, S.P.; Nomura, T.; Torrie, L.S.; Warbrick, E.; Gartner, U.; Wood, G.; McLean, W.H. A lack of premature termination codon read-through efficacy of PTC124 (Ataluren) in a diverse array of reporter assays. PLoS Biol. 2013, 11, e1001593. [Google Scholar] [CrossRef] [PubMed]
- Bushby, K.; Finkel, R.; Wong, B.; Barohn, R.; Campbell, C.; Comi, G.P.; Connolly, A.M.; Day, J.W.; Flanigan, K.M.; Goemans, N.; et al. Ataluren treatment of patients with nonsense mutation dystrophinopathy. Muscle Nerve 2014, 50, 477–487. [Google Scholar] [CrossRef]
- Gómez-Grau, M.; Garrido, E.; Cozar, M.; Rodriguez-Sureda, V.; Domínguez, C.; Arenas, C.; Gatti, R.A.; Cormand, B.; Grinberg, D.; Vilageliu, L. Evaluation of Aminoglycoside and Non-Aminoglycoside Compounds for Stop-Codon Readthrough Therapy in Four Lysosomal Storage Diseases. PLoS ONE 2015, 10, e0135873. [Google Scholar] [CrossRef]
- Ng, M.Y.; Li, H.; Ghelfi, M.D.; Goldman, Y.E.; Cooperman, B.S. Ataluren and aminoglycosides stimulate read-through of nonsense codons by orthogonal mechanisms. Proc. Natl. Acad. Sci. USA 2021, 118, e2020599118. [Google Scholar] [CrossRef]
- Huang, S.; Bhattacharya, A.; Ghelfi, M.D.; Li, H.; Fritsch, C.; Chenoweth, D.M.; Goldman, Y.E.; Cooperman, B.S. Ataluren binds to multiple protein synthesis apparatus sites and competitively inhibits release factor-dependent termination. Nat. Commun. 2022, 13, 2413. [Google Scholar] [CrossRef]
- Leung, A.; Sacristan-Reviriego, A.; Perdigao, P.R.L.; Sai, H.; Georgiou, M.; Kalitzeos, A.; Carr, A.F.; Coffey, P.J.; Michaelides, M.; Bainbridge, J.; et al. Investigation of PTC124-mediated translational readthrough in a retinal organoid model of AIPL1-associated Leber congenital amaurosis. Stem Cell Rep. 2022, 17, 2187–2202. [Google Scholar] [CrossRef]
- Wang, D.; Xue, X.; Gunn, G.; Du, M.; Siddiqui, A.; Weetall, M.; Keeling, K.M. Ataluren suppresses a premature termination codon in an MPS I-H mouse. J. Mol. Med. 2022, 100, 1223–1235. [Google Scholar] [CrossRef]
- Wu, C.; Iyer, S.; Wolfe, S.A.; Jacobson, A. Functional restoration of mouse Nf1 nonsense alleles in differentiated cultured neurons. J. Hum. Genet. 2022, 67, 661–668. [Google Scholar] [CrossRef]
- Konstan, M.W.; VanDevanter, D.R.; Rowe, S.M.; Wilschanski, M.; Kerem, E.; Sermet-Gaudelus, I.; DiMango, E.; Melotti, P.; McIntosh, J.; De Boeck, K. Efficacy and safety of ataluren in patients with nonsense-mutation cystic fibrosis not receiving chronic inhaled aminoglycosides: The international, randomized, double-blind, placebo-controlled Ataluren Confirmatory Trial in Cystic Fibrosis (ACT CF). J. Cyst. Fibros. 2020, 19, 595–601. [Google Scholar] [CrossRef] [PubMed]
- Kerem, E.; Konstan, M.W.; De Boeck, K.; Accurso, F.J.; Sermet-Gaudelus, I.; Wilschanski, M.; Elborn, J.S.; Melotti, P.; Bronsveld, I.; Fajac, I.; et al. Ataluren for the treatment of nonsense-mutation cystic fibrosis: A randomised, double-blind, placebo-controlled phase 3 trial. Lancet Respir. Med. 2014, 2, 539–547. [Google Scholar] [CrossRef] [PubMed]
- McDonald, C.M.; Muntoni, F.; Penematsa, V.; Jiang, J.; Kristensen, A.; Bibbiani, F.; Goodwin, E.; Gordish-Dressman, H.; Morgenroth, L.; Werner, C.; et al. Ataluren delays loss of ambulation and respiratory decline in nonsense mutation Duchenne muscular dystrophy patients. J. Comp. Eff. Res. 2022, 11, 139–155. [Google Scholar] [CrossRef] [PubMed]
- Baradaran-Heravi, A.; Bauer, C.C.; Pickles, I.B.; Hosseini-Farahabadi, S.; Balgi, A.D.; Choi, K.; Linley, D.M.; Beech, D.J.; Roberge, M.; Bon, R.S. Nonselective TRPC channel inhibition and suppression of aminoglycoside-induced premature termination codon readthrough by the small molecule AC1903. J. Biol. Chem. 2022, 298, 101546. [Google Scholar] [CrossRef]
- Bidou, L.; Bugaud, O.; Merer, G.; Coupet, M.; Hatin, I.; Chirkin, E.; Karri, S.; Demais, S.; François, P.; Cintrat, J.C.; et al. 2-Guanidino-quinazoline promotes the readthrough of nonsense mutations underlying human genetic diseases. Proc. Natl. Acad. Sci. USA 2022, 119, e2122004119. [Google Scholar] [CrossRef]
- Palomar-Siles, M.; Heldin, A.; Zhang, M.; Strandgren, C.; Yurevych, V.; van Dinter, J.T.; Engels, S.A.G.; Hofman, D.A.; Ohlin, S.; Meineke, B.; et al. Translational readthrough of nonsense mutant TP53 by mRNA incorporation of 5-Fluorouridine. Cell Death Dis. 2022, 13, 997. [Google Scholar] [CrossRef] [PubMed]
- Morrill, C.; Friesen, W.J.; Babu, S.; Baiazitov, R.Y.; Du, W.; Karloff, D.B.; Lee, C.S.; Moon, Y.C.; Ren, H.; Sierra, J.; et al. Guanidino quinazolines and pyrimidines promote readthrough of premature termination codons in cells with native nonsense mutations. Bioorg. Med. Chem. Lett. 2022, 76, 128989. [Google Scholar] [CrossRef]
- Carnes, J.; Jacobson, M.; Leinwand, L.; Yarus, M. Stop codon suppression via inhibition of eRF1 expression. RNA 2003, 9, 648–653. [Google Scholar] [CrossRef] [PubMed]
- Baradaran-Heravi, A.; Balgi, A.D.; Hosseini-Farahabadi, S.; Choi, K.; Has, C.; Roberge, M. Effect of small molecule eRF3 degraders on premature termination codon readthrough. Nucleic Acids Res. 2021, 49, 3692–3708. [Google Scholar] [CrossRef] [PubMed]
- Sharma, J.; Du, M.; Wong, E.; Mutyam, V.; Li, Y.; Chen, J.; Wangen, J.; Thrasher, K.; Fu, L.; Peng, N.; et al. A small molecule that induces translational readthrough of CFTR nonsense mutations by eRF1 depletion. Nat. Commun. 2021, 12, 4358. [Google Scholar] [CrossRef]
- Lee, R.E.; Lewis, C.A.; He, L.; Bulik-Sullivan, E.C.; Gallant, S.C.; Mascenik, T.M.; Dang, H.; Cholon, D.M.; Gentzsch, M.; Morton, L.C.; et al. Small-molecule eRF3a degraders rescue CFTR nonsense mutations by promoting premature termination codon readthrough. J. Clin. Investig. 2022, 132, e154571. [Google Scholar] [CrossRef]
- Bauer, J.W.; Brandl, C.; Haubenreisser, O.; Wimmer, B.; Weber, M.; Karl, T.; Klausegger, A.; Breitenbach, M.; Hintner, H.; von der Haar, T.; et al. Specialized yeast ribosomes: A customized tool for selective mRNA translation. PLoS ONE 2013, 8, e67609. [Google Scholar] [CrossRef]
- Rathner, A.; Rathner, P.; Friedrich, A.; Wiessner, M.; Kitzler, C.M.; Schernthaner, J.; Karl, T.; Krauss, J.; Lottspeich, F.; Mewes, W.; et al. Drug Development for Target Ribosomal Protein rpL35/uL29 for Repair of LAMB3R635X in Rare Skin Disease Epidermolysis Bullosa. Skin. Pharmacol. Physiol. 2021, 34, 167–182. [Google Scholar] [CrossRef]
- Barna, M.; Karbstein, K.; Tollervey, D.; Ruggero, D.; Brar, G.; Greer, E.L.; Dinman, J.D. The promises and pitfalls of specialized ribosomes. Mol. Cell 2022, 82, 2179–2184. [Google Scholar] [CrossRef]
- Xue, S.; Barna, M. Specialized ribosomes: A new frontier in gene regulation and organismal biology. Nat. Rev. Mol. Cell. Biol. 2012, 13, 355–369. [Google Scholar] [CrossRef]
- Bowling, A.; Eastman, A.; Merlo, C.; Lin, G.; West, N.; Patel, S.; Cutting, G.; Sharma, N. Downstream Alternate Start Site Allows N-Terminal Nonsense Variants to Escape NMD and Results in Functional Recovery by Readthrough and Modulator Combination. J. Pers. Med. 2022, 12, 1448. [Google Scholar] [CrossRef]
- Beryozkin, A.; Samanta, A.; Gopalakrishnan, P.; Khateb, S.; Banin, E.; Sharon, D.; Nagel-Wolfrum, K. Translational Read-Through Drugs (TRIDs) Are Able to Restore Protein Expression and Ciliogenesis in Fibroblasts of Patients with Retinitis Pigmentosa Caused by a Premature Termination Codon in FAM161A. Int. J. Mol. Sci. 2022, 23, 3541. [Google Scholar] [CrossRef]
- Bezzerri, V.; Lentini, L.; Api, M.; Busilacchi, E.M.; Cavalieri, V.; Pomilio, A.; Diomede, F.; Pegoraro, A.; Cesaro, S.; Poloni, A.; et al. Novel Translational Read-through-Inducing Drugs as a Therapeutic Option for Shwachman-Diamond Syndrome. Biomedicines 2022, 10, 886. [Google Scholar] [CrossRef]
- Omachi, K.; Kai, H.; Roberge, M.; Miner, J.H. NanoLuc reporters identify COL4A5 nonsense mutations susceptible to drug-induced stop codon readthrough. iScience 2022, 25, 103891. [Google Scholar] [CrossRef]
- Abreu, R.B.V.; Gomes, T.T.; Nepomuceno, T.C.; Li, X.; Fuchshuber-Moraes, M.; De Gregoriis, G.; Suarez-Kurtz, G.; Monteiro, A.N.A.; Carvalho, M.A. Functional Restoration of BRCA1 Nonsense Mutations by Aminoglycoside-Induced Readthrough. Front. Pharmacol. 2022, 13, 935995. [Google Scholar] [CrossRef] [PubMed]
- Has, C.; Sayar, S.B.; Zheng, S.; Chacón-Solano, E.; Condrat, I.; Yadav, A.; Roberge, M.; Larcher Laguzzi, F. Read-Through for Nonsense Mutations in Type XVII Collagen-Deficient Junctional Epidermolysis Bullosa. J. Investig. Dermatol. 2022, 142, 1227–1230.e4. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Santamaría, L.; Maseda, R.; de Arriba, M.D.C.; Membrilla, J.A.; Sigüenza, A.I.; Mascías, J.; García, M.; Quintana, L.; Esteban-Rodríguez, I.; Hernández-Fernández, C.P.; et al. Evaluation of Systemic Gentamicin as Translational Readthrough Therapy for a Patient With Epidermolysis Bullosa Simplex With Muscular Dystrophy Owing to PLEC1 Pathogenic Nonsense Variants. JAMA Dermatol. 2022, 158, 439–443. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wagner, R.N.; Wießner, M.; Friedrich, A.; Zandanell, J.; Breitenbach-Koller, H.; Bauer, J.W. Emerging Personalized Opportunities for Enhancing Translational Readthrough in Rare Genetic Diseases and Beyond. Int. J. Mol. Sci. 2023, 24, 6101. https://doi.org/10.3390/ijms24076101
Wagner RN, Wießner M, Friedrich A, Zandanell J, Breitenbach-Koller H, Bauer JW. Emerging Personalized Opportunities for Enhancing Translational Readthrough in Rare Genetic Diseases and Beyond. International Journal of Molecular Sciences. 2023; 24(7):6101. https://doi.org/10.3390/ijms24076101
Chicago/Turabian StyleWagner, Roland N., Michael Wießner, Andreas Friedrich, Johanna Zandanell, Hannelore Breitenbach-Koller, and Johann W. Bauer. 2023. "Emerging Personalized Opportunities for Enhancing Translational Readthrough in Rare Genetic Diseases and Beyond" International Journal of Molecular Sciences 24, no. 7: 6101. https://doi.org/10.3390/ijms24076101
APA StyleWagner, R. N., Wießner, M., Friedrich, A., Zandanell, J., Breitenbach-Koller, H., & Bauer, J. W. (2023). Emerging Personalized Opportunities for Enhancing Translational Readthrough in Rare Genetic Diseases and Beyond. International Journal of Molecular Sciences, 24(7), 6101. https://doi.org/10.3390/ijms24076101





