Potential Pathogenic Impact of Cow’s Milk Consumption and Bovine Milk-Derived Exosomal MicroRNAs in Diffuse Large B-Cell Lymphoma
Abstract
1. Introduction
2. Epidemiological Evidence for Milk Intake and DLBCL Risk
3. Potential Milk-Related Factors Promoting DLBCL
3.1. Insulin-Like Growth Factor 1 Signaling in DLBCL
3.2. Milk-Induced IGF-1- and Amino Acid-Mediated mTORC1 Signaling
3.2.1. Milk-Derived Essential Branched-Chain Amino Acids
3.2.2. L-Type Neutral Amino Acid Transporter 1 in DLBCL
3.2.3. Glutaminolysis in DLBCL
3.2.4. Milk Proteins: A Rich Source of Glutamine
3.3. Activation of mTORC1 in DLBCL
3.3.1. Milk-Induced Activation of mTORC1
3.3.2. B-Cell Receptor Activation in DLBCL
3.4. Milk Peptide-Induced B-Cell Receptor Activation
3.5. Estrogen Receptor-β Signaling in DLBCL
3.6. Milk-Derived Estrogens
3.6.1. Bisphenol A in Lymphomagenesis
3.6.2. Contamination of Commercial Milk with Bisphenol A
3.7. Viral Agents in DLBCL
3.8. Bovine Meat and Milk Factors
3.9. Bovine Leukemia Virus
4. Exosomal MicroRNAs in the Pathogenesis of DLBCL
4.1. MicroRNA-Mediated Transcriptional Regulation in DLBCL
4.1.1. MicroRNA-let-7 Over-Expression in DLBCL
4.1.2. MicroRNA-125b Over-Expression in DLBCL
4.1.3. MicroRNA-21 Over-Expression in DLBCL
4.1.4. Over-Expressed MicroRNA-155 in DLBCL
4.1.5. MicroRNA-148a Maintains Survival of Immature B Cells
4.2. Potential Uptake of Bovine Milk-Derived Exosomes by B Cells
4.3. Bovine Milk Exosome-Derived MicroRNAs
4.3.1. MicroRNAs of the Let-7 Family
4.3.2. MicroRNA-125a and MicroRNA-125b
4.3.3. MicroRNA-21
4.3.4. Exosomal MicroRNA-21 Exposure and M2 Macrophage Polarization
4.3.5. MicroRNA-29b
4.3.6. MicroRNA-155
4.3.7. MicroRNA-148a
5. Discussion
6. Conclusions
7. Limitations
8. Materials and Methods
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ABC | activated B-cell like |
| AICDA | activation-induced cytidine deaminase |
| AMPK | AMP-activated protein kinase |
| ATF4 | activating transcription factor 4 |
| BACH2 | BTB and CNC homology 2 |
| BCAA | branched-chain amino acid |
| BCL6 | B cell lymphoma 6 |
| BCL10 | B cell CLL/lymphoma 10 |
| BCL11B | B cell lymphoma/leukemia 11B |
| BCR | B cell receptor |
| BIC | B cell integration cluster |
| BIM | BCL2-like 11 |
| BIRC5 | baculoviral IAP repeat-containing protein 5 |
| BLIMP1 | B cell differentiation B lymphocyte-induced maturation protein 1 |
| BLV | bovine leukemia virus |
| BMMF | bovine meat and milk factor |
| BPA | bisphenol A |
| CARD11 | caspase recruitment domain-containing protein 11 |
| CDKN1A | cyclin-dependent kinase inhibitor 1A |
| C-MYK | MYC protooncogene |
| CRESS | circular replicase-encoding single-stranded |
| CTNNB1 | catenin β1 |
| DEPTOR | DEP domain-containing mTOR-interacting protein |
| DLBCL | diffuse large B cell lymphoma |
| DNMT1 | DNA methyltransferase 1 |
| EBNA2 | EBV nuclear antigen 2 |
| E1 | estrone |
| E2 | estradiol |
| E3 | estriol |
| EBV | Epstein-Barr virus |
| E2F1 | E2F transcription factor 1 |
| eIF4B | eukaryotic translation initiation factor 4B |
| eIF4E | eukaryotic translation initiation factor 4E |
| ERβ | estrogen receptor-β |
| EV | extracellular vesicle |
| FcRn | neonatal Fc receptor |
| FoxO1A | forkhead box O1A |
| FoxO3A | forkhead box O3A |
| GADD45A | DNA-damage-inducible gene α |
| GC | germinal center |
| GCB | germinal center B-cell like |
| GH | growth hormone |
| GHR | growth hormone receptor |
| GPR30 | G protein-coupled estrogen receptor 1 |
| HBV | hepatitis B virus |
| HCV | hepatitis C virus |
| HHV8 | human herpes virus 8 |
| HIF-1 | hypoxia-inducible factor 1 |
| HIV | human immunodeficiency virus |
| HPV | human papilloma virus |
| HTLV1 | human T-cell leukemia virus type 1 |
| IGF-1 | insulin-like growth factor 1 |
| IGF1R | insulin-like growth factor 1 receptor |
| IGFPB3 | insulin-like growth factor binding protein 3 |
| IKK | inhibitor of kB kinase |
| INPP5 | inositol polyphosphate-5-phosphatase, 145-KD (SHIP1) |
| IRF4 | interferon regulatory factor 4 |
| KSHV | Kaposi sarcoma-associated herpesvirus |
| LAT1 | L-type neutral amino acid transporter 1 |
| MALT1 | mucosa-associated lymphoid tissue lymphoma translocation gene 1 |
| MAX | MAX protein |
| MDE | milk-derived exosome |
| MDM2 | MDM2 protooncogene |
| MDR1 | ATP-binding cassette, subfamily B, member 1 (ABCB1) |
| MDSC | myeloid-derived suppressor cells |
| miR | micro-ribonucleic acid |
| MITF | microphthalmia-associated transcription factor |
| MSC | mesenchymal stem cell |
| MYC | MYC protooncogene, bHLH transcription factor |
| MYD88 | MYD88 innate immune signal transduction adaptor |
| mTORC1 | mechanistic target of rapamycin complex 1 |
| MXD1 | MAX dimerization protein 1 |
| MXD4 | MAX dimerization protein 4 |
| NF-κB | nuclear factor kappa B |
| NHL | non-Hodgkin lymphoma |
| PBMC | peripheral blood mononuclear cell |
| PDCD4 | programmed cell death 4 |
| PDK1 | phosphoinositide-dependent kinase 1 |
| PI3K | phosphatidylinositol-3 kinase |
| PIK3IP1 | phosphatidylinositol 3-kinase-interacting protein 1 |
| PIK3R1 | PI3K regulatory subunit 1 |
| PPP | picropodophyllin |
| PRDM1 | PR domain-containing protein 1 (BLIMP1) |
| PTEN | phosphatase and tensin homolog |
| RAG | Ragulator |
| R-CHOP | rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone regimen |
| RISC | RNA-induced silencing complex |
| RUNX2 | runt-related transcription factor 2 |
| SIRT3 | sirtuin 3 |
| SMO | Smoothened |
| SOS | Son of Sevenless |
| STAT3 | signal transducer and activator of transcriptin 3 |
| SV40 | Simian virus 40 |
| TAM | tumor-associated macrophage |
| TCA | tricarboxylic acid |
| TCL1 | T-cell leukemia gene 1 |
| TLR | Toll-like receptor |
| TME | tumor microenvironment |
| TNFAIP3 | tumor necrosis factor-α induced protein 3 |
| TNFRSF10B | tumor necrosis factor receptor superfamily, member 10B |
| TP53 | tumor protein p53 |
| TRAF2 | TNF receptor-associated factor 2 |
| TSC2 | tuberin |
| TWIST1 | TWIST family bHLH transcription factor 1 |
| UHT | ultraheat treatment |
| UTR | untranslated region |
| VHL | von Hippel–Lindau tumor suppressor |
| YAP | Hippo-Yes-associated protein |
References
- Martelli, M.; Ferreri, A.J.; Agostinelli, C.; Di Rocco, A.; Pfreundschuh, M.; Pileri, S.A. Diffuse large B-cell lymphoma. Crit. Rev. Oncol. Hematol. 2013, 87, 146–171. [Google Scholar] [PubMed]
- Li, S.; Young, K.H.; Medeiros, L.J. Diffuse large B-cell lymphoma. Pathology 2018, 50, 74–87. [Google Scholar] [PubMed]
- Wilcox, R.A. Cutaneous B-cell lymphomas: 2019 update on diagnosis, risk stratification, and management. Am. J. Hematol. 2018, 93, 1427–1430. [Google Scholar] [PubMed]
- Stadler, R.; Stranzenbach, R. Molecular pathogenesis of cutaneous lymphomas. Exp. Dermatol. 2018, 27, 1078–1083. [Google Scholar] [PubMed]
- Dippel, E.; Assaf, C.; Becker, J.C.; von Bergwelt-Baildon, M.; Bernreiter, S.; Cozzio, A.; Eich, H.T.; Elsayad, K.; Follmann, M.; Grabbe, S.; et al. S2k-Guidelines-Cutaneous lymphomas (ICD10 C82–C86): Update 2021. J. Dtsch. Dermatol. Ges. 2022, 20, 537–554. [Google Scholar]
- Chihara, D.; Johnston, K.; Bolatova, T.; Szabo, S.; Kalsekar, A.; Mutebi, A.; Yang, H.; Liu, Y.; Attinson, D.; Hutchings, M. An epidemiological model to estimate the prevalence of diffuse large B-cell lymphoma in the United States. Clin. Lymphoma Myeloma Leuk. 2022, 22, e1092–e1099. [Google Scholar]
- Kanas, G.; Ge, W.; Quek, R.G.W.; Keeven, K.; Nersesyan, K.; Arnason, J.E. Epidemiology of diffuse large B-cell lymphoma (DLBCL) and follicular lymphoma (FL) in the United States and Western Europe: Population-level projections for 2020–2025. Leuk. Lymphoma 2022, 63, 54–63. [Google Scholar]
- Alizadeh, A.A.; Eisen, M.B.; Davis, R.E.; Ma, C.; Lossos, I.S.; Rosenwald, A.; Boldrick, J.C.; Sabet, H.; Tran, T.; Yu, X.; et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 2000, 403, 503–511. [Google Scholar]
- Caro, P.; Kishan, A.U.; Norberg, E.; Stanley, I.A.; Chapuy, B.; Ficarro, S.B.; Polak, K.; Tondera, D.; Gounarides, J.; Yin, H.; et al. Metabolic signatures uncover distinct targets in molecular subsets of diffuse large B cell lymphoma. Cancer Cell 2012, 22, 547–560. [Google Scholar]
- Miao, Y.; Medeiros, L.J.; Li, Y.; Li, J.; Young, K.H. Genetic alterations and their clinical implications in DLBCL. Nat. Rev. Clin. Oncol. 2019, 16, 634–652. [Google Scholar]
- Liu, Y.; Barta, S.K. Diffuse large B-cell lymphoma: 2019 update on diagnosis, risk stratification, and treatment. Am. J. Hematol. 2019, 94, 604–616. [Google Scholar] [CrossRef]
- Morin, R.D.; Mungall, K.; Pleasance, E.; Mungall, A.J.; Goya, R.; Huff, R.D.; Scott, D.W.; Ding, J.; Roth, A.; Chiu, R.; et al. Mutational and structural analysis of diffuse large B-cell lymphoma using whole-genome sequencing. Blood 2013, 122, 1256–1265. [Google Scholar] [CrossRef]
- Ando, M.; Sato, Y.; Takata, K.; Nomoto, J.; Nakamura, S.; Ohshima, K.; Takeuchi, T.; Orita, Y.; Kobayashi, Y.; Yoshino, T. A20 (TNFAIP3) deletion in Epstein-Barr virus-associated lymphoproliferative disorders/lymphomas. PLoS ONE 2013, 8, e56741. [Google Scholar]
- Davis, R.E.; Ngo, V.N.; Lenz, G.; Tolar, P.; Young, R.M.; Romesser, P.B.; Kohlhammer, H.; Lamy, L.; Zhao, H.; Yang, Y.; et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature 2010, 463, 88–92. [Google Scholar]
- Ngo, V.N.; Young, R.M.; Schmitz, R.; Jhavar, S.; Xiao, W.; Lim, K.H.; Kohlhammer, H.; Xu, W.; Yang, Y.; Zhao, H.; et al. Oncogenically active MYD88 mutations in human lymphoma. Nature 2011, 470, 115–119. [Google Scholar]
- Pasqualucci, L.; Dominguez-Sola, D.; Chiarenza, A.; Fabbri, G.; Grunn, A.; Trifonov, V.; Kasper, L.H.; Lerach, S.; Tang, H.; Ma, J.; et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature 2011, 471, 189–195. [Google Scholar]
- Zhang, J.; Grubor, V.; Love, C.L.; Banerjee, A.; Richards, K.L.; Mieczkowski, P.A.; Dunphy, C.; Choi, W.; Au, W.Y.; Srivastava, G.; et al. Genetic heterogeneity of diffuse large B-cell lymphoma. Proc. Natl. Acad. Sci. USA 2013, 110, 1398–1403. [Google Scholar]
- Pasqualucci, L.; Dalla-Favera, R. Genetics of diffuse large B-cell lymphoma. Blood 2018, 131, 2307–2319. [Google Scholar]
- Schmitz, R.; Wright, G.W.; Huang, D.W.; Johnson, C.A.; Phelan, J.D.; Wang, J.Q.; Roulland, S.; Kasbekar, M.; Young, R.M.; Shaffer, A.L.; et al. Genetics and pathogenesis of diffuse large B-cell lymphoma. N. Engl. J. Med. 2018, 378, 1396–1407. [Google Scholar] [CrossRef]
- De, S.; Shaknovich, R.; Elemento, O.; Geng, H.; Kormaksson, M.; Jiang, Y.; Woolcock, B.; Johnson, N.; Polo, J.M. Aberration in DNA methylation in B-cell lymphomas has a complex origin and increases with disease severity. PLoS Genet. 2013, 9, e1003137. [Google Scholar] [CrossRef]
- Jiang, Y.; Melnick, A. The epigenetic basis of diffuse large B-cell lymphoma. Semin. Hematol. 2015, 52, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Dominguez, P.M.; Melnick, A.M. The many layers of epigenetic dysfunction in B-cell lymphomas. Curr. Opin. Hematol. 2016, 23, 377–384. [Google Scholar] [CrossRef] [PubMed]
- Frazzi, R.; Zanetti, E.; Pistoni, M.; Tamagnini, I.; Valli, R.; Braglia, L.; Merli, F. Methylation changes of SIRT1, KLF4, DAPK1 and SPG20 in B-lymphocytes derived from follicular and diffuse large B-cell lymphoma. Leuk. Res. 2017, 57, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Bakhshi, T.J.; Georgel, P.T. Genetic and epigenetic determinants of diffuse large B-cell lymphoma. Blood Cancer J. 2020, 10, 123. [Google Scholar]
- Oricchio, E. Epigenetic balance in DLBCL. Blood 2021, 138, 355–356. [Google Scholar] [CrossRef]
- Isshiki, Y.; Melnick, A. Epigenetic mechanisms of therapy resistance in diffuse large B cell lymphoma (DLBCL). Curr. Cancer Drug Targets. 2021, 21, 274–282. [Google Scholar] [CrossRef]
- Heward, J.; Konali, L.; D’Avola, A.; Close, K.; Yeomans, A.; Philpott, M.; Dunford, J.; Rahim, T.; Al Seraihi, A.F.; Wang, J.; et al. KDM5 inhibition offers a novel therapeutic strategy for the treatment of KMT2D mutant lymphomas. Blood 2021, 138, 370–381. [Google Scholar] [CrossRef]
- Jiao, J.; Lv, Z.; Zhang, P.; Wang, Y.; Yuan, M.; Yu, X.; Otieno Odhiambo, W.; Zheng, M.; Zhang, H.; Ma, Y.; et al. AID assists DNMT1 to attenuate BCL6 expression through DNA methylation in diffuse large B-cell lymphoma cell lines. Neoplasia 2020, 22, 142–153. [Google Scholar] [CrossRef]
- Shen, H.M.; Peters, A.; Baron, B.; Zhu, X.; Storb, U. Mutation of BCL-6 gene in normal B cells by the process of somatic hypermutation of Ig genes. Science 1998, 280, 1750–1752. [Google Scholar] [CrossRef]
- Thandra, K.C.; Barsouk, A.; Saginala, K.; Padala, S.A.; Barsouk, A.; Rawla, P. Epidemiology of non-Hodgkin’s lymphoma. Med. Sci. 2021, 9, 5. [Google Scholar] [CrossRef]
- Zhang, Y.; Sanjose, S.D.; Bracci, P.M.; Morton, L.M.; Wang, R.; Brennan, P.; Hartge, P.; Boffetta, P.; Becker, N.; Maynadie, M.; et al. Personal use of hair dye and the risk of certain subtypes of non-Hodgkin lymphoma. Am. J. Epidemiol. 2008, 167, 1321–1331. [Google Scholar] [CrossRef]
- Guo, H.; Bassig, B.A.; Lan, Q.; Zhu, Y.; Zhang, Y.; Holford, T.R.; Leaderer, B.; Boyle, P.; Qin, Q.; Zhu, C.; et al. Polymorphisms in DNA repair genes, hair dye use, and the risk of non-Hodgkin lymphoma. Cancer Causes Control 2014, 25, 1261–1270. [Google Scholar] [CrossRef]
- Kleinstern, G.; Abu Seir, R.; Perlman, R.; Khatib, A.; Abdeen, Z.; Elyan, H.; Nirel, R.; Amir, G.; Ramlawi, A.; Sabatin, F.; et al. Ethnic variation in medical and lifestyle risk factors for B cell non-Hodgkin lymphoma: A case-control study among Israelis and Palestinians. PLoS ONE 2017, 12, e0171709. [Google Scholar] [CrossRef]
- Eriksson, M.; Hardell, L.; Carlberg, M.; Akerman, M. Pesticide exposure as risk factor for non-Hodgkin lymphoma including histopathological subgroup analysis. Int. J. Cancer 2008, 123, 1657–1663. [Google Scholar] [CrossRef]
- Boffetta, P.; Ciocan, C.; Zunarelli, C.; Pira, E. Exposure to glyphosate and risk of non-Hodgkin lymphoma: An updated meta-analysis. Med. Lav. 2021, 112, 119–194. [Google Scholar]
- Pahwa, M.; Beane Freeman, L.E.; Spinelli, J.J.; Blair, A.; McLaughlin, J.R.; Zahm, S.H.; Cantor, K.P.; Weisenburger, D.D.; Punam Pahwa, P.P.; Dosman, J.A.; et al. Glyphosate use and associations with non-Hodgkin lymphoma major histological sub-types: Findings from the North American Pooled Project. Scand. J. Work Environ. Health 2019, 45, 600–609. [Google Scholar] [CrossRef]
- Chen, Y.K.; Tan, Y.Y.; Yao, M.; Lin, H.C.; Tsai, M.H.; Li, Y.Y.; Hsu, Y.J.; Huang, T.T.; Chang, C.W.; Cheng, C.M.; et al. Bisphenol A-induced DNA damages promote to lymphoma progression in human lymphoblastoid cells through aberrant CTNNB1 signaling pathway. iScience 2021, 24, 102888. [Google Scholar] [CrossRef]
- Sapkota, S.; Shaikh, H. Non-Hodgkin lymphoma. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Franceschi, S.; Serraino, D.; Carbone, A.; Talamini, R.; La Vecchia, C. Dietary factors and non-Hodgkin’s lymphoma: A case-control study in the northeastern part of Italy. Nutr. Cancer 1989, 12, 333–341. [Google Scholar] [CrossRef]
- Ward, M.H.; Zahm, S.H.; Weisenburger, D.D.; Gridley, G.; Cantor, K.P.; Saal, R.C.; Blair, A. Dietary factors and non-Hodgkin’s lymphoma in Nebraska (United States). Cancer Causes Control 1994, 5, 422–432. [Google Scholar]
- Chang, E.T.; Smedby, K.E.; Zhang, S.M.; Hjalgrim, H.; Melbye, M.; Ost, A.; Glimelius, B.; Wolk, A.; Adami, H.O. Dietary factors and risk of non-Hodgkin lymphoma in men and women. Cancer Epidemiol. Biomark. Prev. 2005, 14, 512–520. [Google Scholar] [CrossRef]
- Skibola, C.F. Obesity, diet and risk of non-Hodgkin lymphoma. Cancer Epidemiol. Biomark. Prev. 2007, 16, 392–395. [Google Scholar] [CrossRef] [PubMed]
- Balasubramaniam, G.; Saoba, S.; Sarade, M.; Pinjare, S. Case-control study of risk factors for non-Hodgkin lymphoma in Mumbai, India. Asian Pac. J. Cancer Prev. 2013, 14, 775–780. [Google Scholar] [CrossRef] [PubMed]
- Chiu, B.C.; Cerhan, J.R.; Folsom, A.R.; Sellers, T.A.; Kushi, L.H.; Wallace, R.B.; Zheng, W.; Potter, J.D. Diet and risk of non-Hodgkin lymphoma in older women. JAMA 1996, 275, 1315–1321. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.M.; Hunter, D.J.; Rosner, B.A.; Giovannucci, E.L.; Colditz, G.A.; Speizer, F.E.; Willett, W.C. Intakes of fruits, vegetables, and related nutrients and the risk of non-Hodgkin’s lymphoma among women. Cancer Epidemiol. Biomark. Prev. 2000, 9, 477–485. [Google Scholar]
- Thompson, C.A.; Habermann, T.M.; Wang, A.H.; Vierkant, R.A.; Folsom, A.R.; Ross, J.A.; Cerhan, J.R. Antioxidant intake from fruits, vegetables and other sources and risk of non-Hodgkin’s lymphoma: The Iowa Women’s Health Study. Int. J. Cancer 2010, 126, 992–1003. [Google Scholar] [CrossRef]
- Ali, A.; Al-Belushi, B.S.; Waly, M.I.; Al-Moundhri, M.; Burney, I.A. Dietary and lifestyle factors and risk of non-Hodgkin’s lymphoma in Oman. Asian Pac. J. Cancer Prev. 2013, 14, 841–848. [Google Scholar] [CrossRef]
- Chiu, B.C.; Kwon, S.; Evens, A.M.; Surawicz, T.; Smith, S.M.; Weisenburger, D.D. Dietary intake of fruit and vegetables and risk of non-Hodgkin lymphoma. Cancer Causes Control 2011, 22, 1183–1195. [Google Scholar] [CrossRef]
- Wang, J.; Li, X.; Zhang, D. Dairy product consumption and risk of non-Hodgkin lymphoma: A meta-analysis. Nutrients 2016, 8, 120. [Google Scholar] [CrossRef]
- Kakkoura, M.G.; Du, H.; Guo, Y.; Yu, C.; Yang, L.; Pei, P.; Chen, Y.; Sansome, S.; Chan, W.C.; Yang, X.; et al. China Kadoorie Biobank (CKB) Collaborative Group. Dairy consumption and risks of total and site-specific cancers in Chinese adults: An 11-year prospective study of 0.5 million people. BMC Med. 2022, 20, 134. [Google Scholar] [CrossRef]
- Saberi Hosnijeh, F.; Casabonne, D.; Nieters, A.; Solans, M.; Naudin, S.; Ferrari, P.; Mckay, J.D.; Benavente, Y.; Weiderpass, E.; Freisling, H.; et al. Association between anthropometry and lifestyle factors and risk of B-cell lymphoma: An exposome-wide analysis. Int. J. Cancer 2021, 148, 2115–2128. [Google Scholar] [CrossRef]
- Melnik, B.C. Lifetime impact of cow’s milk on overactivation of mTORC1: From fetal to childhood overgrowth, acne, diabetes, cancers, and neurodegeneration. Biomolecules 2021, 11, 404. [Google Scholar] [CrossRef]
- Melnik, B.C.; John, S.M.; Schmitz, G. Milk is not just food but most likely a genetic transfection system activating mTORC1 signaling for postnatal growth. Nutr. J. 2013, 12, 103. [Google Scholar]
- Melnik, B.C. Milk—A nutrient system of mammalian evolution promoting mTORC1-dependent translation. Int. J. Mol. Sci. 2015, 16, 17048–17087. [Google Scholar] [CrossRef]
- Sergentanis, T.N.; Ntanasis-Stathopoulos, I.; Tzanninis, I.G.; Gavriatopoulou, M.; Sergentanis, I.N.; Dimopoulos, M.A.; Psaltopoulou, T. Meat, fish, dairy products and risk of hematological malignancies in adults—A systematic review and meta-analysis of prospective studies. Leuk. Lymphoma 2019, 60, 1978–1990. [Google Scholar] [CrossRef]
- Iso, H.; Kubota, Y.; Japan Collaborative Cohort Study for Evaluation of Cancer. Nutrition and disease in the Japan Collaborative Cohort Study for Evaluation of Cancer (JACC). Asian Pac. J. Cancer Prev. 2007, 8, 35–80. [Google Scholar]
- Rohrmann, S.; Linseisen, J.; Jakobsen, M.U.; Overvad, K.; Raaschou-Nielsen, O.; Tjonneland, A.; Boutron-Ruault, M.C.; Kaaks, R.; Becker, N.; Bergmann, M.; et al. Consumption of meat and dairy and lymphoma risk in the European Prospective Investigation into Cancer and Nutrition. Int. J. Cancer 2011, 128, 623–634. [Google Scholar] [CrossRef]
- Nwabo Kamdje, A.H.; Seke Etet, P.F.; Kipanyula, M.J.; Vecchio, L.; Tagne Simo, R.; Njamnshi, A.K.; Lukong, K.E.; Mimche, P.N. Insulin-like growth factor-1 signaling in the tumor microenvironment: Carcinogenesis, cancer drug resistance, and therapeutic potential. Front. Endocrinol. 2022, 13, 927390. [Google Scholar] [CrossRef]
- Kasprzak, A.; Kwasniewski, W.; Adamek, A.; Gozdzicka-Jozefiak, A. Insulin-like growth factor (IGF) axis in cancerogenesis. Mutat. Res. Rev. Mutat. Res. 2017, 772, 78–104. [Google Scholar]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar]
- Strömberg, T.; Feng, X.; Delforoush, M.; Berglund, M.; Lin, Y.; Axelson, M.; Larsson, O.; Georgii-Hemming, P.; Lennartsson, J.; Enblad, G. Picropodophyllin inhibits proliferation and survival of diffuse large B-cell lymphoma cells. Med. Oncol. 2015, 32, 188. [Google Scholar] [CrossRef]
- Ravindra, V.M.; Raheja, A.; Corn, H.; Driscoll, M.; Welt, C.; Simmons, D.L.; Couldwell, W.T. Primary pituitary diffuse large B-cell lymphoma with somatotroph hyperplasia and acromegaly: Case report. J. Neurosurg. 2017, 126, 1725–1730. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Zhang, Y.; Li, Y.; Xu, Y.; Zhang, L.; Li, Y.; Wang, X. Klotho suppresses tumor progression via inhibiting IGF-1R signaling in T-cell lymphoma. Oncol. Rep. 2017, 38, 967–974. [Google Scholar] [PubMed]
- Zhou, X.; Fang, X.; Jiang, Y.; Geng, L.; Li, X.; Li, Y.; Lu, K.; Li, P.; Lv, X.; Wang, X. Klotho, an anti-aging gene, acts as a tumor suppressor and inhibitor of IGF-1R signaling in diffuse large B cell lymphoma. J. Hematol. Oncol. 2017, 10, 37. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Chen, N.; Xu, H.; Zhou, X.; Wang, J.; Fang, X.; Zhang, Y.; Li, Y.; Yang, J.; Wang, X. Regulation of Hippo-YAP signaling by insulin-like growth factor-1 receptor in the tumorigenesis of diffuse large B-cell lymphoma. J. Hematol. Oncol. 2020, 13, 77. [Google Scholar] [PubMed]
- Agarwal, N.K.; Kim, C.H.; Kunkalla, K.; Vaghefi, A.; Sanchez, S.; Manuel, S.; Bilbao, D.; Vega, F.; Landgraf, R. Smoothened (SMO) regulates insulin-like growth factor 1 receptor (IGF1R) levels and protein kinase B (AKT) localization and signaling. Lab. Investig. 2022, 102, 401–410. [Google Scholar] [CrossRef]
- Qin, L.Q.; He, K.; Xu, J.Y. Milk consumption and circulating insulin-like growth factor-I level: A systematic literature review. Int. J. Food Sci. Nutr. 2009, 60 (Suppl. S7), 330–340. [Google Scholar]
- Rich-Edwards, J.W.; Ganmaa, D.; Pollak, M.N.; Nakamoto, E.K.; Kleinman, K.; Tserendolgor, U.; Willett, W.C.; Frazier, A.L. Milk consumption and the prepubertal somatotropic axis. Nutr. J. 2007, 6, 28. [Google Scholar]
- Hoppe, C.; Mølgaard, C.; Juul, A.; Michaelsen, K.F. High intakes of skimmed milk, but not meat, increase serum IGF-I and IGFBP-3 in eight-year-old boys. Eur. J. Clin. Nutr. 2004, 58, 1211–1216. [Google Scholar]
- Hoppe, C.; Mølgaard, C.; Dalum, C.; Vaag, A.; Michaelsen, K.F. Differential effects of casein versus whey on fasting plasma levels of insulin, IGF-1 and IGF-1/IGFBP-3: Results from a randomized 7-day supplementation study in prepubertal boys. Eur. J. Clin. Nutr. 2009, 63, 1076–1083. [Google Scholar] [CrossRef]
- Barrea, L.; Di Somma, C.; Macchia, P.E.; Falco, A.; Savanelli, M.C.; Orio, F.; Colao, A.; Savastano, S. Influence of nutrition on somatotropic axis: Milk consumption in adult individuals with moderate-severe obesity. Clin. Nutr. 2017, 36, 293–301. [Google Scholar]
- Rogers, I.; Emmett, P.; Gunnell, D.; Dunger, D.; Holly, J.; ALSPAC Study Tteam. Milk as a food for growth? The insulin-like growth factors link. Public Health Nutr. 2006, 9, 359–368. [Google Scholar] [CrossRef]
- Norat, T.; Dossus, L.; Rinaldi, S.; Overvad, K.; Grønbaek, H.; Tjønneland, A.; Olsen, A.; Clavel-Chapelon, F.; Boutron-Ruault, M.C.; Boeing, H.; et al. Diet, serum insulin-like growth factor-I and IGF-binding protein-3 in European women. Eur. J. Clin. Nutr. 2007, 61, 91–98. [Google Scholar]
- Crowe, F.L.; Key, T.J.; Allen, N.E.; Appleby, P.N.; Roddam, A.; Overvad, K.; Grønbaek, H.; Tjønneland, A.; Halkjaer, J.; Dossus, L.; et al. The association between diet and serum concentrations of IGF-I, IGFBP-1, IGFBP-2, and IGFBP-3 in the European Prospective Investigation into Cancer and Nutrition. Cancer Epidemiol. Biomark. Prev. 2009, 18, 1333–1340. [Google Scholar] [CrossRef]
- Romo Ventura, E.; Konigorski, S.; Rohrmann, S.; Schneider, H.; Stalla, G.K.; Pischon, T.; Linseisen, J.; Nimptsch, K. Association of dietary intake of milk and dairy products with blood concentrations of insulin-like growth factor 1 (IGF-1) in Bavarian adults. Eur. J. Nutr. 2020, 59, 1413–1420. [Google Scholar] [CrossRef]
- Lovell, A.L.; Milne, T.; Matsuyama, M.; Hill, R.J.; Davies, P.S.W.; Grant, C.C.; Wall, C.R. Protein intake, IGF-1 concentrations, and growth in the second year of life in children receiving Growing Up Milk—Lite (GUMLi) or cow’s milk (CM) intervention. Front. Nutr. 2021, 8, 666228. [Google Scholar] [CrossRef]
- Honegger, A.; Humbel, R.E. Insulin-like growth factors I and II in fetal and adult bovine serum. Purification, primary structures, and immunological cross-reactivities. J. Biol. Chem. 1986, 261, 569–575. [Google Scholar]
- Wheelhouse, N.M.; Stubbs, A.K.; Lomax, M.A.; MacRae, J.C.; Hazlerigg, D.G. Growth hormone and amino acid supply interact synergistically to control insulin-like growth factor-I production and gene expression in cultured ovine hepatocytes. J. Endocrinol. 1999, 163, 353–361. [Google Scholar] [CrossRef]
- Stubbs, A.K.; Wheelhouse, N.M.; Lomax, M.A.; Hazlerigg, D.G. Nutrient-hormone interaction in the ovine liver: Methionine supply selectively modulates growth hormone-induced IGF-I gene expression. J. Endocrinol. 2002, 174, 335–341. [Google Scholar]
- Tsugawa, Y.; Handa, H.; Imai, T. Arginine induces IGF-1 secretion from the endoplasmic reticulum. Biochem. Biophys. Res. Commun. 2019, 514, 1128–1132. [Google Scholar] [CrossRef]
- Wan, X.; Wang, S.; Xu, J.; Zhuang, L.; Xing, K.; Zhang, M.; Zhu, X.; Wang, L.; Gao, P.; Xi, Q.; et al. Dietary protein-induced hepatic IGF-1 secretion mediated by PPARγ activation. PLoS ONE 2017, 12, e0173174. [Google Scholar] [CrossRef]
- Melnik, B.C.; Schmitz, G.; John, S.M. Health risks related to milk consumption: A critical evaluation from the medical perspective. MMW Fortschr. Med. 2021, 163 (Suppl. S4), 3–9. [Google Scholar] [CrossRef] [PubMed]
- Bounous, G.; Kongshavn, P.A. Differential effect of dietary protein type on the B-cell and T-cell immune responses in mice. J. Nutr. 1985, 115, 1403–1408. [Google Scholar] [CrossRef] [PubMed]
- Bounous, G.; Shenouda, N.; Kongshavn, P.A.; Osmond, D.G. Mechanism of altered B-cell response induced by changes in dietary protein type in mice. J. Nutr. 1985, 115, 1409–1417. [Google Scholar] [CrossRef] [PubMed]
- Cross, M.L.; Gill, H.S. Modulation of immune function by a modified bovine whey protein concentrate. Immunol. Cell Biol. 1999, 77, 345–350. [Google Scholar] [CrossRef] [PubMed]
- Jewell, J.L.; Kim, Y.C.; Russell, R.C.; Yu, F.X.; Park, H.W.; Plouffe, S.W.; Tagliabracci, V.S.; Guan, K.L. Metabolism. Differential regulation of mTORC1 by leucine and glutamine. Science 2015, 347, 194–198. [Google Scholar] [CrossRef]
- Moro, T.; Brightwell, C.R.; Velarde, B.; Fry, C.S.; Nakayama, K.; Sanbongi, C.; Volpi, E.; Rasmussen, B.B. Whey protein hydrolysate increases amino acid uptake, mTORC1 signaling, and protein synthesis in skeletal muscle of healthy young men in a randomized crossover trial. J. Nutr. 2019, 149, 1149–1158. [Google Scholar] [CrossRef]
- Shimobayashi, M.; Hall, M.N. Multiple amino acid sensing inputs to mTORC1. Cell Res. 2016, 26, 7–20. [Google Scholar] [CrossRef]
- Condon, K.J.; Sabatini, D.M. Nutrient regulation of mTORC1 at a glance. J. Cell Sci. 2019, 132, jcs222570. [Google Scholar] [CrossRef]
- Melick, C.H.; Jewell. J.L. Regulation of mTORC1 by upstream stimuli. Genes 2020, 11, 989. [Google Scholar] [CrossRef]
- Raybuck, A.L.; Lee, K.; Cho, S.H.; Li, J.; Thomas, J.W.; Boothby, M.R. mTORC1 as a cell-intrinsic rheostat that shapes development, preimmune repertoire, and function of B lymphocytes. FASEB J. 2019, 33, 13202–13215. [Google Scholar] [CrossRef]
- Davis, T.A.; Nguyen, H.V.; Garcia-Bravo, R.; Fiorotto, M.L.; Jackson, E.M.; Lewis, D.S.; Lee, D.R.; Reeds, P.J. Amino acid composition of human milk is not unique. J. Nutr. 1994, 124, 1126–1132. [Google Scholar] [CrossRef]
- Millward, D.J.; Layman, D.K.; Tomé, D.; Schaafsma, G. Protein quality assessment: Impact of expanding understanding of protein and amino acid needs for optimal health. Am. J. Clin. Nutr. 2008, 87, 1576S–1581S. [Google Scholar]
- Boirie, Y.; Dangin, M.; Gachon, P.; Vasson, M.P.; Maubois, J.L.; Beaufrère, B. Slow and fast dietary proteins differently modulate postprandial protein accretion. Proc. Natl. Acad. Sci. USA 1997, 94, 14930–14935. [Google Scholar] [CrossRef]
- He, T.; Giuseppin, M.L. Slow and fast dietary proteins differentially modulate postprandial metabolism. Int. J. Food Sci. Nutr. 2014, 65, 386–390. [Google Scholar] [CrossRef]
- Boutrou, R.; Gaudichon, C.; Dupont, D.; Jardin, J.; Airinei, G.; Marsset-Baglieri, A.; Benamouzig, R.; Tomé, D.; Leonil, J. Sequential release of milk protein-derived bioactive peptides in the jejunum in healthy humans. Am. J. Clin. Nutr. 2013, 97, 1314–1323. [Google Scholar] [CrossRef]
- Mahé, S.; Roos, N.; Benamouzig, R.; Davin, L.; Luengo, C.; Gagnon, L.; Gaussergès, N.; Rautureau, J.; Tomé, D. Gastrojejunal kinetics and the digestion of [15N]beta-lactoglobulin and casein in humans: The influence of the nature and quantity of the protein. Am. J. Clin. Nutr. 1996, 63, 546–552. [Google Scholar] [CrossRef]
- Kanai, Y.; Segawa, H.; Miyamoto, K.; Uchino, H.; Takeda, E.; Endou, H. Expression cloning and characterization of a transporter for large neutral amino acids activated by the heavy chain of 4F2 antigen (CD98). J. Biol. Chem. 1998, 273, 23629–23632. [Google Scholar] [CrossRef]
- Jigjidkhorloo, N.; Kanekura, K.; Matsubayashi, J.; Akahane, D.; Fujita, K.; Oikawa, K.; Kurata, A.; Takanashi, M.; Endou, H.; Nagao, T.; et al. Expression of L-type amino acid transporter 1 is a poor prognostic factor for non-Hodgkin’s lymphoma. Sci. Rep. 2021, 11, 21638. [Google Scholar] [CrossRef]
- Le, A.; Lane, A.N.; Hamaker, M.; Bose, S.; Gouw, A.; Barbi, J.; Tsukamoto, T.; Rojas, C.J.; Slusher, B.S.; Zhang, H.; et al. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab. 2012, 15, 110–121. [Google Scholar]
- Li, M.; Chiang, Y.L.; Lyssiotis, C.A.; Teater, M.R.; Hong, J.Y.; Shen, H.; Wang, L.; Hu, J.; Jing, H.; Chen, Z.; et al. Non-oncogene addiction to SIRT3 plays a critical role in lymphomagenesis. Cancer Cell. 2019, 35, 916–931. [Google Scholar] [CrossRef]
- Durán, R.V.; Hall, M.N. Glutaminolysis feeds mTORC1. Cell Cycle 2012, 11, 4107–4108. [Google Scholar] [CrossRef] [PubMed]
- Wei, P.; Bott, A.J.; Cluntun, A.A.; Morgan, J.T.; Cunningham, C.N.; Schell, J.C.; Ouyang, Y.; Ficarro, S.B.; Marto, J.A.; Danial, N.N.; et al. Mitochondrial pyruvate supports lymphoma proliferation by fueling a glutamate pyruvate transaminase 2-dependent glutaminolysis pathway. Sci. Adv. 2022, 8, eabq0117. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Teater, M.R.; Hong, J.Y.; Park, N.R.; Duy, C.; Shen, H.; Wang, L.; Chen, Z.; Cerchietti, L.; Davidson, S.M.; et al. Translational activation of ATF4 through mitochondrial anaplerotic metabolic pathways is required for DLBCL growth and survival. Blood Cancer Discov. 2022, 3, 50–65. [Google Scholar] [CrossRef] [PubMed]
- Durán, R.V.; Oppliger, W.; Robitaille, A.M.; Heiserich, L.; Skendaj, R.; Gottlieb, E.; Hall, M.N. Glutaminolysis activates Rag-mTORC1 signaling. Mol. Cell. 2012, 47, 349–358. [Google Scholar] [CrossRef]
- Lenders, C.M.; Liu, S.; Wilmore, D.W.; Sampson, L.; Dougherty, L.W.; Spiegelman, D.; Willett, W.C. Evaluation of a novel food composition database that includes glutamine and other amino acids derived from gene sequencing data. Eur. J. Clin. Nutr. 2009, 63, 1433–1439. [Google Scholar] [CrossRef]
- Baxter, J.H.; Phillips, R.R.; Dowlati, L.; Johns, P.W. Glutamine in commercial liquid nutritional products. J. Agric. Food Chem. 2004, 52, 4963–4968. [Google Scholar] [CrossRef]
- Ricci, J.E.; Chiche, J. Metabolic reprogramming of Non-Hodgkin’s B-cell lymphomas and potential therapeutic strategies. Front. Oncol. 2018, 8, 556. [Google Scholar] [CrossRef]
- Mlynarczyk, C.; Fontán, L.; Melnick, A. Germinal center-derived lymphomas: The darkest side of humoral immunity. Immunol. Rev. 2019, 288, 214–239. [Google Scholar] [CrossRef]
- Fontan, L.; Goldstein, R.; Casalena, G.; Durant, M.; Teater, M.R.; Wilson, J.; Phillip, J.; Xia, M.; Shah, S.; Us, I.; et al. Identification of MALT1 feedback mechanisms enables rational design of potent antilymphoma regimens for ABC-DLBCL. Blood 2021, 137, 788–800. [Google Scholar] [CrossRef]
- Ferch, U.; Kloo, B.; Gewies, A.; Pfänder, V.; Düwel, M.; Peschel, C.; Krappmann, D.; Ruland, J. Inhibition of MALT1 protease activity is selectively toxic for activated B cell-like diffuse large B cell lymphoma cells. J. Exp. Med. 2009, 206, 2313–2320. [Google Scholar] [CrossRef]
- Hailfinger, S.; Lenz, G.; Ngo, V.; Posvitz-Fejfar, A.; Rebeaud, F.; Guzzardi, M.; Penas, E.M.; Dierlamm, J.; Chan, W.C.; Staudt, L.M.; et al. Essential role of MALT1 protease activity in activated B cell-like diffuse large B-cell lymphoma. Proc. Natl. Acad. Sci. USA 2009, 106, 19946–19951, Erratum in: Proc. Natl. Acad. Sci. USA 2013, 110, 2677. [Google Scholar] [CrossRef]
- Phelan, J.D.; Young, R.M.; Webster, D.E.; Roulland, S.; Wright, G.W.; Kasbekar, M.; Shaffer, A.L.; Ceribelli, M.; Wang, J.Q.; Schmitz, R.; et al. A multiprotein supercomplex controlling oncogenic signalling in lymphoma. Nature 2018, 560, 387–391. [Google Scholar] [CrossRef]
- Horvilleur, E.; Sbarrato, T.; Hill, K.; Spriggs, R.V.; Screen, M.; Goodrem, P.J.; Sawicka, K.; Chaplin, L.C.; Touriol, C.; Packham, G.; et al. A role for eukaryotic initiation factor 4B overexpression in the pathogenesis of diffuse large B-cell lymphoma. Leukemia 2014, 28, 1092–1102. [Google Scholar] [CrossRef]
- Ruggero, D.; Montanaro, L.; Ma, L.; Xu, W.; Londei, P.; Cordon-Cardo, C.; Pandolfi, P.P. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat. Med. 2004, 10, 484–486. [Google Scholar] [CrossRef]
- Wanner, K.; Hipp, S.; Oelsner, M.; Ringshausen, I.; Bogner, C.; Peschel, C.; Decker, T. Mammalian target of rapamycin inhibition induces cell cycle arrest in diffuse large B cell lymphoma (DLBCL) cells and sensitises DLBCL cells to rituximab. Br. J. Haematol. 2006, 134, 475–484. [Google Scholar] [CrossRef]
- Zhao, X.F.; Gartenhaus, R.B. Phospho-p70S6K and cdc2/cdk1 as therapeutic targets for diffuse large B-cell lymphoma. Expert Opin. Ther. Targets 2009, 13, 1085–1093. [Google Scholar] [CrossRef]
- Xu, Z.Z.; Xia, Z.G.; Wang, A.H.; Wang, W.F.; Liu, Z.Y.; Chen, L.Y.; Li, J.M. Activation of the PI3K/AKT/mTOR pathway in diffuse large B cell lymphoma: Clinical significance and inhibitory effect of rituximab. Ann. Hematol. 2013, 92, 1351–1358. [Google Scholar] [CrossRef]
- Xu, Z.Z.; Wang, W.F.; Fu, W.B.; Wang, A.H.; Liu, Z.Y.; Chen, L.Y.; Guo, P.; Li, J.M. Combination of rituximab and mammalian target of rapamycin inhibitor everolimus (RAD001) in diffuse large B-cell lymphoma. Leuk. Lymphoma 2014, 55, 1151–1157. [Google Scholar] [CrossRef]
- Majchrzak, A.; Witkowska, M.; Smolewski, P. Inhibition of the PI3K/Akt/mTOR signaling pathway in diffuse large B-cell lymphoma: Current knowledge and clinical significance. Molecules 2014, 19, 14304–14315. [Google Scholar] [CrossRef]
- Merli, M.; Ferrario, A.; Maffioli, M.; Arcaini, L.; Passamonti, F. Everolimus in diffuse large B-cell lymphomas. Future Oncol. 2015, 11, 373–383. [Google Scholar] [CrossRef]
- Mao, Y.; Xu, L.; Wang, J.; Zhang, L.; Hou, N.; Xu, J.; Wang, L.; Yang, S.; Chen, Y.; Xiong, L.; et al. ROR1 associates unfavorable prognosis and promotes lymphoma growth in DLBCL by affecting PI3K/Akt/mTOR signaling pathway. Biofactors 2019, 45, 416–426. [Google Scholar] [PubMed]
- Taylor, J.; Yeomans, A.M.; Packha, G. Targeted inhibition of mRNA translation initiation factors as a novel therapeutic strategy for mature B-cell neoplasms. Explor. Target Antitumor Ther. 2020, 1, 3–25. [Google Scholar] [PubMed]
- Hu, K.; Li, B.; Ma, R.; Yi, H.; Xu, Z.; Peng, Y.; Yu, D.; Wu, H.; Cheng, T.; Lu, Y.; et al. Anti-DLBCL efficacy of DCZ0825 in vitro and in vivo: Involvement of the PI3K–AKT–mTOR/JNK pathway. Acta Biochim. Biophys. Sin. 2021, 53, 575–583. [Google Scholar] [CrossRef] [PubMed]
- Yamin, H.; Barnea, M.; Genzer, Y.; Chapnik, N.; Froy, O. Long-term commercial cow’s milk consumption and its effects on metabolic parameters associated with obesity in young mice. Mol. Nutr. Food Res. 2014, 58, 1061–1068. [Google Scholar] [CrossRef] [PubMed]
- D’Hulst, G.; Masschelein, E.; De Bock, K. Dampened muscle mTORC1 response following ingestion of high-quality plant-based protein and insect protein compared to whey. Nutrients 2021, 13, 1396. [Google Scholar] [CrossRef]
- Tanaka, S.; Baba, Y. B cell receptor signaling. Adv. Exp. Med. Biol. 2020, 1254, 23–36. [Google Scholar]
- Küppers, R. Mechanisms of B-cell lymphoma pathogenesis. Nat. Rev. Cancer. 2005, 5, 251–262. [Google Scholar] [CrossRef]
- Thurner, L.; Hartmann, S.; Neumann, F.; Hoth, M.; Stilgenbauer, S.; Küppers, R.; Preuss, K.D.; Bewarder, M. Role of specific B-cell receptor antigens in lymphomagenesis. Front. Oncol. 2020, 10, 604685. [Google Scholar]
- Thurner, L.; Hartmann, S.; Bewarder, M.; Fadle, N.; Regitz, E.; Schormann, C.; Quiroga, N.; Kemele, M.; Klapper, W.; Rosenwald, A.; et al. Identification of the atypically modified autoantigen Ars2 as the target of B-cell receptors from activated B-cell-type diffuse large B-cell lymphoma. Haematologica 2021, 106, 2224–2232. [Google Scholar]
- Wilson, W.H.; Young, R.M.; Schmitz, R.; Yang, Y.; Pittaluga, S.; Wright, G.; Lih, C.J.; Williams, P.M.; Shaffer, A.L.; Gerecitano, J.; et al. Targeting B cell receptor signaling with ibrutinib in diffuse large B cell lymphoma. Nat. Med. 2015, 21, 922–926. [Google Scholar] [CrossRef]
- Xu, W.; Berning, P.; Lenz, G. Targeting B-cell receptor and PI3K signaling in diffuse large B-cell lymphoma. Blood 2021, 138, 1110–1119. [Google Scholar]
- Wal, J.M. Cow‘s milk proteins/allergens. Ann. Allergy Asthma Immunol. 2002, 89, 3–10. [Google Scholar]
- Wal, J.M. Bovine milk allergenicity. Ann. Allergy Asthma Immunol. 2004, 93, S2–S11. [Google Scholar]
- Caira, S.; Pinto, G.; Picariello, G.; Vitaglione, P.; De Pascale, S.; Scaloni, A.; Addeo, F. In vivo absorptomics: Identification of bovine milk-derived peptides in human plasma after milk intake. Food Chem. 2022, 385, 132663. [Google Scholar]
- Mousan, G.; Kamat, D. Cow’s milk protein allergy. Clin. Pediatr. 2016, 55, 1054–1063. [Google Scholar]
- Tedner, S.G.; Asarnoj, A.; Thulin, H.; Westman, M.; Konradsen, J.R.; Nilsson, C. Food allergy and hypersensitivity reactions in children and adults—A review. J. Intern. Med. 2022, 291, 283–302. [Google Scholar]
- Lam, H.Y.; van Hoffen, E.; Michelsen, A.; Guikers, K.; van der Tas, C.H.; Bruijnzeel-Koomen, C.A.; Knulst, A.C. Cow’s milk allergy in adults is rare but severe: Both casein and whey proteins are involved. Clin. Exp. Allergy 2008, 38, 995–1002. [Google Scholar]
- Laugesen, M.; Elliott, R. Ischaemic heart disease, type 1 diabetes, and cow milk A1 beta-casein. N. Z. Med. J. 2003, 116, U295. [Google Scholar]
- Neyestani, T.R.; Djalali, M.; Pezeshki, M.; Siassi, F.; Eshraghian, M.R.; Rajab, A.; Keshavarz, A. Serum antibodies to the major proteins found in cow’s milk of Iranian patients with type 1 diabetes mellitus. Diabetes Nutr. Metab. 2004, 17, 76–83. [Google Scholar]
- Monetini, L.; Cavallo, M.G.; Manfrini, S.; Stefanini, L.; Picarelli, A.; Di Tola, M.; Petrone, A.; Bianchi, M.; La Presa, M.; Di Giulio, C.; et al. Antibodies to bovine beta-casein in diabetes and other autoimmune diseases. Horm. Metab. Res. 2002, 34, 455–459. [Google Scholar]
- Lamb, M.M.; Miller, M.; Seifert, J.A.; Frederiksen, B.; Kroehl, M.; Rewers, M.; Norris, J.M. The effect of childhood cow’s milk intake and HLA-DR genotype on risk of islet autoimmunity and type 1 diabetes: The Diabetes Autoimmunity Study in the Young. Pediatr. Diabetes 2015, 16, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Vieira Borba, V.; Lerner, A.; Matthias, T.; Shoenfeld, Y. Bovine milk proteins as a trigger for autoimmune diseases: Myth or reality? Int. J. Celiac Dis. 2020, 8, 10–21. [Google Scholar]
- Wang, Y.; Liu, X.; Yan, P.; Bi, Y.; Liu, Y.; Zhang, Z.J. Association between type 1 and type 2 diabetes and risk of non-Hodgkin’s lymphoma: A meta-analysis of cohort studies. Diabetes Metab. 2020, 46, 8–19. [Google Scholar] [PubMed]
- Lidén, M.; Kristjánsson, G.; Valtysdottir, S.; Venge, P.; Hällgren, R. Cow’s milk protein sensitivity assessed by the mucosal patch technique is related to irritable bowel syndrome in patients with primary Sjögren’s syndrome. Clin. Exp. Allergy 2008, 38, 929–935. [Google Scholar] [PubMed]
- Smedby, K.; Vajdic, C.M.; Falster, M.; Engels, E.A.; Martínez-Maza, O.; Turner, J.; Hjalgrim, H.; Vineis, P.; Seniori Costantini, A.; Bracci, P.M.; et al. Autoimmune disorders and risk of non-Hodgkin lymphoma subtypes: A pooled analysis within the InterLymph Consortium. Blood 2008, 111, 4029–4038. [Google Scholar]
- Järvinen, K.M.; Beyer, K.; Vila, L.; Chatchatee, P.; Busse, P.J.; Sampson, H.A. B-cell epitopes as a screening instrument for persistent cow’s milk allergy. J. Allergy Clin. Immunol. 2002, 110, 293–297. [Google Scholar]
- Chatchatee, P.; Järvinen, K.M.; Bardina, L.; Vila, L.; Beyer, K.; Sampson, H.A. Identification of IgE and IgG binding epitopes on beta- and kappa-casein in cow’s milk allergic patients. Clin. Exp. Allergy 2001, 31, 1256–1262. [Google Scholar] [CrossRef]
- Enomoto, A.; Shon, D.H.; Aoki, Y.; Yamauchi, K.; Kaminogawa, S. Antibodies raised against peptide fragments of bovine alpha s1-casein cross-react with the intact protein only when the peptides contain both B and T cell determinants. Mol. Immunol. 1990, 27, 581–586. [Google Scholar]
- Shon, D.H.; Enomoto, A.; Yamauchi, K.; Kaminogawa, S. Antibodies raised against peptide fragments of bovine alpha s1-casein cross-react with the native protein, but recognize sites distinct from the determinants on the protein. Eur. J. Immunol. 1991, 21, 1475–1480. [Google Scholar] [CrossRef]
- Fuc, E.; Złotkowska, D.; Stachurska, E.; Wróblewska, B. Immunoreactive properties of α-casein and κ-casein: Ex vivo and in vivo studies. J. Dairy Sci. 2018, 101, 10703–10713. [Google Scholar]
- Barati, M.; Jabbari, M.; Fathollahi, M.; Fathollahi, A.; Khaki, V.; Javanmardi, F.; Jazayeri, S.M.H.M.; Shabani, M.; Davoodi, S.H.; Huseyn, E.; et al. Evaluation of different types of milk proteins-derived epitopes using in-silico tools: A primarily study to propose a new definition for bioactive peptides. Food Sci. Technol. Camp. 2022, 42, e102821. [Google Scholar] [CrossRef]
- Lozano-Ojalvo, D.; Larré, C.; Klueber, J.; Gelencser, E.; Bueno-Diaz, C.; Diaz-Perales, A.; Benedé, S.; Bavaro, S.L.; Kuehn, A.; Hoffmann-Sommergruber, K.; et al. Are physicochemical properties shaping the allergenic potency of animal allergens? Clin. Rev. Allergy Immunol. 2022, 62, 1–36. [Google Scholar]
- Bogahawaththa, D.; Ashraf, R.; Chandrapala, J.; Donkor, O.; Vasiljevic, T. In vitro immunogenicity of various native and thermally processed bovine milk proteins and their mixtures. J. Dairy Sci. 2018, 101, 8726–8736. [Google Scholar] [CrossRef]
- Morisawa, Y.; Kitamura, A.; Ujihara, T.; Zushi, N.; Kuzume, K.; Shimanouchi, Y.; Tamura, S.; Wakiguchi, H.; Saito, H.; Matsumoto, K. Effect of heat treatment and enzymatic digestion on the B cell epitopes of cow’s milk proteins. Clin. Exp. Allergy 2009, 39, 918–925. [Google Scholar] [CrossRef]
- Teras, L.R.; Patel, A.V.; Hildebrand, J.S.; Gapstur, S.M. Postmenopausal unopposed estrogen and estrogen plus progestin use and risk of non-Hodgkin lymphoma in the American Cancer Society Cancer Prevention Study-II Cohort. Leuk. Lymphoma 2013, 54, 720–725. [Google Scholar]
- Hasni, M.S.; Berglund, M.; Yakimchuk, K.; Guan, J.; Linderoth, J.; Amini, R.M.; Enblad, G.; Okret, S. Estrogen receptor β1 in diffuse large B-cell lymphoma growth and as a prognostic biomarker. Leuk. Lymphoma 2017, 58, 418–427. [Google Scholar] [CrossRef]
- Faknuam, S.; Assanasen, T.; Ruangvejvorachai, P.; Hanvivadhanakul, P.; Intragumtornchai, T.; Rojnuckarin, P. Estrogen receptor beta expression and prognosis of diffuse large B cell lymphoma. Hematology 2018, 23, 235–241. [Google Scholar]
- Langendonk, M.; de Jong, M.R.W.; Smit, N.; Seiler, J.; Reitsma, B.; Ammatuna, E.; Glaudemans, A.W.J.M.; van den Berg, A.; Huls, G.A.; Visser, L.; et al. Identification of the estrogen receptor beta as a possible new tamoxifen-sensitive target in diffuse large B-cell lymphoma. Blood Cancer J. 2022, 12, 36. [Google Scholar] [CrossRef]
- Katzenellenbogen, B.S. Biology and receptor interactions of estriol and estriol derivatives in vitro and in vivo. J. Steroid Biochem. 1984, 20, 1033–1037. [Google Scholar] [CrossRef]
- Barkhem, T.; Carlsson, B.; Nilsson, Y.; Enmark, E.; Gustafsson, J.; Nilsson, S. Differential response of estrogen receptor alpha and estrogen receptor beta to partial estrogen agonists/antagonists. Mol. Pharmacol. 1998, 54, 105–112. [Google Scholar] [CrossRef]
- Paterni, I.; Granchi, C.; Katzenellenbogen, J.A.; Minutolo, F. Estrogen receptors alpha (ERα) and beta (ERβ): Subtype-selective ligands and clinical potential. Steroids 2014, 90, 13–29. [Google Scholar] [CrossRef] [PubMed]
- Perkins, M.S.; Louw-du Toit, R.; Africander, D. A comparative characterization of estrogens used in hormone therapy via estrogen receptor (ER)-α and -β. J. Steroid Biochem. Mol. Biol. 2017, 174, 27–39. [Google Scholar] [CrossRef] [PubMed]
- Mal, R.; Magner, A.; David, J.; Datta, J.; Vallabhaneni, M.; Kassem, M.; Manouchehri, J.; Willingham, N.; Stover, D.; Vandeusen, J.; et al. Estrogen receptor beta (ERβ): A ligand activated tumor suppressor. Front. Oncol. 2020, 10, 587386. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Liao, Y.; Chen, G.; Xu, L.; Zhang, C.; Ju, S.; Zhou, S. Estrogen upregulates the IGF-1 signaling pathway in lung cancer through estrogen receptor-β. Med. Oncol. 2012, 29, 2640–2648. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Liao, Y.; Xu, L.; Zhang, C.; Liu, Z.; Deng, Y.; Jiang, Z.; Fu, S.; Chen, Z.; Zhou, S. Estrogen and insulin-like growth factor 1 synergistically promote the development of lung adenocarcinoma in mice. Int. J. Cancer 2013, 133, 2473–2482. [Google Scholar] [CrossRef]
- Heap, R.B.; Hamon, M. Oestrone sulphate in milk as an indicator of a viable conceptus in cows. Br. Vet. J. 1979, 135, 355–363. [Google Scholar] [CrossRef]
- Snoj, T.; Zuzek, M.C.; Cebulj-Kadunc, N.; Majdic, G. Short communication: Heat treatment and souring do not affect milk estrone and 17β-estradiol concentrations. J. Dairy Sci. 2018, 101, 61–65. [Google Scholar] [CrossRef]
- Maruyama, K.; Oshima, T.; Ohyama, K. Exposure to exogenous estrogen through intake of commercial milk produced from pregnant cows. Pediatr. Int. 2010, 52, 28–33. [Google Scholar] [CrossRef]
- Farlow, D.W.; Xu, X.; Veenstra, T.D. Comparison of estrone and 17β-estradiol levels in commercial goat and cow milk. J. Dairy Sci. 2012, 95, 1699–1708, Erratum in: J. Dairy Sci. 2012, 95, 4732. [Google Scholar] [CrossRef]
- Goyon, A.; Cai, J.Z.; Kraehenbuehl, K.; Hartmann, C.; Shao, B.; Mottier, P. Determination of steroid hormones in bovine milk by LC-MS/MS and their levels in Swiss Holstein cow milk. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 2016, 33, 804–816. [Google Scholar] [CrossRef]
- Pape-Zambito, D.A.; Roberts, R.F.; Kensinger, R.S. Estrone and 17beta-estradiol concentrations in pasteurized-homogenized milk and commercial dairy products. J. Dairy Sci. 2010, 93, 2533–2540. [Google Scholar] [CrossRef]
- Pape-Zambito, D.A.; Magliaro, A.L.; Kensinger, R.S. Concentrations of 17beta-estradiol in Holstein whole milk. J. Dairy Sci. 2007, 90, 3308–3313. [Google Scholar] [CrossRef]
- Hartmann, S.; Lacorn, M.; Steinhart, H. Natural occurrence of steroid hormones in food. Food Chem. 1989, 62, 7–20. [Google Scholar] [CrossRef]
- Cavaliere, C.; Capriotti, A.L.; Foglia, P.; Piovesana, S.; Samperi, R.; Ventura, S.; Laganà, A. Natural estrogens in dairy products: Determination of free and conjugated forms by ultra high performance liquid chromatography with tandem mass spectrometry. J. Sep. Sci. 2015, 38, 3599–3606. [Google Scholar] [CrossRef]
- Fenichel, P.; Chevalier, N.; Brucker-Davis, F. Bisphenol A: An endocrine and metabolic disruptor. Ann. Endocrinol. 2013, 74, 211–220. [Google Scholar] [CrossRef]
- Rochester, J.R. Bisphenol A and human health: A review of the literature. Reprod. Toxicol. 2013, 42, 132–155. [Google Scholar] [CrossRef]
- Abraham, A.; Chakraborty, P. A review on sources and health impacts of bisphenol A. Rev. Environ. Health 2020, 35, 201–210. [Google Scholar] [CrossRef]
- Rogers, J.A.; Metz, L.; Yong, V.W. Review: Endocrine disrupting chemicals and immune responses: A focus on bisphenol-A and its potential mechanisms. Mol. Immunol. 2013, 53, 421–430. [Google Scholar]
- Kimber, I. Bisphenol A and immunotoxic potential: A commentary. Regul. Toxicol. Pharmacol. 2017, 90, 358–363. [Google Scholar] [CrossRef]
- Segovia-Mendoza, M.; Nava-Castro, K.E.; Palacios-Arreola, M.I.; Garay-Canales, C.; Morales-Montor, J. How microplastic components influence the immune system and impact on children health: Focus on cancer. Birth Defects Res. 2020, 12, 1341–1361. [Google Scholar] [CrossRef]
- Ge, X.; Lv, X.; Feng, L.; Liu, X.; Wang, X. High expression and nuclear localization of β-catenin in diffuse large B-cell lymphoma. Mol. Med. Rep. 2012, 5, 1433–1437. [Google Scholar] [PubMed]
- Zhang, D.Y.; Wang, H.J.; Tan, Y.Z. Wnt/β-catenin signaling induces the aging of mesenchymal stem cells through the DNA damage response and the p53/p21 pathway. PLoS ONE 2011, 6, e21397. [Google Scholar] [CrossRef] [PubMed]
- Derenzini, E.; Agostinelli, C.; Imbrogno, E.; Iacobucci, I.; Casadei, B.; Brighenti, E.; Righi, S.; Fuligni, F.; Ghelli Luserna Di Rorà, A.; Ferrari, A.; et al. Constitutive activation of the DNA damage response pathway as a novel therapeutic target in diffuse large B-cell lymphoma. Oncotarget 2015, 6, 6553–6569. [Google Scholar] [CrossRef] [PubMed]
- Dupont, J.; Fernandez, A.M.; Glackin, C.A.; Helman, L.; LeRoith, D. Insulin-like growth factor 1 (IGF-1)-induced twist expression is involved in the anti-apoptotic effects of the IGF-1 receptor. J. Biol. Chem. 2001, 276, 26699–26707. [Google Scholar] [CrossRef]
- Lemma, S.; Karihtala, P.; Haapasaari, K.M.; Jantunen, E.; Soini, Y.; Bloigu, R.; Pasanen, A.K.; Turpeenniemi-Hujanen, T.; Kuittinen, O. Biological roles and prognostic values of the epithelial-mesenchymal transition-mediating transcription factors Twist, ZEB1 and Slug in diffuse large B-cell lymphoma. Histopathology 2013, 62, 326–333. [Google Scholar] [CrossRef]
- Davis, R.E.; Brown, K.D.; Siebenlist, U.; Staudt, L.M. Constitutive nuclear factor kappaB activity is required for survival of activated B cell-like diffuse large B cell lymphoma cells. J. Exp. Med. 2001, 194, 1861–1874. [Google Scholar] [CrossRef]
- Compagno, M.; Lim, W.K.; Grunn, A.; Nandula, S.V.; Brahmachary, M.; Shen, Q.; Bertoni, F.; Ponzoni, M.; Scandurra, M.; Califano, A.; et al. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature 2009, 459, 717–721. [Google Scholar] [CrossRef]
- Nagel, D.; Vincendeau, M.; Eitelhuber, A.C.; Krappmann, D. Mechanisms and consequences of constitutive NF-κB activation in B-cell lymphoid malignancies. Oncogene 2014, 33, 5655–5665. [Google Scholar] [CrossRef]
- Zhu, J.; Jiang, L.; Liu, Y.; Qian, W.; Liu, J.; Zhou, J.; Gao, R.; Xiao, H.; Wang, J. MAPK and NF-κB pathways are involved in bisphenol A-induced TNF-α and IL-6 production in BV2 microglial cells. Inflammation 2015, 38, 637–648. [Google Scholar] [CrossRef]
- Jalal, N.; Surendranath, A.R.; Pathak, J.L.; Yu, S.; Chung, C.Y. Bisphenol A (BPA) the mighty and the mutagenic. Toxicol. Rep. 2017, 5, 76–84. [Google Scholar] [CrossRef]
- Yu, M.; Xu, Y.; Li, M.; Li, D.; Lu, Y.; Yu, D.; Du, W. Bisphenol A accelerates meiotic progression in embryonic chickens via the estrogen receptor β signaling pathway. Gen. Comp. Endocrinol. 2018, 259, 66–75. [Google Scholar] [CrossRef]
- Burkhardt, F.; Schulz, S.D.; Hellwig, E.; Vach, K.; Tomakidi, P.; Polydorou, O. Low-dose bisphenol A and its analogues bisphenol F and S activate estrogen receptor ß and slightly modulate genes in human gingival keratinocytes. Dent. Mater. 2021, 37, 625–635. [Google Scholar] [CrossRef]
- Wen, X.; Zhu, M.; Li, Z.; Li, T.; Xu, X. Dual effects of bisphenol A on wound healing, involvement of estrogen receptor β. Ecotoxicol. Environ. Saf. 2022, 231, 113207. [Google Scholar] [CrossRef]
- Zhang, Y.; Wei, F.; Zhang, J.; Hao, L.; Jiang, J.; Dang, L.; Mei, D.; Fan, S.; Yu, Y.; Jiang, L. Bisphenol A and estrogen induce proliferation of human thyroid tumor cells via an estrogen-receptor-dependent pathway. Arch. Biochem. Biophys. 2017, 633, 29–39. [Google Scholar] [CrossRef]
- Farahani, M.; Rezaei-Tavirani, M.; Arjmand, B. A systematic review of microRNA expression studies with exposure to bisphenol A. J. Appl. Toxicol. 2021, 41, 4–19. [Google Scholar] [CrossRef]
- Ferrante, M.; Cristaldi, A.; Oliveri Conti, G. Oncogenic role of miRNA in environmental exposure to plasticizers: A systematic review. J. Pers. Med. 2021, 11, 500. [Google Scholar] [CrossRef]
- Hui, L.; Li, H.; Lu, G.; Chen, Z.; Sun, W.; Shi, Y.; Fu, Z.; Huang, B.; Zhu, X.; Lu, W.; et al. Low dose of bisphenol A modulates ovarian cancer gene expression profile and promotes epithelial to mesenchymal transition via canonical Wnt pathway. Toxicol. Sci. 2018, 164, 527–538. [Google Scholar] [CrossRef]
- Sabry, R.; Saleh, A.C.; Stalker, L.; LaMarre, J.; Favetta, L.A. Effects of bisphenol A and bisphenol S on microRNA expression during bovine (Bos taurus) oocyte maturation and early embryo development. Reprod. Toxicol. 2021, 99, 96–108. [Google Scholar] [CrossRef]
- Oldenburg, J.; Fürhacker, M.; Hartmann, C.; Steinbichl, P.; Banaderakhshan, R.; Haslberger, A. Different bisphenols induce non-monotonous changes in miRNA expression and LINE-1 methylation in two cell lines. Environ. Epigenet. 2021, 7, dvab011. [Google Scholar] [CrossRef]
- Mercogliano, R.; Santonicola, S. Investigation on bisphenol A levels in human milk and dairy supply chain: A review. Food Chem. Toxicol. 2018, 114, 98–107. [Google Scholar] [CrossRef]
- Herrero, L.; Quintanilla-López, J.E.; Fernández, M.A.; Gómara, B. Plasticisers and preservatives in commercial milk products: A comprehensive study on packages used in the Spanish market. Food Chem. 2021, 338, 128031. [Google Scholar] [PubMed]
- Santonicola, S.; Ferrante, M.C.; Murru, N.; Gallo, P.; Mercogliano, R. Hot topic: Bisphenol A in cow milk and dietary exposure at the farm level. J. Dairy Sci. 2019, 102, 1007–1013. [Google Scholar] [PubMed]
- Mercogliano, R.; Santonicola, S.; Albrizio, S.; Ferrante, M.C. Occurrence of bisphenol A in the milk chain: A monitoring model for risk assessment at a dairy company. J. Dairy Sci. 2021, 104, 5125–5132. [Google Scholar] [PubMed]
- Farooq, M.U.; Jalees, M.I.; Qurat-Ul-Ain, H.G.; Anis, M.; Islam, U. Health risk assessment of endocrine disruptor bisphenol A leaching from plastic bottles of milk and soft drinks. Environ. Sci. Pollut. Res. Int. 2021, 28, 57090–57098. [Google Scholar] [PubMed]
- Högfeldt, T.; Jaing, C.; Loughlin, K.M.; Thissen, J.; Gardner, S.; Bahnassy, A.A.; Gharizadeh, B.; Lundahl, J.; Österborg, A.; Porwit, A.; et al. Differential expression of viral agents in lymphoma tissues of patients with ABC diffuse large B-cell lymphoma from high and low endemic infectious disease regions. Oncol. Lett. 2016, 12, 2782–2788. [Google Scholar] [CrossRef]
- Bilajac, E.; Mahmutović, L.; Lundstrom, K.; Glamočlija, U.; Šutković, J.; Sezer, A.; Hromić-Jahjefendić, A. Viral agents as potential drivers of diffuse large B-Cell lymphoma tumorigenesis. Viruses 2022, 14, 2105. [Google Scholar]
- Hong, J.Y.; Ryu, K.J.; Park, C.; Hong, M.; Ko, Y.H.; Kim, W.S.; Kim, S.J. Clinical impact of serum survivin positivity and tissue expression of EBV-encoded RNA in diffuse large B-cell lymphoma patients treated with rituximab-CHOP. Oncotarget 2017, 8, 13782–13791. [Google Scholar]
- Castillo, J.J.; Beltran, B.E.; Miranda, R.N.; Young, K.H.; Chavez, J.C.; Sotomayor, E.M. EBV-positive diffuse large B-cell lymphoma, not otherwise specified: 2018 update on diagnosis, risk-stratification and management. Am. J. Hematol. 2018, 93, 953–962. [Google Scholar]
- Beltran, B.E.; Castro, D.; Paredes, S.; Miranda, R.N.; Castillo, J.J. EBV-positive diffuse large B-cell lymphoma, not otherwise specified: 2020 update on diagnosis, risk-stratification and management. Am. J. Hematol. 2020, 95, 435–445. [Google Scholar]
- Marques-Piubelli, M.L.; Salas, Y.I.; Pachas, C.; Becker-Hecker, R.; Vega, F.; Miranda, R.N. Epstein-Barr virus-associated B-cell lymphoproliferative disorders and lymphomas: A review. Pathology 2020, 52, 40–52. [Google Scholar]
- Shibusawa, M.; Kidoguchi, K.; Tanimoto, T. Chapter 2: Epstein-Barr Virus-Positive Diffuse Large B Cell Lymphoma. In Lymphoma; Gallamini, A., Juweid, M., Eds.; Exon Publications: Brisbane, Australia, 2021. [Google Scholar]
- Zhao, C.X.; Wen, J.J.; Fu, D.; Xu, P.P.; Cheng, S.; Wang, L.; Wang, C.F.; Fei, X.C.; Wang, X.; Zhou, J.F.; et al. Clinical and molecular features of Epstein-Barr virus-positive diffuse large B-cell lymphoma: Results in a multi-center trial. Clin. Transl. Med. 2021, 11, e539. [Google Scholar] [CrossRef]
- Chabay, P. Advances in the pathogenesis of EBV-associated diffuse large B cell lymphoma. Cancers 2021, 13, 2717. [Google Scholar]
- Satou, A.; Nakamura, S. EBV-positive B-cell lymphomas and lymphoproliferative disorders: Review from the perspective of immune escape and immunodeficiency. Cancer Med. 2021, 10, 6777–6785. [Google Scholar]
- Malpica, L.; Marques-Piubelli, M.L.; Beltran, B.E.; Chavez, J.C.; Miranda, R.N.; Castillo, J.J. EBV-positive diffuse large B-cell lymphoma, not otherwise specified: 2022 update on diagnosis, risk-stratification, and management. Am. J. Hematol. 2022, 97, 951–965. [Google Scholar]
- Carbone, A.; Cesarman, E.; Spina, M.; Gloghini, A.; Schulz, T.F. HIV-associated lymphomas and gamma-herpesviruses. Blood 2009, 113, 1213–1224. [Google Scholar]
- Gloghini, A.; Dolcetti, R.; Carbone, A. Lymphomas occurring specifically in HIV-infected patients: From pathogenesis to pathology. Semin. Cancer Biol. 2013, 23, 457–467. [Google Scholar]
- Baptista, M.J.; Garcia, O.; Morgades, M.; Gonzalez-Barca, E.; Miralles, P.; Lopez-Guillermo, A.; Abella, E.; Moreno, M.; Sancho, J.M.; Feliu, E.; et al. HIV-infection impact on clinical-biological features and outcome of diffuse large B-cell lymphoma treated with R-CHOP in the combination antiretroviral therapy era. AIDS 2015, 29, 811–818. [Google Scholar]
- Besson, C.; Lancar, R.; Prevot, S.; Algarte-Genin, M.; Delobel, P.; Bonnet, F.; Meyohas, M.C.; Partisani, M.; Oberic, L.; Gabarre, J.; et al. Outcomes for HIV-associated diffuse large B-cell lymphoma in the modern combined antiretroviral therapy era. AIDS 2017, 31, 2493–2501. [Google Scholar]
- Wu, J.; Miao, Y.; Qian, C.; Tao, P.; Wang, X.; Dong, X.; Li, X.; Lou, J.; Liang, J.; Xu, W.; et al. Clinical characteristics and outcomes in HIV-associated diffuse large B-cell lymphoma in China: A retrospective single-center study. J. Cancer 2021, 12, 2903–2911. [Google Scholar] [CrossRef]
- Ren, W.; Ye, X.; Su, H.; Li, W.; Liu, D.; Pirmoradian, M.; Wang, X.; Zhang, B.; Zhang, Q.; Chen, L.; et al. Genetic landscape of hepatitis B virus-associated diffuse large B-cell lymphoma. Blood 2018, 131, 2670–2681, Erratum: Blood 2019, 133, 620. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, H.; Pan, S.; Hu, T.; Shen, J.; Zheng, H.; Xie, S.; Xie, Y.; Lu, R.; Guo, L. Capable infection of hepatitis B virus in diffuse large B-cell lymphoma. J. Cancer 2018, 9, 1575–1581. [Google Scholar] [CrossRef] [PubMed]
- Visentini, M.; Casato, M. HBV messing with the B-cell genome leads to DLBCL. Blood 2018, 131, 2602–2603. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Wuchter, P.; Egerer, G.; Kriegsmann, M.; Kommoss, F.K.F.; Witzens-Harig, M.; Kriegsmann, K. Serological hepatitis B virus (HBV) activity in patients with HBV infection and B-cell non-Hodgkin’s lymphoma. Eur. J. Haematol. 2020, 104, 469–475. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Wang, L.; Nie, M.; Wang, Y.; Dong, S.; Ren, W.; Li, G.; Li, Z.M.; Wu, K.; Pan-Hammarström, Q. A single-cell atlas of diffuse large B cell lymphoma. Cell Rep. 2022, 39, 110713. [Google Scholar] [CrossRef] [PubMed]
- Visco, C.; Finotto, S. Hepatitis C virus and diffuse large B-cell lymphoma: Pathogenesis, behavior and treatment. World J. Gastroenterol. 2014, 20, 11054–11061. [Google Scholar] [CrossRef]
- Saleh, L.M.; Canioni, D.; Shamaa, S.; El-Zaafarany, M.; Emarah, Z.; Abdel-Aziz, S.; Eladle, E.; Abdelaziz, A.; Hermine, O.; Besson, C.; et al. High prevalence of hepatitis C virus among B-cell non Hodgkin lymphoma patients in Mansoura Region (Egypt), ANRS 12263 Study. Mediterr. J. Hematol. Infect. Dis. 2019, 11, e2019011. [Google Scholar] [CrossRef]
- Merli, M.; Frigeni, M.; Alric, L.; Visco, C.; Besson, C.; Mannelli, L.; Di Rocco, A.; Ferrari, A.; Farina, L.; Pirisi, M.; et al. Direct-acting antivirals in hepatitis C virus-associated diffuse large B-cell lymphomas. Oncologist 2019, 24, e720–e729. [Google Scholar] [CrossRef]
- Beltran, B.E.; Quiñones, P.; Morales, D.; Revilla, J.C.; Alva, J.C.; Castillo, J.J. Diffuse large B-cell lymphoma in human T-lymphotropic virus type 1 carriers. Leuk. Res. Treat. 2012, 2012, 262363. [Google Scholar] [CrossRef]
- Amara, K.; Trimeche, M.; Ziadi, S.; Laatiri, A.; Hachana, M.; Sriha, B.; Mokni, M.; Korbi, S. Presence of simian virus 40 DNA sequences in diffuse large B-cell lymphomas in Tunisia correlates with aberrant promoter hypermethylation of multiple tumor suppressor genes. Int. J. Cancer 2007, 121, 2693–2702. [Google Scholar] [CrossRef]
- Amara, K.; Trimeche, M.; Ziadi, S.; Laatiri, A.; Mestiri, S.; Sriha, B.; Mokni, M.; Korbi, S. Presence of simian virus 40 in diffuse large B-cell lymphomas in Tunisia correlates with germinal center B-cell immunophenotype, t(14;18) translocation, and P53 accumulation. Mod. Pathol. 2008, 21, 282–296. [Google Scholar] [CrossRef]
- Samaka, R.M.; Aiad, H.A.; Kandil, M.A.; Asaad, N.Y.; Holah, N.S. The prognostic role and relationship between E2F1 and SV40 in diffuse large B-cell lymphoma of Egyptian patients. Anal. Cell Pathol. 2015, 2015, 919834. [Google Scholar] [CrossRef]
- Calabrò, M.L.; Sarid, R. Human herpesvirus 8 and lymphoproliferative disorders. Mediterr. J. Hematol. Infect. Dis. 2018, 10, e2018061. [Google Scholar] [CrossRef]
- Wen, K.W.; Wang, L.; Menke, J.R.; Damania, B. Cancers associated with human gammaherpesviruses. FEBS J. 2022, 289, 7631–7669. [Google Scholar] [CrossRef]
- Fenu, E.M.; Beaty, M.W.; O’Neill, T.E.; O’Neill, S.S. Cardiac involvement by human herpesvirus 8-positive diffuse large B-cell lymphoma: An unusual presentation in a patient with human immunodeficiency virus. Case Rep. Pathol. 2022, 2022, 1298121. [Google Scholar] [CrossRef]
- Sreehari, S. High risk HPV, HSIL and primary diffuse large B cell lymphoma of cervix: An unsual case. IJCSMB 2019, 5, 555666. [Google Scholar] [CrossRef]
- Ren, X.; Cheng, Y.; Wu, S.; Zeng, X.; Shi, X.; Ling, Q.; Li, Z.; Liang, Z.; Wang, B. Primary non-Hodgkin lymphoma of the tongue base: The clinicopathology of seven cases and evaluation of HPV and EBV status. Diagn. Pathol. 2020, 15, 30. [Google Scholar] [CrossRef]
- Swerdlow, S.H.; Campo, E.; Pileri, S.A.; Harris, N.L.; Stein, H.; Siebert, R.; Advani, R.; Ghielmini, M.; Salles, G.A.; Zelenetz, A.D.; et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood 2016, 127, 2375–2390. [Google Scholar] [CrossRef]
- Linnstaedt, S.D.; Gottwein, E.; Skalsky, R.L.; Luftig, M.A.; Culle, B.R. Virally induced cellular microRNA miR-155 plays a key role in B-cell immortalization by Epstein-Barr virus. J. Virol. 2010, 84, 11670–11678. [Google Scholar] [CrossRef]
- Wood, C.D.; Carvell, T.; Gunnell, A.; Ojeniyi, O.O.; Osborne, C.; West, M.J. Enhancer control of microRNA miR-155 expression in Epstein-Barr virus-infected B cells. J. Virol. 2018, 92, e00716–e00718, Erratum in: J. Virol. 2019, 93, 3. [Google Scholar] [CrossRef]
- Gottwein, E.; Mukherjee, N.; Sachse, C.; Frenzel, C.; Majoros, W.H.; Chi, J.T.; Braich, R.; Manoharan, M.; Soutschek, J.; Ohler, U.; et al. A viral microRNA functions as an orthologue of cellular miR-155. Nature 2007, 450, 1096–1099. [Google Scholar] [CrossRef]
- Sekar, D.; Hairul Islam, V.I.; Thirugnanasambantham, K.; Saravanan, S. Relevance of miR-21 in HIV and non-HIV-related lymphomas. Tumour Biol. 2014, 35, 8387–8393. [Google Scholar] [CrossRef] [PubMed]
- Thapa, D.R.; Bhatia, K.; Bream, J.H.; D’Souza, G.; Rinaldo, C.R.; Wolinsky, S.; Detels, R.; Martínez-Maza, O. B-cell activation induced microRNA-21 is elevated in circulating B cells preceding the diagnosis of AIDS-related non-Hodgkin lymphomas. AIDS 2012, 26, 1177–1180. [Google Scholar] [CrossRef] [PubMed]
- He, G.; Ding, J.; Zhang, Y.; Cai, M.; Yang, J.; Cho, W.C.; Zheng, Y. microRNA-21: A key modulator in oncogenic viral infections. RNA Biol. 2021, 18, 809–817. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Yang, J.; Ding, J.; Guo, X.; Zhu, X.Q.; Zheng, Y. Exosomes in virus-associated cancer. Cancer Lett. 2018, 438, 44–51. [Google Scholar] [CrossRef]
- Krupovic, M.; Varsani, A.; Kazlauskas, D.; Breitbart, M.; Delwart, E.; Rosario, K.; Yutin, N.; Wolf, Y.I.; Harrach, B.; Zerbini, F.M.; et al. Cressdnaviricota: A virus phylum unifying seven families of Rep-encoding viruses with single-stranded, circular DNA genomes. J. Virol. 2020, 94, e00582-20. [Google Scholar] [CrossRef]
- Zur Hausen, H.; de Villiers, E.M. Dairy cattle serum and milk factors contributing to the risk of colon and breast cancers. Int. J. Cancer 2015, 137, 959–967. [Google Scholar] [CrossRef]
- Zur Hausen, H.; Bund, T.; de Villiers, E.M. Infectious agents in bovine red meat and milk and their potential role in cancer and other chronic diseases. Curr. Top. Microbiol. Immunol. 2017, 407, 83–116. [Google Scholar]
- Zur Hausen, H.; Bund, T.; de Villiers, E.M. Specific nutritional infections early in life as risk factors for human colon and breast cancers several decades later. Int. J. Cancer 2019, 144, 1574–1583. [Google Scholar] [CrossRef]
- Bund, T.; Nikitina, E.; Chakraborty, D.; Ernst, C.; Gunst, K.; Boneva, B.; Tessmer, C.; Volk, N.; Brobeil, A.; Weber, A.; et al. Analysis of chronic inflammatory lesions of the colon for BMMF Rep antigen expression and CD68 macrophage interactions. Proc. Natl. Acad. Sci. USA 2021, 118, e2025830118. [Google Scholar] [CrossRef]
- Nikitina, E.; Alikhanyan, K.; Neßling, M.; Richter, K.; Kaden, S.; Ernst, C.; Seitz, S.; Chuprikova, L.; Häfele, L.; Gunst, K.; et al. Structural expression of bovine milk and meat factors in tissues of colorectal, lung and pancreatic cancer patients. Int. J. Cancer 2022. [Google Scholar] [CrossRef]
- de Villiers, E.M.; Gunst, K.; Chakraborty, D.; Ernst, C.; Bund, T.; Zur Hausen, H. A specific class of infectious agents isolated from bovine serum and dairy products and peritumoral colon cancer tissue. Emerg. Microbes Infect. 2019, 8, 1205–1218. [Google Scholar] [CrossRef]
- König, M.T.; Fux, R.; Link, E.; Sutter, G.; Märtlbauer, E.; Didier, A. Identification and characterization of circular single-stranded DNA genomes in sheep and goat milk. Viruses 2021, 13, 2176. [Google Scholar] [CrossRef]
- König, M.T.; Fux, R.; Link, E.; Sutter, G.; Märtlbauer, E.; Didier, A. Circular Rep-encoding single-stranded DNA sequences in milk from water buffaloes (Bubalus arnee f. bubalis). Viruses 2021, 13, 1088. [Google Scholar] [CrossRef]
- de Villiers, E.M.; Zur Hausen, H. Bovine meat and milk factors (BMMFs): Their proposed role in common human cancers and type 2 diabetes mellitus. Cancers 2021, 13, 5407. [Google Scholar] [CrossRef]
- Ashrafi, F.; Nassiri, M.; Javadmanesh, A.; Rahimi, H.; Rezaee, S.A. Epigenetics evaluation of the oncogenic mechanisms of two closely related bovine and human deltaretroviruses: A system biology study. Microb. Pathog. 2020, 139, 103845. [Google Scholar] [CrossRef]
- Marawan, M.A.; Alouffi, A.; El Tokhy, S.; Badawy, S.; Shirani, I.; Dawood, A.; Guo, A.; Almutairi, M.M.; Alshammari, F.A.; Selim, A. Bovine leukaemia virus: Current epidemiological circumstance and future prospective. Viruses 2021, 13, 2167. [Google Scholar] [CrossRef]
- Ferrer, J.F.; Piper, C.E. An evaluation of the role of milk in the natural transmission of BLV. Ann. Rech. Vet. 1978, 9, 803–807. [Google Scholar]
- Ferrer, J.F.; Piper, C.E. Role of colostrum and milk in the natural transmission of the bovine leukemia virus. Cancer Res. 1981, 41, 4906–4909. [Google Scholar]
- Corredor-Figueroa, A.P.; Olaya-Galán, N.N.; Velandia-Álvarez, S.; Muñoz, M.; Salas-Cárdenas, S.P.; Ibáñez-Pinilla, M.; Patarroyo, M.A.; Gutiérrez, M.F. Co-circulation of bovine leukemia virus haplotypes among humans, animals, and food products: New insights of its zoonotic potential. Int. J. Environ. Res. Public Health 2021, 18, 4883. [Google Scholar] [CrossRef]
- Olaya-Galán, N.N.; Corredor-Figueroa, A.P.; Guzmán-Garzón, T.C.; Ríos-Hernandez, K.S.; Salas-Cárdenas, S.P.; Patarroyo, M.A.; Gutierrez, M.F. Bovine leukaemia virus DNA in fresh milk and raw beef for human consumption. Epidemiol. Infect. 2017, 145, 3125–3130. [Google Scholar] [CrossRef]
- Buehring, G.C.; DeLaney, A.; Shen, H.; Chu, D.L.; Razavian, N.; Schwartz, D.A.; Demkovich, Z.R.; Bates, M.N. Bovine leukemia virus discovered in human blood. BMC Infect. Dis. 2019, 19, 297. [Google Scholar] [CrossRef] [PubMed]
- Buehring, G.C.; Shen, H.M.; Jensen, H.M.; Choi, K.Y.; Sun, D.; Nuovo, G. Bovine leukemia virus DNA in human breast tissue. Emerg. Infect. Dis. 2014, 20, 772–782. [Google Scholar] [CrossRef] [PubMed]
- Khan, Z.; Abubakar, M.; Arshed, M.J.; Aslam, R.; Sattar, S.; Shah, N.A.; Javed, S.; Tariq, A.; Bostan, N.; Manzoor, S. Molecular investigation of possible relationships concerning bovine leukemia virus and breast cancer. Sci. Rep. 2022, 12, 4161. [Google Scholar] [CrossRef] [PubMed]
- Kincaid, R.P.; Burke, J.M.; Sullivan, C.S. RNA virus microRNA that mimics a B-cell oncomiR. Proc. Natl. Acad. Sci. USA 2012, 109, 3077–3082. [Google Scholar] [CrossRef]
- Souza, O.F.; Popi, A.F. Role of microRNAs in B-cell compartment: Development, proliferation and hematological diseases. Biomedicine 2022, 10, 2004. [Google Scholar] [CrossRef]
- Silva, M.; Melo, S.A. Non-coding RNAs in exosomes: New players in cancer biology. Curr. Genomics 2015, 16, 295–303. [Google Scholar] [CrossRef]
- Hanahan, D. Hallmarks of cancer: New dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Coffre, M.; Koralov, S.B. miRNAs in B cell development and lymphomagenesis. Trends Mol. Med. 2017, 23, 721–736. [Google Scholar] [CrossRef]
- Fuertes, T.; Salgado, I.; de Yébenes, V.G. microRNA fine-tuning of the germinal center response. Front. Immunol. 2021, 12, 660450. [Google Scholar] [CrossRef]
- Shi, Y.; Ding, D.; Qu, R.; Tang, Y.; Hao, S. Non-coding RNAs in diffuse large B-cell lymphoma. Onco Targets Ther. 2020, 13, 12097–12112. [Google Scholar] [CrossRef]
- Yazdanparast, S.; Huang, Z.; Keramat, S.; Izadirad, M.; Li, Y.D.; Bo, L.; Gharehbaghian, A.; Chen, Z.S. The roles of exosomal microRNAs in diffuse large B-cell lymphoma: Diagnosis, prognosis, clinical application, and biomolecular mechanisms. Front. Oncol. 2022, 12, 904637. [Google Scholar] [CrossRef]
- Rutherford, S.C.; Fachel, A.A.; Li, S.; Sawh, S.; Muley, A.; Ishii, J.; Saxena, A.; Dominguez, P.M.; Caldas Lopes, E.; Agirre, X.; et al. Extracellular vesicles in DLBCL provide abundant clues to aberrant transcriptional programming and genomic alterations. Blood 2018, 132, e13–e23. [Google Scholar] [CrossRef]
- Alderton, G.K. Metastasis. Exosomes drive premetastatic niche formation. Nat. Rev. Cancer 2012, 12, 447. [Google Scholar] [CrossRef]
- Ghosh, A.K.; Secreto, C.R.; Knox, T.R.; Ding, W.; Mukhopadhyay, D.; Kay, N.E. Circulating microvesicles in B-cell chronic lymphocytic leukemia can stimulate marrow stromal cells: Implications for disease progression. Blood 2010, 115, 1755–1764. [Google Scholar] [CrossRef]
- Peinado, H.; Alečković, M.; Lavotshkin, S.; Matei, I.; Costa-Silva, B.; Moreno-Bueno, G.; Hergueta-Redondo, M.; Williams, C.; García-Santos, G.; Ghajar, C.; et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 2012, 18, 883–891, Erratum in: Nat. Med. 2016, 22, 1502. [Google Scholar] [CrossRef]
- Melnik, B.C.; John, S.M.; Carrera-Bastos, P.; Schmitz, G. MicroRNA-21-enriched exosomes as epigenetic regulators in melanomagenesis and melanoma progression: The impact of Western lifestyle factors. Cancers 2020, 12, 2111. [Google Scholar] [CrossRef]
- Costa-Silva, B.; Aiello, N.M.; Ocean, A.J.; Singh, S.; Zhang, H.; Thakur, B.K.; Becker, A.; Hoshino, A.; Mark, M.T.; Molina, H.; et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 2015, 17, 816–826. [Google Scholar] [CrossRef]
- Lin, Q.; Zhou, C.R.; Bai, M.J.; Zhu, D.; Chen, J.W.; Wang, H.F.; Li, M.A.; Wu, C.; Li, Z.R.; Huang, M.S. Exosome-mediated miRNA delivery promotes liver cancer EMT and metastasis. Am. J. Transl. Res. 2020, 12, 1080–1095. [Google Scholar]
- Melnik, B.C. Dairy consumption and hepatocellular carcinoma risk. Ann. Transl. Med. 2021, 9, 736. [Google Scholar] [CrossRef]
- Yuan, X.; Qian, N.; Ling, S.; Li, Y.; Sun, W.; Li, J.; Du, R.; Zhong, G.; Liu, C.; Yu, G.; et al. Breast cancer exosomes contribute to pre-metastatic niche formation and promote bone metastasis of tumor cells. Theranostics 2021, 11, 1429–1445. [Google Scholar] [CrossRef]
- Chen, X.; Feng, J.; Chen, W.; Shao, S.; Chen, L.; Wan, H. Small extracellular vesicles: From promoting pre-metastatic niche formation to therapeutic strategies in breast cancer. Cell Commun. Signal. 2022, 20, 141. [Google Scholar] [CrossRef] [PubMed]
- Saber, S.H.; Ali, H.E.A.; Gaballa, R.; Gaballah, M.; Ali, H.I.; Zerfaoui, M.; Abd Elmageed, Z.Y. Exosomes are the driving force in preparing the soil for the metastatic seeds: Lessons from the prostate cancer. Cells 2020, 9, 564. [Google Scholar] [CrossRef] [PubMed]
- Melnik, B.C.; John, S.M.; Weiskirchen, R.; Schmitz, G. The endocrine and epigenetic impact of persistent cow milk consumption on prostate carcinogenesis. J. Transl. Genet. Genom. 2022, 6, 1–45. [Google Scholar] [CrossRef]
- Caner, V.; Cetin, G.O.; Hacioglu, S.; Baris, I.C.; Tepeli, E.; Turk, N.S.; Bagci, G.; Yararbas, K.; Cagliyan, G. The miRNA content of circulating exosomes in DLBCL patients and in vitro influence of DLBCL-derived exosomes on miRNA expression of healthy B-cells from peripheral blood. Cancer Biomark. 2021, 32, 519–529. [Google Scholar] [CrossRef]
- Feng, Y.; Zhong, M.; Zeng, S.; Wang, L.; Liu, P.; Xiao, X.; Liu, Y. Exosome-derived miRNAs as predictive biomarkers for diffuse large B-cell lymphoma chemotherapy resistance. Epigenomics 2019, 11, 35–51. [Google Scholar] [CrossRef]
- Zhang, L.; Zhou, S.; Zhou, T.; Li, X.; Tang, J. Potential of the tumor-derived extracellular vesicles carrying the miR-125b-5p target TNFAIP3 in reducing the sensitivity of diffuse large B cell lymphoma to rituximab. Int. J. Oncol. 2021, 58, 31. [Google Scholar] [CrossRef]
- Pujari, R.; Hunte, R.; Khan, W.N.; Shembade, N. A20-mediated negative regulation of canonical NF-κB signaling pathway. Immunol. Res. 2013, 57, 166–171. [Google Scholar] [CrossRef]
- Zetoune, F.S.; Murthy, A.R.; Shao, Z.; Hlaing, T.; Zeidler, M.G.; Li, Y.; Vincenz, C. A20 inhibits NF-kappa B activation downstream of multiple Map3 kinases and interacts with the I kappa B signalosome. Cytokine 2001, 15, 282–298. [Google Scholar] [CrossRef]
- Bu, R.; Bavi, P.; Abubaker, J.; Jehan, Z.; Al-Haqawi, W.; Ajarim, D.; Al-Dayel, F.; Uddin, S.; Al-Kuraya, K.S. Role of nuclear factor-κB regulators TNFAIP3 and CARD11 in Middle Eastern diffuse large B-cell lymphoma. Leuk. Lymphoma 2012, 53, 1971–1977. [Google Scholar] [CrossRef]
- Gamboa-Cedeño, A.M.; Castillo, M.; Xiao, W.; Waldmann, T.A.; Ranuncolo, S.M. Alternative and canonical NF-kB pathways DNA-binding hierarchies networks define Hodgkin lymphoma and Non-Hodgkin diffuse large B Cell lymphoma respectively. J. Cancer Res. Clin. Oncol. 2019, 145, 1437–1448. [Google Scholar] [CrossRef]
- Wenzl, K.; Manske, M.K.; Sarangi, V.; Asmann, Y.W.; Greipp, P.T.; Schoon, H.R.; Braggio, E.; Maurer, M.J.; Feldman, A.L.; Witzig, T.E.; et al. Loss of TNFAIP3 enhances MYD88L265P-driven signaling in non-Hodgkin lymphoma. Blood Cancer J. 2018, 8, 97. [Google Scholar] [CrossRef]
- Shaffer, A.L.; Lin, K.I.; Kuo, T.C.; Yu, X.; Hurt, E.M.; Rosenwald, A.; Giltnane, J.M.; Yang, L.; Zhao, H.; Calame, K.; et al. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity 2002, 17, 51–62. [Google Scholar] [CrossRef]
- Nadeau, S.; Martins, G.A. Conserved and unique functions of Blimp1 in immune cells. Front. Immunol. 2022, 12, 805260. [Google Scholar] [CrossRef]
- Dent, A.L.; Shaffer, A.L.; Yu, X.; Allman, D.; Staudt, L.M. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science 1997, 276, 589–592. [Google Scholar] [CrossRef]
- Ye, B.H.; Cattoretti, G.; Shen, Q.; Zhang, J.; Hawe, N.; de Waard, R.; Leung, C.; Nouri-Shirazi, M.; Orazi, A.; Chaganti, R.S.; et al. The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nat. Genet. 1997, 16, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, T.; Yoshida, T.; Okada, S.; Hatano, M.; Miki, T.; Ishibashi, K.; Okabe, S.; Koseki, H.; Hirosawa, S.; Taniguchi, M.; et al. Disruption of the Bcl6 gene results in an impaired germinal center formation. J. Exp. Med. 1997, 186, 439–448. [Google Scholar] [CrossRef] [PubMed]
- Ci, W.; Polo, J.M.; Cerchietti, L.; Shaknovich, R.; Wang, L.; Yang, S.N.; Ye, K.; Farinha, P.; Horsman, D.E.; Gascoyne, R.D.; et al. The BCL6 transcriptional program features repression of multiple oncogenes in primary B cells and is deregulated in DLBCL. Blood 2009, 113, 5536–5548. [Google Scholar] [CrossRef] [PubMed]
- Parekh, S.; Privé, G.; Melnick, A. Therapeutic targeting of the BCL6 oncogene for diffuse large B-cell lymphomas. Leuk. Lymphoma 2008, 49, 874–882. [Google Scholar] [CrossRef]
- Parekh, S.; Polo, J.M.; Shaknovich, R.; Juszczynski, P.; Lev, P.; Ranuncolo, S.M.; Yin, Y.; Klein, U.; Cattoretti, G.; Dalla Favera, R.; et al. BCL6 programs lymphoma cells for survival and differentiation through distinct biochemical mechanisms. Blood 2007, 110, 2067–2074. [Google Scholar] [CrossRef]
- Hatzi, K.; Melnick, A. Breaking bad in the germinal center: How deregulation of BCL6 contributes to lymphomagenesis. Trends Mol. Med. 2014, 20, 343–352. [Google Scholar] [CrossRef]
- Phan, R.T.; Dalla-Favera, R. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature 2004, 432, 635–639. [Google Scholar] [CrossRef]
- Reljic, R.; Wagner, S.D.; Peakman, L.J.; Fearon, D.T. Suppression of signal transducer and activator of transcription 3-dependent B lymphocyte terminal differentiation by BCL-6. J. Exp. Med. 2000, 192, 1841–1848. [Google Scholar]
- Xu-Monette, Z.Y.; Wu, L.; Visco, C.; Tai, Y.C.; Tzankov, A.; Liu, W.M.; Montes-Moreno, S.; Dybkaer, K.; Chiu, A.; Orazi, A.; et al. Mutational profile and prognostic significance of TP53 in diffuse large B-cell lymphoma patients treated with R-CHOP: Report from an International DLBCL Rituximab-CHOP Consortium Program Study. Blood 2012, 120, 3986–3996. [Google Scholar] [CrossRef]
- Pascual, M.; Mena-Varas, M.; Robles, E.F.; Garcia-Barchino, M.J.; Panizo, C.; Hervas-Stubbs, S.; Alignani, D.; Sagardoy, A.; Martinez-Ferrandis, J.I.; Bunting, K.L.; et al. PD-1/PD-L1 immune checkpoint and p53 loss facilitate tumor progression in activated B-cell diffuse large B-cell lymphomas. Blood 2019, 133, 2401–2412. [Google Scholar] [CrossRef]
- Pasqualucci, L.; Compagno, M.; Houldsworth, J.; Monti, S.; Grunn, A.; Nandula, S.V.; Aster, J.C.; Murty, V.V.; Shipp, M.A.; Dalla-Favera, R. Inactivation of the PRDM1/BLIMP1 gene in diffuse large B cell lymphoma. J. Exp. Med. 2006, 203, 311–317. [Google Scholar] [CrossRef]
- Tam, W.; Gomez, M.; Chadburn, A.; Lee, J.W.; Chan, W.C.; Knowles, D.M. Mutational analysis of PRDM1 indicates a tumor-suppressor role in diffuse large B-cell lymphomas. Blood 2006, 107, 4090–4100. [Google Scholar] [CrossRef]
- Kallies, A.; Nutt, S.L. Terminal differentiation of lymphocytes depends on Blimp-1. Curr. Opin. Immunol. 2007, 19, 156–162. [Google Scholar] [CrossRef]
- Nutt, S.L.; Fairfax, K.A.; Kallies, A. BLIMP1 guides the fate of effector B and T cells. Nat. Rev. Immunol. 2007, 7, 923–927. [Google Scholar] [CrossRef]
- Lossos, I.S. BLIMP1 against lymphoma: The verdict is reached. Cancer Cell 2010, 18, 537–539. [Google Scholar] [CrossRef]
- Hangaishi, A.; Kurokawa, M. Blimp-1 is a tumor suppressor gene in lymphoid malignancies. Int. J. Hematol. 2010, 91, 46–53. [Google Scholar] [CrossRef]
- Mandelbaum, J.; Bhagat, G.; Tang, H.; Mo, T.; Brahmachary, M.; Shen, Q.; Chadburn, A.; Rajewsky, K.; Tarakhovsky, A.; Pasqualucci, L.; et al. BLIMP1 is a tumor suppressor gene frequently disrupted in activated B cell-like diffuse large B cell lymphoma. Cancer Cell 2010, 18, 568–579. [Google Scholar] [CrossRef] [PubMed]
- Calado, D.P.; Zhang, B.; Srinivasan, L.; Sasaki, Y.; Seagal, J.; Unitt, C.; Rodig, S.; Kutok, J.; Tarakhovsky, A.; Schmidt-Supprian, M.; et al. Constitutive canonical NF-κB activation cooperates with disruption of BLIMP1 in the pathogenesis of activated B cell-like diffuse large cell lymphoma. Cancer Cell 2010, 18, 580–589. [Google Scholar] [CrossRef]
- Boi, M.; Zucca, E.; Inghirami, G.; Bertoni, F. PRDM1/BLIMP1: A tumor suppressor gene in B and T cell lymphomas. Leuk. Lymphoma 2015, 56, 1223–1228. [Google Scholar] [CrossRef] [PubMed]
- Nong, L.; Zheng, Y.; Li, X.; Li, D.; Liang, L.; Wang, W.; Li, T. The genetic deletion and protein expression of PRDM1 and its clinical implications in diffuse large B cell lymphoma: A retrospective cohort study in China. Pathol. Res. Pract. 2022, 233, 153860. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Xu-Monette, Z.Y.; Tzankov, A.; Li, X.; Manyam, G.C.; Murty, V.; Bhagat, G.; Zhang, S.; Pasqualucci, L.; Visco, C.; et al. Loss of PRDM1/BLIMP-1 function contributes to poor prognosis of activated B-cell-like diffuse large B-cell lymphoma. Leukemia 2017, 31, 625–636. [Google Scholar] [CrossRef]
- Qing, K.; Jin, Z.; Xu, Z.; Wang, W.; Li, X.; Zhang, Y.; Wang, L.; Zhu, H.; Xiang, R.; Wu, S.; et al. Dysregulated MDR1 by PRDM1/Blimp1 is involved in the doxorubicin resistance of non-germinal center B-cell-like diffuse large B-cell lymphoma. Chemotherapy 2022, 67, 12–23. [Google Scholar] [CrossRef]
- Lin, Y.; Wong, K.; Calame, K. Repression of c-myc transcription by Blimp-1, an inducer of terminal B cell differentiation. Science 1997, 276, 596–599. [Google Scholar] [CrossRef]
- Zhang, X.Y.; Ma, Z.P.; Cui, W.L.; Pang, X.L.; Chen, R.; Wang, L.; Zhang, W.; Li, X.X. Impact of PRDM1 gene inactivation on C-MYC regulation in diffuse large B-cell lymphoma. Zhonghua Bing Li Xue Za Zhi 2018, 47, 25–31. [Google Scholar]
- Asirvatham, A.J.; Gregorie, C.J.; Hu, Z.; Magner, J.; Tomasi, T.B. MicroRNA targets in immune genes and the Dicer/Argonaute and ARE machinery components. Mol. Immunol. 2008, 45, 1995–2006. [Google Scholar] [CrossRef]
- Ma, J.; Nie, K.; Redmond, D.; Liu, Y.; Elemento, O.; Knowles, D.M.; Tam, W. EBV-miR-BHRF1-2 targets PRDM1/Blimp1: Potential role in EBV lymphomagenesis. Leukemia 2016, 30, 594–604. [Google Scholar] [CrossRef]
- Alsaadi, M.; Khan, M.Y.; Dalhat, M.H.; Bahashwan, S.; Khan, M.U.; Albar, A.; Almehdar, H.; Qadri, I. Dysregulation of miRNAs in DLBCL: Causative factor for pathogenesis, diagnosis and prognosis. Diagnostics 2021, 11, 1739. [Google Scholar] [CrossRef]
- Nie, K.; Gomez, M.; Landgraf, P.; Garcia, J.F.; Liu, Y.; Tan, L.H.; Chadburn, A.; Tuschl, T.; Knowles, D.M.; Tam, W. MicroRNA-mediated down-regulation of PRDM1/Blimp-1 in Hodgkin/Reed-Sternberg cells: A potential pathogenetic lesion in Hodgkin lymphomas. Am. J. Pathol. 2008, 173, 242–252. [Google Scholar] [CrossRef]
- Nie, K.; Zhang, T.; Allawi, H.; Gomez, M.; Liu, Y.; Chadburn, A.; Wang, Y.L.; Knowles, D.M.; Tam, W. Epigenetic down-regulation of the tumor suppressor gene PRDM1/Blimp-1 in diffuse large B cell lymphomas: A potential role of the microRNA let-7. Am. J. Pathol. 2010, 177, 1470–1479. [Google Scholar] [CrossRef]
- Malumbres, R.; Sarosiek, K.A.; Cubedo, E.; Ruiz, J.W.; Jiang, X.; Gascoyne, R.D.; Tibshirani, R.; Lossos, I.S. Differentiation stage-specific expression of microRNAs in B lymphocytes and diffuse large B-cell lymphomas. Blood 2009, 113, 3754–3764. [Google Scholar] [CrossRef]
- Lu, T.X.; Young, K.H.; Xu, W.; Li, J.Y. TP53 dysfunction in diffuse large B-cell lymphoma. Crit. Rev. Oncol. Hematol. 2016, 97, 47–55. [Google Scholar] [CrossRef]
- Le, M.T.; Shyh-Chang, N.; Khaw, S.L.; Chin, L.; The, C.; Tay, J.; O’Day, E.; Korzh, V.; Yang, H.; Lal, A.; et al. Conserved regulation of p53 network dosage by microRNA-125b occurs through evolving miRNA-target gene pairs. PLoS Genet. 2011, 7, e1002242. [Google Scholar] [CrossRef]
- Kumar, M.; Lu, Z.; Takwi, A.A.; Chen, W.; Callander, N.S.; Ramos, K.S.; Young, K.H.; Li, Y. Negative regulation of the tumor suppressor p53 gene by microRNAs. Oncogene 2011, 30, 843–853. [Google Scholar] [CrossRef]
- Li, Y.; Gordon, M.W.; Xu-Monette, Z.Y.; Visco, C.; Tzankov, A.; Zou, D.; Qiu, L.; Montes-Moreno, S.; Dybkaer, K.; Orazi, A.; et al. Single nucleotide variation in the TP53 3’ untranslated region in diffuse large B-cell lymphoma treated with rituximab-CHOP: A report from the International DLBCL Rituximab-CHOP Consortium Program. Blood 2013, 121, 4529–4540. [Google Scholar] [CrossRef]
- Manfè, V.; Biskup, E.; Willumsgaard, A.; Skov, A.G.; Palmieri, D.; Gasparini, P.; Laganá, A.; Woetmann, A.; Ødum, N.; Croce, C.M.; et al. cMyc/miR-125b-5p signalling determines sensitivity to bortezomib in preclinical model of cutaneous T-cell lymphomas. PLoS ONE 2013, 8, e59390. [Google Scholar] [CrossRef]
- Lawrie, C.H.; Gal, S.; Dunlop, H.M.; Pushkaran, B.; Liggins, A.P.; Pulford, K.; Banham, A.H.; Pezzella, F.; Boultwood, J.; Wainscoat, J.S.; et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br. J. Haematol. 2008, 141, 672–675. [Google Scholar] [CrossRef]
- Chen, W.; Wang, H.; Chen, H.; Liu, S.; Lu, H.; Kong, D.; Huang, X.; Kong, Q.; Lu, Z. Clinical significance and detection of microRNA-21 in serum of patients with diffuse large B-cell lymphoma in Chinese population. Eur. J. Haematol. 2014, 92, 407–412. [Google Scholar] [CrossRef] [PubMed]
- Inada, K.; Okoshi, Y.; Cho, Y.; Saito, H.; Iijima, T.; Hori, M.; Kojima, H. Availability of circulating microRNAs as a biomarker for early diagnosis of diffuse large B-cell lymphoma. Open J. Blood Dis. 2015, 5, 48–58. [Google Scholar] [CrossRef]
- Li, J.; Fu, R.; Yang, L.; Tu, W. miR-21 expression predicts prognosis in diffuse large B-cell lymphoma. Int. J. Clin. Exp. Pathol. 2015, 8, 15019–15024. [Google Scholar] [PubMed]
- Larrabeiti-Etxebarria, A.; Lopez-Santillan, M.; Santos-Zorrozua, B.; Lopez-Lopez, E.; Garcia-Orad, A. Systematic review of the potential of microRNAs in diffuse large B cell lymphoma. Cancers 2019, 11, 144. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Santillan, M.; Larrabeiti-Etxebarria, A.; Arzuaga-Mendez, J.; Lopez-Lopez, E.; Garcia-Orad, A. Circulating miRNAs as biomarkers in diffuse large B-cell lymphoma: A systematic review. Oncotarget 2018, 9, 22850–22861. [Google Scholar] [CrossRef]
- Abou Elnour, E.S.; El Sayed, I.E.T.; Mohamed Abd Elbary, H.; Sohaib, A.; Amin Mohammed Atia, S.; El Sayed Ramadan Genena, S. Biochemical and clinical impacts of miR-150 and miR-21 expression levels in diffuse large B cell lymphoma. J. Immunoass. Immunochem. 2022, 43, 648–664. [Google Scholar] [CrossRef]
- Roehle, A.; Hoefig, K.P.; Repsilber, D.; Thorns, C.; Ziepert, M.; Wesche, K.O.; Thiere, M.; Loeffler, M.; Klapper, W.; Pfreundschuh, M.; et al. MicroRNA signatures characterize diffuse large B-cell lymphomas and follicular lymphomas. Br. J. Haematol. 2008, 142, 732–744. [Google Scholar]
- Lawrie, C.H.; Soneji, S.; Marafioti, T.; Cooper, C.D.; Palazzo, S.; Paterson, J.C.; Cattan, H.; Enver, T.; Mager, R.; Boultwood, J.; et al. MicroRNA expression distinguishes between germinal center B cell-like and activated B cell-like subtypes of diffuse large B cell lymphoma. Int. J. Cancer 2007, 121, 1156–1161. [Google Scholar] [CrossRef]
- Musilova, K.; Mraz, M. MicroRNAs in B-cell lymphomas: How a complex biology gets more complex. Leukemia 2015, 29, 1004–1017. [Google Scholar] [CrossRef]
- Barnes, N.A.; Stephenson, S.; Cocco, M.; Tooze, R.M.; Doody, G.M. BLIMP-1 and STAT3 counterregulate microRNA-21 during plasma cell differentiation. J. Immunol. 2012, 189, 253–260. [Google Scholar] [CrossRef]
- Munch-Petersen, H.D.; Ralfkiaer, U.; Sjö, L.D.; Hother, C.; Asmar, F.; Nielsen, B.S.; Brown, P.; Ralfkiaer, E.; Grønbæk, K. Differential expression of miR-155 and miR-21 in tumor and stroma cells in diffuse large B-cell lymphoma. Appl. Immunohistochem. Mol. Morphol. 2015, 23, 188–195. [Google Scholar] [CrossRef]
- Cao, D.; Cao, X.; Jiang, Y.; Xu, J.; Zheng, Y.; Kang, D.; Xu, C. Circulating exosomal microRNAs as diagnostic and prognostic biomarkers in patients with diffuse large B-cell lymphoma. Hematol. Oncol. 2022, 40, 172–180. [Google Scholar] [CrossRef]
- Li, S.; He, X.; Gan, Y.; Zhang, J.; Gao, F.; Lin, L.; Qiu, X.; Yu, T.; Zhang, X.; Chen, P.; et al. Targeting miR-21 with NL101 blocks c-Myc/Mxd1 loop and inhibits the growth of B cell lymphoma. Theranostics 2021, 11, 3439–3451. [Google Scholar] [CrossRef]
- Chen, L.; Zhan, C.Z.; Wang, T.; You, H.; Yao, R. Curcumin inhibits the proliferation, migration, invasion, and apoptosis of diffuse large B-cell lymphoma cell line by regulating miR-21/VHL Axis. Yonsei Med. J. 2020, 61, 20–29. [Google Scholar] [CrossRef]
- Chen, J.; Xu, T.; Chen, C. The critical roles of miR-21 in anti-cancer effects of curcumin. Ann. Transl. Med. 2015, 3, 330. [Google Scholar]
- Choudhury, S.N.; Li, Y. miR-21 and let-7 in the Ras and NF-κB pathways. Microrna 2012, 1, 65–69. [Google Scholar] [CrossRef]
- Bai, D.; Ueno, L.; Vogt, P.K. Akt-mediated regulation of NFkappaB and the essentialness of NFkappaB for the oncogenicity of PI3K and Akt. Int. J. Cancer 2009, 125, 2863–2870. [Google Scholar] [CrossRef]
- Go, H.; Jang, J.Y.; Kim, P.J.; Kim, Y.G.; Nam, S.J.; Paik, J.H.; Kim, T.M.; Heo, D.S.; Kim, C.W.; Jeon, Y.K. MicroRNA-21 plays an oncogenic role by targeting FOXO1 and activating the PI3K/AKT pathway in diffuse large B-cell lymphoma. Oncotarget 2015, 6, 15035–15049. [Google Scholar] [CrossRef]
- Song, J.; Shao, Q.; Li, C.; Liu, H.; Li, J.; Wang, Y.; Song, W.; Li, L.; Wang, G.; Shao, Z.; et al. Effects of microRNA-21 on apoptosis by regulating the expression of PTEN in diffuse large B-cell lymphoma. Medicine 2017, 96, e7952. [Google Scholar] [CrossRef]
- Szydlowski, M.; Kiliszek, P.; Sewastianik, T.; Jablonska, E.; Bialopiotrowicz, E.; Gorniak, P.; Polak, A.; Markowicz, S.; Nowak, E.; Grygorowicz, M.A.; et al. FOXO1 activation is an effector of SYK and AKT inhibition in tonic BCR signal-dependent diffuse large B-cell lymphomas. Blood 2016, 127, 739–748. [Google Scholar] [CrossRef]
- Bouvy, C.; Wannez, A.; George, F.; Graux, C.; Chatelain, C.; Dogné, J.M. Circulating microRNAs as biomarkers in diffuse large B-cell lymphoma: A pilot prospective longitudinal clinical study. Biomark. Cancer 2018, 10, 1179299X18781095. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.; Song, G.; Chen, L.; Nie, Z.; He, B.; Pan, Y.; Xu, Y.; Li, R.; Gao, T.; Cho, W.C.; et al. Inhibition of miR-21 induces biological and behavioral alterations in diffuse large B-cell lymphoma. Acta Haematol. 2013, 130, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Li, X.; Zhu, Y.; Gu, W.; Xie, X.; Jiang, J. Prognostic significance of miRNA in patients with diffuse large B-cell lymphoma: A meta-analysis. Cell. Physiol. Biochem. 2016, 39, 1891–1904. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Zheng, Z.; Wang, L.; Cheng, S.; Shi, Q.; Qu, B.; Fu, D.; Leboeuf, C.; Zhao, Y.; Ye, J.; et al. A novel prognostic model based on four circulating miRNA in diffuse large B-cell lymphoma: Implications for the roles of MDSC and Th17 cells in lymphoma progression. Mol. Oncol. 2021, 15, 246–261. [Google Scholar] [CrossRef]
- Fuertes, T.; Ramiro, A.R.; de Yebenes, V.G. miRNA-based therapies in B cell non-Hodgkin lymphoma. Trends Immunol. 2020, 41, 932–947. [Google Scholar] [CrossRef]
- Thai, T.H.; Calado, D.P.; Casola, S.; Ansel, K.M.; Xiao, C.; Xue, Y.; Murphy, A.; Frendewey, D.; Valenzuela, D.; Kutok, J.L.; et al. Regulation of the germinal center response by microRNA-155. Science 2007, 316, 604–608. [Google Scholar] [CrossRef]
- Elton, T.S.; Selemon, H.; Elton, S.M.; Parinandi, N.L. Regulation of the MIR155 host gene in physiological and pathological processes. Gene 2013, 532, 1–12. [Google Scholar] [CrossRef]
- Eis, P.S.; Tam, W.; Sun, L.; Chadburn, A.; Li, Z.; Gomez, M.F.; Lund, E.; Dahlberg, J.E. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc. Natl. Acad. Sci. USA 2005, 102, 3627–3632. [Google Scholar] [CrossRef]
- Anastasiadou, E.; Seto, A.G.; Beatty, X.; Hermreck, M.; Gilles, M.E.; Stroopinsky, D.; Pinter-Brown, L.C.; Pestano, L.; Marchese, C.; Avigan, D.; et al. Cobomarsen, an oligonucleotide inhibitor of miR-155, slows DLBCL tumor cell growth in vitro and in vivo. Clin. Cancer Res. 2021, 27, 1139–1149. [Google Scholar] [CrossRef]
- Zhong, H.; Xu, L.; Zhong, J.H.; Xiao, F.; Liu, Q.; Huang, H.H.; Chen, F.Y. Clinical and prognostic significance of miR-155 and miR-146a expression levels in formalin-fixed/paraffin-embedded tissue of patients with diffuse large B-cell lymphoma. Exp. Ther. Med. 2012, 3, 763–770. [Google Scholar] [CrossRef]
- Havranek, O.; Xu, J.; Köhrer, S.; Wang, Z.; Becker, L.; Comer, J.M.; Henderson, J.; Ma, W.; Man Chun Ma, J.; Westin, J.R.; et al. Tonic B-cell receptor signaling in diffuse large B-cell lymphoma. Blood 2017, 130, 995–1006. [Google Scholar] [CrossRef]
- Huang, X.; Shen, Y.; Liu, M.; Bi, C.; Jiang, C.; Iqbal, J.; McKeithan, T.W.; Chan, W.C.; Ding, S.J.; Fu, K. Quantitative proteomics reveals that miR-155 regulates the PI3K-AKT pathway in diffuse large B-cell lymphoma. Am. J. Pathol. 2012, 181, 26–33. [Google Scholar] [CrossRef]
- Fu, X.; Wen, H.; Jing, L.; Yang, Y.; Wang, W.; Liang, X.; Nan, K.; Yao, Y.; Tian, T. MicroRNA-155-5p promotes hepatocellular carcinoma progression by suppressing PTEN through the PI3K/Akt pathway. Cancer Sci. 2017, 108, 620–631. [Google Scholar] [CrossRef]
- Sun, J.F.; Zhang, D.; Gao, C.J.; Zhang, Y.W.; Dai, Q.S. Exosome-mediated miR-155 transfer contributes to hepatocellular carcinoma cell proliferation by targeting PTEN. Med. Sci. Monit. Basic Res. 2019, 25, 218–228. [Google Scholar] [CrossRef]
- Jabłońska, E.; Białopiotrowicz, E.; Szydłowski, M.; Prochorec-Sobieszek, M.; Juszczyński, P.; Szumera-Ciećkiewicz, A. DEPTOR is a microRNA-155 target regulating migration and cytokine production in diffuse large B-cell lymphoma cells. Exp. Hematol. 2020, 88, 56–67. [Google Scholar] [CrossRef]
- Morales-Martinez, M.; Lichtenstein, A.; Vega, M.I. Function of Deptor and its roles in hematological malignancies. Aging (Albany NY) 2021, 13, 1528–1564. [Google Scholar] [CrossRef]
- Peterson, T.R.; Laplante, M.; Thoreen, C.C.; Sancak, Y.; Kang, S.A.; Kuehl, W.M.; Gray, N.S.; Sabatini, D.M. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell 2009, 137, 873–886. [Google Scholar] [CrossRef]
- Fu, W.; Hall, M.N. Regulation of mTORC2 signaling. Genes 2020, 11, 1045. [Google Scholar] [CrossRef]
- Huang, W.T.; Kuo, S.H.; Kuo, Y.C.; Lin, C.W. miR-155-regulated mTOR and Toll-like receptor 5 in gastric diffuse large B-cell lymphoma. Cancer Med. 2022, 11, 555–570. [Google Scholar] [CrossRef]
- O’Connell, R.M.; Chaudhuri, A.A.; Rao, D.S.; Baltimore, D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc. Natl. Acad. Sci. USA 2009, 106, 7113–7118. [Google Scholar] [CrossRef]
- Damen, J.E.; Liu, L.; Rosten, P.; Humphries, R.K.; Jefferson, A.B.; Majerus, P.W.; Krystal, G. The 145-kDa protein induced to associate with Shc by multiple cytokines is an inositol tetraphosphate and phosphatidylinositol 3,4,5-triphosphate 5-phosphatase. Proc. Natl. Acad. Sci. USA 1996, 93, 1689–1693. [Google Scholar] [CrossRef] [PubMed]
- Brauweiler, A.; Tamir, I.; Dal Porto, J.; Benschop, R.J.; Helgason, C.D.; Humphries, R.K.; Freed, J.H.; Cambier, J.C. Differential regulation of B cell development, activation, and death by the src homology 2 domain-containing 5’ inositol phosphatase (SHIP). J. Exp. Med. 2000, 191, 1545–1554. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, I.M.; Otero, D.; Kao, E.; Miletic, A.V.; Hother, C.; Ralfkiaer, E.; Rickert, R.C.; Gronbaek, K.; David, M. Onco-miR-155 targets SHIP1 to promote TNFalpha-dependent growth of B cell lymphomas. EMBO Mol. Med. 2009, 1, 288–295. [Google Scholar] [CrossRef] [PubMed]
- Lemm, E.A.; Valle-Argos, B.; Smith, L.D.; Richter, J.; Gebreselassie, Y.; Carter, M.J.; Karolova, J.; Svaton, M.; Helman, K.; Weston-Bell, N.J.; et al. Preclinical evaluation of a novel SHIP1 phosphatase activator for inhibition of PI3K signaling in malignant B cells. Clin. Cancer Res. 2020, 26, 1700–1711. [Google Scholar] [CrossRef]
- Ahmadvand, M.; Eskandari, M.; Pashaiefar, H.; Yaghmaie, M.; Manoochehrabadi, S.; Khakpour, G.; Sheikhsaran, F.; Montazer Zohour, M. Over expression of circulating miR-155 predicts prognosis in diffuse large B-cell lymphoma. Leuk. Res. 2018, 70, 45–48. [Google Scholar] [CrossRef]
- Zare, N.; Haghjooy Javanmard, S.; Mehrzad, V.; Eskandari, N.; Kefayat, A. Evaluation of exosomal miR-155, let-7g and let-7i levels as a potential noninvasive biomarker among refractory/relapsed patients, responsive patients and patients receiving R-CHOP. Leuk. Lymphoma 2019, 60, 1877–1889. [Google Scholar] [CrossRef]
- Seda, V.; Mraz, M. B-cell receptor signalling and its crosstalk with other pathways in normal and malignant cells. Eur. J. Haematol. 2015, 94, 193–205. [Google Scholar] [CrossRef]
- Gonzalez-Martin, A.; Adams, B.D.; Lai, M.; Shepherd, J.; Salvador-Bernaldez, M.; Salvador, J.M.; Lu, J.; Nemazee, D.; Xiao, C. The microRNA miR-148a functions as a critical regulator of B cell tolerance and autoimmunity. Nat. Immunol. 2016, 17, 433–440. [Google Scholar] [CrossRef]
- Liebermann, D.A.; Hoffman, B. Gadd45 in stress signaling. J. Mol. Signal. 2008, 3, 15. [Google Scholar] [CrossRef]
- Hollander, M.C.; Fornace, A.J., Jr. Genomic instability, centrosome amplification, cell cycle checkpoints and Gadd45a. Oncogene 2002, 21, 6228–6233. [Google Scholar] [CrossRef]
- Bhalla, S.; Evens, A.M.; Dai, B.; Prachand, S.; Gordon, L.I.; Gartenhaus, R.B. The novel anti-MEK small molecule AZD6244 induces BIM-dependent and AKT-independent apoptosis in diffuse large B-cell lymphoma. Blood 2011, 118, 1052–1061. [Google Scholar] [CrossRef]
- Porstner, M.; Winkelmann, R.; Daum, P.; Schmid, J.; Pracht, K.; Côrte-Real, J.; Schreiber, S.; Haftmann, C.; Brandl, A.; Mashreghi, M.F.; et al. miR-148a promotes plasma cell differentiation and targets the germinal center transcription factors Mitf and Bach2. Eur. J. Immunol. 2015, 45, 1206–1215. [Google Scholar] [CrossRef]
- Ichikawa, S.; Fukuhara, N.; Katsushima, H.; Takahashi, T.; Yamamoto, J.; Yokoyama, H.; Sasaki, O.; Fukuhara, O.; Nomura, J.; Ishizawa, K.; et al. Association between BACH2 expression and clinical prognosis in diffuse large B-cell lymphoma. Cancer Sci. 2014, 105, 437–444. [Google Scholar] [CrossRef]
- Pracht, K.; Meinzinger, J.; Schulz, S.R.; Daum, P.; Côrte-Real, J.; Hauke, M.; Roth, E.; Kindermann, D.; Mielenz, D.; Schuh, W.; et al. miR-148a controls metabolic programming and survival of mature CD19-negative plasma cells in mice. Eur. J. Immunol. 2021, 51, 1089–1109. [Google Scholar] [CrossRef]
- Melnik, B.C.; Schmitz, G. Pasteurized non-fermented cow’s milk but not fermented milk is a promoter of mTORC1-driven aging and increased mortality. Ageing Res. Rev. 2021, 67, 101270. [Google Scholar] [CrossRef]
- International Dairy Federation. Heat Treatment of Milk—Overview, 001/2018-02. IDF Factsheet. 2018. Available online: http://www.ukidf.org/documents/Factsheet-001_2018_Heat-treatment-1-2_001.pdf (accessed on 20 January 2023).
- Kleinjan, M.; van Herwijnen, M.J.; Libregts, S.F.; van Neerven, R.J.; Feitsma, A.L.; Wauben, M.H. Regular industrial processing of bovine milk impacts the integrity and molecular composition of extracellular vesicles. J. Nutr. 2021, 151, 1416–1425. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, Q.; Hou, J.; Huang, G.; Zhao, S.; Zheng, N.; Wang, J. Loss of bioactive microRNAs in cow’s milk by ultra-high-temperature treatment but not by pasteurization treatment. J. Sci. Food Agric. 2022, 102, 2676–2685. [Google Scholar] [CrossRef]
- Ngu, A.; Wang, S.; Wang, H.; Khanam, A.; Zempleni, J. Milk exosomes in nutrition and drug delivery. Am. J. Physiol. Cell Physiol. 2022, 322, C865–C874. [Google Scholar] [CrossRef]
- Benmoussa, A.; Lee, C.H.; Laffont, B.; Savard, P.; Laugier, J.; Boilard, E.; Gilbert, C.; Fliss, I.; Provost, P. Commercial dairy cow milk microRNAs resist digestion under simulated gastrointestinal tract conditions. J. Nutr. 2016, 146, 2206–2215. [Google Scholar] [CrossRef]
- Rani, P.; Vashisht, M.; Golla, N.; Vashisht, M.; Shandilya, S. Milk miRNAs encapsulated in exosomes are stable to human digestion and permeable to intestinal barrier in vitro. J. Funct. Foods 2017, 34, 431–439. [Google Scholar] [CrossRef]
- Wolf, T.; Baier, S.R.; Zempleni, J. The intestinal transport of bovine milk exosomes is mediated by endocytosis in human colon carcinoma Caco-2 cells and rat small intestinal IEC-6 cells. J. Nutr. 2015, 145, 2201–2206. [Google Scholar] [CrossRef] [PubMed]
- Kusuma, R.J.; Manca, S.; Friemel, T.; Sukreet, S.; Nguyen, C.; Zempleni, J. Human vascular endothelial cells transport foreign exosomes from cow’s milk by endocytosis. Am. J. Physiol. Cell Physiol. 2016, 310, C800–C807. [Google Scholar] [CrossRef]
- González-Sarrías, A.; Iglesias-Aguirre, C.E.; Cortés-Martín, A.; Vallejo, F.; Cattivelli, A.; Del Pozo-Acebo, L.; Del Saz, A.; López de Las Hazas, M.C.; Dávalos, A.; Espín, J.C. Milk-derived exosomes as nanocarriers to deliver curcumin and resveratrol in breast tissue and enhance their anticancer activity. Int. J. Mol. Sci. 2022, 23, 2860. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Sadri, M.; Giraud, D.; Zempleni, J. RNase H2-dependent polymerase chain reaction and elimination of confounders in sample collection, storage, and analysis strengthen evidence that microRNAs in bovine milk are bioavailable in humans. J. Nutr. 2018, 148, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Chen, T.; Xie, M.; Li, M.; Zeng, B.; Sun, R.; Zhu, Y.; Ye, D.; Wu, J.; Sun, J.; et al. Oral administration of bovine and porcine milk exosome alter miRNAs profiles in piglet serum. Sci. Rep. 2020, 10, 6983. [Google Scholar] [CrossRef]
- Feng, X.; Chen, X.; Zheng, X.; Zhu, H.; Qi, Q.; Liu, S.; Zhang, H.; Che, J. Latest trend of milk derived exosomes: Cargos, functions, and applications. Front. Nutr. 2021, 8, 747294. [Google Scholar] [CrossRef]
- López de Las Hazas, M.C.; Del Pozo-Acebo, L.; Hansen, M.S.; Gil-Zamorano, J.; Mantilla-Escalante, D.C.; Gómez-Coronado, D.; Marín, F.; Garcia-Ruiz, A.; Rasmussen, J.T.; Dávalos, A. Dietary bovine milk miRNAs transported in extracellular vesicles are partially stable during GI digestion, are bioavailable and reach target tissues but need a minimum dose to impact on gene expression. Eur. J. Nutr. 2022, 61, 1043–1056. [Google Scholar] [CrossRef]
- Manca, S.; Upadhyaya, B.; Mutai, E.; Desaulniers, A.T.; Cederberg, R.A.; White, B.R.; Zempleni, J. Milk exosomes are bioavailable and distinct microRNA cargos have unique tissue distribution patterns. Sci. Rep. 2018, 8, 11321. [Google Scholar] [CrossRef]
- Betker, J.L.; Angle, B.M.; Graner, M.W.; Anchordoquy, T.J. The potential of exosomes from cow milk for oral delivery. J. Pharm. Sci. 2019, 108, 1496–1505. [Google Scholar] [CrossRef]
- Aguilar-Lozano, A.; Baier, S.; Grove, R.; Shu, J.; Giraud, D.; Leiferman, A.; Mercer, K.E.; Cui, J.; Badger, T.M.; Adamec, J.; et al. Concentrations of purine metabolites are elevated in fluids from adults and infants and in livers from mice fed diets depleted of bovine milk exosomes and their RNA cargos. J. Nutr. 2018, 148, 1886–1894. [Google Scholar] [CrossRef]
- Sadri, M.; Shu, J.; Kachman, S.D.; Cui, J.; Zempleni, J. Milk exosomes and miRNA cross the placenta and promote embryo survival in mice. Reproduction 2020, 160, 501–509. [Google Scholar] [CrossRef]
- Sanwlani, R.; Fonseka, P.; Chitti, S.V.; Mathivanan, S. Milk-derived extracellular vesicles in inter-organism, cross-species communication and drug delivery. Proteomes 2020, 8, 11. [Google Scholar] [CrossRef]
- Sanwlani, R.; Fonseka, P.; Mathivanan, S. Are dietary extracellular vesicles bioavailable and functional in consuming organisms? Subcell. Biochem. 2021, 97, 509–521. [Google Scholar]
- Askenase, P.W. Ancient evolutionary origin and properties of universally produced natural exosomes contribute to their therapeutic superiority compared to artificial nanoparticles. Int. J. Mol. Sci. 2021, 22, 1429. [Google Scholar] [CrossRef]
- Samuel, M.; Fonseka, P.; Sanwlani, R.; Gangoda, L.; Chee, S.H.; Keerthikumar, S.; Spurling, A.; Chitti, S.V.; Zanker, D.; Ang, C.S.; et al. Oral administration of bovine milk-derived extracellular vesicles induces senescence in the primary tumor but accelerates cancer metastasis. Nat. Commun. 2021, 12, 3950. [Google Scholar] [CrossRef]
- Baier, S.R.; Nguyen, C.; Xie, F.; Wood, J.R.; Zempleni, J. MicroRNAs are absorbed in biologically meaningful amounts from nutritionally relevant doses of cow milk and affect gene expression in peripheral blood mononuclear cells, HEK-293 kidney cell cultures, and mouse livers. J. Nutr. 2014, 144, 1495–1500. [Google Scholar] [CrossRef]
- Lässer, C.; Alikhani, V.S.; Ekström, K.; Eldh, M.; Paredes, P.T.; Bossios, A.; Sjöstrand, M.; Gabrielsson, S.; Lötvall, J.; Valadi, H. Human saliva, plasma and breast milk exosomes contain RNA: Uptake by macrophages. J. Transl. Med. 2011, 9, 9. [Google Scholar] [CrossRef]
- Izumi, H.; Tsuda, M.; Sato, Y.; Kosaka, N.; Ochiya, T.; Iwamoto, H.; Namba, K.; Takeda, Y. Bovine milk exosomes contain microRNA and mRNA and are taken up by human macrophages. J. Dairy Sci. 2015, 98, 2920–2933. [Google Scholar] [CrossRef]
- He, Y.; He, Z.; Leone, S.; Liu, S. Milk exosomes transfer oligosaccharides into macrophages to modulate immunity and attenuate adherent-invasive E. coli (AIEC) infection. Nutrients 2021, 13, 3198. [Google Scholar] [CrossRef]
- Admyre, C.; Johansson, S.M.; Qazi, K.R.; Filén, J.J.; Lahesmaa, R.; Norman, M.; Neve, E.P.; Scheynius, A.; Gabrielsson, S. Exosomes with immune modulatory features are present in human breast milk. J. Immunol. 2007, 179, 1969–1978. [Google Scholar] [CrossRef]
- Vashisht, M.; Rani, P.; Onteru, S.K.; Singh, D. Curcumin encapsulated in milk exosomes resists human digestion and possesses enhanced intestinal permeability in vitro. Appl. Biochem. Biotechnol. 2017, 183, 993–1007. [Google Scholar] [CrossRef]
- Salisbury, J.L.; Condeelis, J.S.; Satir, P. Role of coated vesicles, microfilaments, and calmodulin in receptor-mediated endocytosis by cultured B lymphoblastoid cells. J. Cell Biol. 1980, 87, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Guagliardi, L.E.; Koppelman, B.; Blum, J.S.; Marks, M.S.; Cresswell, P.; Brodsky, F.M. Co-localization of molecules involved in antigen processing and presentation in an early endocytic compartment. Nature 1990, 343, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Hoogeboom, R.; Tolar, P. Molecular mechanisms of B cell antigen gathering and endocytosis. Curr. Top. Microbiol. Immunol. 2016, 393, 45–63. [Google Scholar] [PubMed]
- Roberts, A.D.; Davenport, T.M.; Dickey, A.M.; Ahn, R.; Sochacki, K.A.; Taraska, J.W. Structurally distinct endocytic pathways for B cell receptors in B lymphocytes. Mol. Biol. Cell. 2020, 31, 2826–2840. [Google Scholar] [CrossRef]
- Malinova, D.; Wasim, L.; Newman, R.; Martínez-Riaño, A.; Engels, N.; Tolar, P. Endophilin A2 regulates B-cell endocytosis and is required for germinal center and humoral responses. EMBO Rep. 2021, 22, e51328. [Google Scholar] [CrossRef]
- McShane, A.N.; Malinova, D. The ins and outs of antigen uptake in B cells. Front. Immunol. 2022, 13, 892169. [Google Scholar] [CrossRef]
- Khanam, A.; Yu, J.; Zempleni, J. Class A scavenger receptor-1/2 facilitates the uptake of bovine milk exosomes in murine bone marrow-derived macrophages and C57BL/6J mice. Am. J. Physiol. Cell Physiol. 2021, 321, C607–C614. [Google Scholar] [CrossRef]
- Baos, S.; Cremades-Jimeno, L.; López-Ramos, M.; de Pedro, M.Á.; Uriarte, S.A.; Sastre, J.; González-Mangado, N.; Rodríguez-Nieto, M.J.; Peces-Barba, G.; Cárdaba, B. Expression of macrophage scavenger receptor (MSR1) in peripheral blood cells from patients with different respiratory diseases: Beyond monocytes. J. Clin. Med. 2022, 11, 1439. [Google Scholar] [CrossRef]
- Nam, S.J.; Kim, S.; Kwon, D.; Kim, H.; Kim, S.; Lee, E.; Kim, T.M.; Heo, D.S.; Park, S.H.; Lim, M.S.; et al. Prognostic implications of tumor-infiltrating macrophages, M2 macrophages, regulatory T-cells, and indoleamine 2,3-dioxygenase-positive cells in primary diffuse large B-cell lymphoma of the central nervous system. Oncoimmunology 2018, 7, e1442164. [Google Scholar] [CrossRef]
- van Bilsen, K.; Bastiaans, J.; Dik, W.A.; De Haas, E.F.; Baarsma, S.G.; Kuipers, R.W.; van Hagen, P.M. The neonatal Fc receptor is expressed by human lymphocytes. J. Transl. Med. 2010, 8 (Suppl. S1), P1. [Google Scholar] [CrossRef]
- Montoyo, H.P.; Vaccaro, C.; Hafner, M.; Ober, R.J.; Mueller, W.; Ward, E.S. Conditional deletion of the MHC class I-related receptor FcRn reveals the sites of IgG homeostasis in mice. Proc. Natl. Acad. Sci. USA 2009, 106, 2788–2793. [Google Scholar] [CrossRef]
- Mi, W.; Wanjie, S.; Lo, S.T.; Gan, Z.; Pickl-Herk, B.; Ober, R.J.; Ward, E.S. Targeting the neonatal fc receptor for antigen delivery using engineered fc fragments. J. Immunol. 2008, 181, 7550–7561. [Google Scholar] [CrossRef]
- Golan-Gerstl, R.; Elbaum Shiff, Y.; Moshayoff, V.; Schecter, D.; Leshkowitz, D.; Reif, S. Characterization and biological function of milk-derived miRNAs. Mol. Nutr. Food Res. 2017, 61, 1700009. [Google Scholar] [CrossRef]
- van Herwijnen, M.J.C.; Driedonks, T.A.P.; Snoek, B.L.; Kroon, A.M.T.; Kleinjan, M.; Jorritsma, R.; Pieterse, C.M.J.; Hoen, E.N.M.N.; Wauben, M.H.M. Abundantly present miRNAs in milk-derived extracellular vesicles are conserved between mammals. Front. Nutr. 2018, 5, 81. [Google Scholar] [CrossRef]
- Oh, S.; Park, M.R.; Son, S.J.; Kim, Y. Comparison of total RNA isolation methods for analysis of immune-related microRNAs in market milks. Korean J. Food Sci. Anim. Resour. 2015, 35, 459–465. [Google Scholar] [CrossRef]
- Benmoussa, A.; Laugier, J.; Beauparlant, C.J.; Lambert, M.; Droit, A.; Provost, P. Complexity of the microRNA transcriptome of cow milk and milk-derived extracellular vesicles isolated via differential ultracentrifugation. J. Dairy Sci. 2020, 103, 16–29. [Google Scholar] [CrossRef]
- Mecocci, S.; Pietrucci, D.; Milanesi, M.; Pascucci, L.; Filippi, S.; Rosato, V.; Chillemi, G.; Capomaccio, S.; Cappelli, K. Transcriptomic characterization of cow, donkey and goat milk extracellular vesicles reveals their anti-inflammatory and immunomodulatory potential. Int. J. Mol. Sci. 2021, 22, 12759. [Google Scholar] [CrossRef]
- Rashidi, M.; Bijari, S.; Khazaei, A.H.; Shojaei-Ghahrizjani, F.; Rezakhani, L. The role of milk-derived exosomes in the treatment of diseases. Front. Genet. 2022, 13, 1009338. [Google Scholar] [CrossRef]
- Rani, P.; Onteru, S.K.; Singh, D. Genome-wide profiling and analysis of microRNA expression in buffalo milk exosomes. Food Biosci. 2020, 38, 100769. [Google Scholar] [CrossRef]
- Chen, Z.; Xie, Y.; Luo, J.; Chen, T.; Xi, Q.; Zhang, Y.; Sun, J. Milk exosome-derived miRNAs from water buffalo are implicated in immune response and metabolism process. BMC Vet. Res. 2020, 16, 123. [Google Scholar] [CrossRef] [PubMed]
- Reif, S.; Atias, A.; Musseri, M.; Koroukhov, N.; Gerstl, R.G. Beneficial effects of milk-derived extracellular vesicles on liver fibrosis progression by inhibiting hepatic stellate cell activation. Nutrients 2022, 14, 4049. [Google Scholar] [CrossRef]
- Baddela, V.S.; Nayan, V.; Rani, P.; Onteru, S.K.; Singh, D. Physicochemical biomolecular insights into Buffalo milk-derived nanovesicles. Appl. Biochem. Biotechnol. 2016, 178, 544–557. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, R.; Hansmann, M.L.; Bohle, V.; Martin-Subero, J.I.; Hartmann, S.; Mechtersheimer, G.; Klapper, W.; Vater, I.; Giefing, M.; Gesk, S.; et al. TNFAIP3 (A20) is a tumor suppressor gene in Hodgkin lymphoma and primary mediastinal B cell lymphoma. J. Exp. Med. 2009, 206, 981–989. [Google Scholar] [CrossRef]
- Kato, M.; Sanada, M.; Kato, I.; Sato, Y.; Takita, J.; Takeuchi, K.; Niwa, A.; Chen, Y.; Nakazaki, K.; Nomoto, J.; et al. Frequent inactivation of A20 in B-cell lymphomas. Nature 2009, 459, 712–716. [Google Scholar] [CrossRef] [PubMed]
- Heyninck, K.; De Valck, D.; Vanden Berghe, W.; Van Criekinge, W.; Contreras, R.; Fiers, W.; Haegeman, G.; Beyaert, R. The zinc finger protein A20 inhibits TNF-induced NF-kappaB-dependent gene expression by interfering with an RIP- or TRAF2-mediated transactivation signal and directly binds to a novel NF-kappaB-inhibiting protein ABIN. J. Cell Biol. 1999, 145, 1471–1482. [Google Scholar] [CrossRef] [PubMed]
- Wertz, I.E.; O’Rourke, K.M.; Zhou, H.; Eby, M.; Aravind, L.; Seshagiri, S.; Wu, P.; Wiesmann, C.; Baker, R.; Boone, D.L.; et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature 2004, 430, 694–699. [Google Scholar] [CrossRef]
- Shembade, N.; Ma, A.; Harhaj, E.W. Inhibition of NF-kappaB signaling by A20 through disruption of ubiquitin enzyme complexes. Science 2010, 327, 1135–1139. [Google Scholar] [CrossRef]
- Kim, S.W.; Ramasamy, K.; Bouamar, H.; Lin, A.P.; Jiang, D.; Aguiar, R.C. MicroRNAs miR-125a and miR-125b constitutively activate the NF-κB pathway by targeting the tumor necrosis factor alpha-induced protein 3 (TNFAIP3, A20). Proc. Natl. Acad. Sci. USA 2012, 109, 7865–7870. [Google Scholar] [CrossRef]
- Shome, S.; Jernigan, R.L.; Beitz, D.C.; Clark, S.; Testroet, E.D. Non-coding RNA in raw and commercially processed milk and putative targets related to growth and immune-response. BMC Genomics 2021, 22, 749. [Google Scholar] [CrossRef]
- Sun, Y.M.; Lin, K.Y.; Chen, Y.Q. Diverse functions of miR-125 family in different cell contexts. J. Hematol. Oncol. 2013, 6, 6. [Google Scholar] [CrossRef] [PubMed]
- Fromm, B.; Tosar, J.P.; Lu, Y.; Halushka, M.K.; Witwer, K.W. Human and cow have identical miR-21-5p and miR-30a-5p sequences, which are likely unsuited to study dietary uptake from cow milk. J. Nutr. 2018, 148, 1506–1507. [Google Scholar] [CrossRef] [PubMed]
- Mutai, E.; Ramer-Tait, A.E.; Zempleni, J. MicroRNAs in bovine milk exosomes are bioavailable in humans but do not elicit a robust pro-inflammatory cytokine response. ExRNA 2020, 2, 2. [Google Scholar] [CrossRef]
- Chen, X.; Gao, C.; Li, H.; Huang, L.; Sun, Q.; Dong, Y.; Tian, C.; Gao, S.; Dong, H.; Guan, D.; et al. Identification and characterization of microRNAs in raw milk during different periods of lactation, commercial fluid, and powdered milk products. Cell Res. 2010, 20, 1128–1137. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Zhao, Z.; Sun, L.; Li, P. Fermentation results in quantitative changes in milk-derived exosomes and different effects on cell growth and survival. J. Agric. Food Chem. 2017, 65, 1220–1228. [Google Scholar] [CrossRef]
- Ding, X.; Jing, N.; Shen, A.; Guo, F.; Song, Y.; Pan, M.; Ma, X.; Zhao, L.; Zhang, H.; Wu, L.; et al. MiR-21-5p in macrophage-derived extracellular vesicles affects podocyte pyroptosis in diabetic nephropathy by regulating A20. J. Endocrinol. Investig. 2021, 44, 1175–1184. [Google Scholar] [CrossRef]
- Xue, Z.; Xi, Q.; Liu, H.; Guo, X.; Zhang, J.; Zhang, Z.; Li, Y.; Yang, G.; Zhou, D.; Yang, H.; et al. miR-21 promotes NLRP3 inflammasome activation to mediate pyroptosis and endotoxic shock. Cell Death Dis. 2019, 10, 461. [Google Scholar] [CrossRef]
- Lei, B.X.; Liu, Z.H.; Li, Z.J.; Li, C.; Deng, Y.F. miR-21 induces cell proliferation and suppresses the chemosensitivity in glioblastoma cells via downregulation of FOXO1. Int. J. Clin. Exp. Med. 2014, 7, 2060–2066. [Google Scholar]
- Song, W.; Wang, L.; Wang, L.; Li, Q. Interplay of miR-21 and FoxO1 modulates growth of pancreatic ductal adenocarcinoma. Tumour Biol. 2015, 36, 4741–4745. [Google Scholar] [CrossRef]
- Song, W.; Li, Q.; Wang, L.; Wang, L. Modulation of FoxO1 expression by miR-21 to promote growth of pancreatic ductal adenocarcinoma. Cell. Physiol. Biochem. 2015, 35, 184–190. [Google Scholar] [CrossRef]
- Liu, H.Y.; Zhang, Y.Y.; Zhu, B.L.; Feng, F.Z.; Yan, H.; Zhang, H.Y.; Zhou, B. miR-21 regulates the proliferation and apoptosis of ovarian cancer cells through PTEN/PI3K/AKT. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 4149–4155. [Google Scholar]
- Wang, Z.; Zhou, H.; Cheng, F.; Zhang, Z.; Long, S. MiR-21 regulates epithelial-mesenchymal transition in intestinal fibrosis of Crohn’s disease by targeting PTEN/mTOR. Dig. Liver Dis. 2022, 54, 1358–1366. [Google Scholar] [CrossRef]
- Xie, X.; Qu, P.; Wu, H.; Liu, P.; Luo, J.; Chi, J.; Liu, X.; Chen, X.; Xu, C. Circulating exosomal miR-21 mediates HUVEC proliferation and migration through PTEN/PI3K/AKT in Crohn’s disease. Ann. Transl. Med. 2022, 10, 258. [Google Scholar] [CrossRef]
- Zhang, L.; Yu, L.; Liu, Y.; Wang, S.; Hou, Z.; Zhou, J. miR-21-5p promotes cell proliferation by targeting BCL11B in Thp-1 cells. Oncol. Lett. 2021, 21, 119. [Google Scholar] [CrossRef]
- Wakabayashi, Y.; Inoue, J.; Takahashi, Y.; Matsuki, A.; Kosugi-Okano, H.; Shinbo, T.; Mishima, Y.; Niwa, O.; Kominami, R. Homozygous deletions and point mutations of the Rit1/Bcl11b gene in gamma-ray induced mouse thymic lymphomas. Biochem. Biophys. Res. Commun. 2003, 301, 598–603. [Google Scholar] [CrossRef]
- Kominami, R. Role of the transcription factor Bcl11b in development and lymphomagenesis. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2012, 88, 72–87. [Google Scholar] [CrossRef]
- Thompson, P.A.; Eam, B.; Young, N.P.; Fish, S.; Chen, J.; Barrera, M.; Howard, H.; Sung, E.; Parra, A.; Staunton, J.; et al. Targeting oncogene mRNA translation in B-cell malignancies with eFT226, a potent and selective inhibitor of eIF4A. Mol. Cancer Ther. 2021, 20, 26–36. [Google Scholar] [CrossRef]
- Dorrello, N.V.; Peschiaroli, A.; Guardavaccaro, D.; Colburn, N.H.; Sherman, N.E.; Pagano, M. S6K1- and betaTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science 2006, 314, 467–471. [Google Scholar] [CrossRef]
- Yang, C.H.; Yue, J.; Pfeffer, S.R.; Fan, M.; Paulus, E.; Hosni-Ahmed, A.; Sims, M.; Qayyum, S.; Davidoff, A.M.; Handorf, C.R.; et al. MicroRNA-21 promotes glioblastoma tumorigenesis by down-regulating insulin-like growth factor-binding protein-3 (IGFBP3). J. Biol. Chem. 2014, 289, 25079–25087. [Google Scholar] [CrossRef]
- Satzger, I.; Mattern, A.; Kuettler, U.; Weinspach, D.; Niebuhr, M.; Kapp, A.; Gutzmer, R. microRNA-21 is upregulated in malignant melanoma and influences apoptosis of melanocytic cells. Exp. Dermatol. 2012, 21, 509–514. [Google Scholar] [CrossRef]
- Melnik, B.C. MiR-21: An environmental driver of malignant melanoma? J. Transl. Med. 2015, 13, 202. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Singh, A.K.; Giri, R.; Kumar, D.; Sharma, R.; Valis, M.; Kuca, K.; Garg, N. The role of microRNA-21 in the onset and progression of cancer. Future Med. Chem. 2021, 13, 1885–1906. [Google Scholar] [CrossRef] [PubMed]
- Cioroianu, A.I.; Stinga, P.I.; Sticlaru, L.; Cioplea, M.D.; Nichita, L.; Popp, C.; Staniceanu, F. Tumor microenvironment in diffuse large B-cell lymphoma: Role and prognosis. Anal. Cell Pathol. 2019, 2019, 8586354. [Google Scholar] [CrossRef] [PubMed]
- Ng, W.L.; Ansell, S.M.; Mondello, P. Insights into the tumor microenvironment of B cell lymphoma. J. Exp. Clin. Cancer Res. 2022, 41, 362. [Google Scholar] [CrossRef] [PubMed]
- Opinto, G.; Vegliante, M.C.; Negri, A.; Skrypets, T.; Loseto, G.; Pileri, S.A.; Guarini, A.; Ciavarella, S. The tumor microenvironment of DLBCL in the computational era. Front. Oncol. 2020, 10, 351. [Google Scholar] [CrossRef]
- Shen, L.; Li, H.; Shi, Y.; Wang, D.; Gong, J.; Xun, J.; Zhou, S.; Xiang, R.; Tan, X. M2 tumour-associated macrophages contribute to tumour progression via legumain remodelling the extracellular matrix in diffuse large B cell lymphoma. Sci. Rep. 2016, 6, 30347. [Google Scholar] [CrossRef]
- Mitteldorf, C.; Berisha, A.; Pfaltz, M.C.; Broekaert, S.M.C.; Schön, M.P.; Kerl, K.; Kempf, W. Tumor microenvironment and checkpoint molecules in primary cutaneous diffuse large B-cell lymphoma-new therapeutic targets. Am. J. Surg. Pathol. 2017, 41, 998–1004. [Google Scholar] [CrossRef]
- Zhu, M.Y.; Liu, W.J.; Wang, H.; Wang, W.D.; Liu, N.W.; Lu, Y. NSE from diffuse large B-cell lymphoma cells regulates macrophage polarization. Cancer Manag. Res. 2019, 11, 4577–4595. [Google Scholar] [CrossRef]
- Huang, Y.H.; Cai, K.; Xu, P.P.; Wang, L.; Huang, C.X.; Fang, Y.; Cheng, S.; Sun, X.J.; Liu, F.; Huang, J.Y.; et al. CREBBP/EP300 mutations promoted tumor progression in diffuse large B-cell lymphoma through altering tumor-associated macrophage polarization via FBXW7-NOTCH-CCL2/CSF1 axis. Signal Transduct. Target Ther. 2021, 6, 10. [Google Scholar] [CrossRef]
- An, Y.; Yang, Q. MiR-21 modulates the polarization of macrophages and increases the effects of M2 macrophages on promoting the chemoresistance of ovarian cancer. Life Sci. 2020, 242, 117162. [Google Scholar] [CrossRef]
- Xue, J.; Xiao, T.; Wei, S.; Sun, J.; Zou, Z.; Shi, M.; Sun, Q.; Dai, X.; Wu, L.; Li, J.; et al. miR-21-regulated M2 polarization of macrophage is involved in arsenicosis-induced hepatic fibrosis through the activation of hepatic stellate cells. J. Cell. Physiol. 2021, 236, 6025–6041. [Google Scholar] [CrossRef]
- Shen, D.; He, Z. Mesenchymal stem cell-derived exosomes regulate the polarization and inflammatory response of macrophages via miR-21-5p to promote repair after myocardial reperfusion injury. Ann. Transl. Med. 2021, 9, 1323. [Google Scholar] [CrossRef]
- Li, J.; Wei, C.; Yang, Y.; Gao, Z.; Guo, Z.; Qi, F. Apoptotic bodies extracted from adipose mesenchymal stem cells carry microRNA-21-5p to induce M2 polarization of macrophages and augment skin wound healing by targeting KLF6. Burns 2022, 48, 1893–1908. [Google Scholar] [CrossRef]
- Xi, J.; Huang, Q.; Wang, L.; Ma, X.; Deng, Q.; Kumar, M.; Zhou, Z.; Li, L.; Zeng, Z.; Young, K.H.; et al. miR-21 depletion in macrophages promotes tumoricidal polarization and enhances PD-1 immunotherapy. Oncogene 2018, 37, 3151–3165. [Google Scholar] [CrossRef]
- Lu, J.; Xie, L.; Sun, S. The inhibitor miR-21 regulates macrophage polarization in an experimental model of chronic obstructive pulmonary disease. Tob. Induc. Dis. 2021, 19, 69. [Google Scholar] [CrossRef]
- Lee, H.Y.; Hur, J.; Kang, J.Y.; Rhee, C.K.; Lee, S.Y. MicroRNA-21 inhibition suppresses alveolar M2 macrophages in an ovalbumin-induced allergic asthma mice model. Allergy Asthma Immunol. Res. 2021, 13, 312–329. [Google Scholar] [CrossRef]
- Chen, Q.; Li, Y.; Gao, W.; Chen, L.; Xu, W.; Zhu, X. Exosome-mediated crosstalk between tumor and tumor-associated macrophages. Front. Mol. Biosci. 2021, 8, 764222. [Google Scholar] [CrossRef]
- Liu, W.; Zhu, M.; Wang, H.; Wang, W.; Lu, Y. Diffuse large B cell lymphoma-derived extracellular vesicles educate macrophages to promote tumours progression by increasing PGC-1β. Scand. J. Immunol. 2020, 91, e12841. [Google Scholar] [CrossRef]
- Ling, H.Y.; Yang, Z.; Wang, P.J.; Sun, Y.; Ju, S.G.; Li, J.; Fu, J.X. Diffuse large B-cell lymphoma-derived exosomes push macrophage polarization toward M2 phenotype via GP130/STAT3 signaling pathway. Chem. Biol. Interact. 2022, 352, 109779. [Google Scholar] [CrossRef]
- Ren, W.; Hou, J.; Yang, C.; Wang, H.; Wu, S.; Wu, Y.; Zhao, X.; Lu, C. Extracellular vesicles secreted by hypoxia pre-challenged mesenchymal stem cells promote non-small cell lung cancer cell growth and mobility as well as macrophage M2 polarization via miR-21-5p delivery. J. Exp. Clin. Cancer Res. 2019, 38, 62. [Google Scholar] [CrossRef]
- Hsieh, C.H.; Tai, S.K.; Yang, M.H. Snail-overexpressing cancer cells promote M2-like polarization of tumor-associated macrophages by delivering miR-21-abundant exosomes. Neoplasia 2018, 20, 775–788. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.; Yin, H.B.; Li, X.Y.; Zhu, G.M.; He, W.Y.; Gou, X. Bladder cancer cell-secreted exosomal miR-21 activates the PI3K/AKT pathway in macrophages to promote cancer progression. Int. J. Oncol. 2020, 56, 151–164. [Google Scholar] [CrossRef] [PubMed]
- Yao, M.; Cui, B.; Zhang, W.; Ma, W.; Zhao, G.; Xing, L. Exosomal miR-21 secreted by IL-1β-primed-mesenchymal stem cells induces macrophage M2 polarization and ameliorates sepsis. Life Sci. 2021, 264, 118658. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Yu, G. Hypoxic lung cancer cell-derived exosomal miR-21 mediates macrophage M2 polarization and promotes cancer cell proliferation through targeting IRF1. World J. Surg. Oncol. 2022, 20, 241. [Google Scholar] [CrossRef] [PubMed]
- Iliopoulos, D.; Jaeger, S.A.; Hirsch, H.A.; Bulyk, M.L.; Struhl, K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol. Cell. 2010, 39, 493–506. [Google Scholar] [CrossRef] [PubMed]
- Kumarswamy, R.; Volkmann, I.; Thum, T. Regulation and function of miRNA-21 in health and disease. RNA Biol. 2011, 8, 706–713. [Google Scholar] [CrossRef] [PubMed]
- van der Fits, L.; van Kester, M.S.; Qin, Y.; Out-Luiting, J.J.; Smit, F.; Zoutman, W.H.; Willemze, R.; Tensen, C.P.; Vermeer, M.H. MicroRNA-21 expression in CD4+ T cells is regulated by STAT3 and is pathologically involved in Sézary syndrome. J. Investig. Dermatol. 2011, 131, 762–768. [Google Scholar] [CrossRef]
- Chuang, H.Y.; Su, Y.K.; Liu, H.W.; Chen, C.H.; Chiu, S.C.; Cho, D.Y.; Lin, S.Z.; Chen, Y.S.; Lin, C.M. Preclinical evidence of STAT3 inhibitor pacritinib overcoming temozolomide resistance via downregulating miR-21-enriched exosomes from M2 glioblastoma-associated macrophages. J. Clin. Med. 2019, 8, 959. [Google Scholar] [CrossRef]
- Lan, J.; Sun, L.; Xu, F.; Liu, L.; Hu, F.; Song, D.; Hou, Z.; Wu, W.; Luo, X.; Wang, J.; et al. M2 macrophage-derived exosomes promote cell migration and invasion in colon cancer. Cancer Res. 2019, 79, 146–158. [Google Scholar] [CrossRef]
- Pu, J.; Xu, Z.; Nian, J.; Fang, Q.; Yang, M.; Huang, Y.; Li, W.; Ge, B.; Wang, J.; Wei, H. M2 macrophage-derived extracellular vesicles facilitate CD8+ T cell exhaustion in hepatocellular carcinoma via the miR-21-5p/YOD1/YAP/β-catenin pathway. Cell Death Discov. 2021, 7, 182. [Google Scholar] [CrossRef]
- Chang, J.; Li, H.; Zhu, Z.; Mei, P.; Hu, W.; Xiong, X.; Tao, J. microRNA-21-5p from M2 macrophage-derived extracellular vesicles promotes the differentiation and activity of pancreatic cancer stem cells by mediating KLF3. Cell Biol. Toxicol. 2022, 38, 577–590. [Google Scholar] [CrossRef]
- Howard, K.M.; Jati Kusuma, R.; Baier, S.R.; Friemel, T.; Markham, L.; Vanamala, J.; Zempleni, J. Loss of miRNAs during processing and storage of cow’s (Bos taurus) milk. J. Agric. Food Chem. 2015, 63, 588–592. [Google Scholar] [CrossRef]
- Li, Z.; Hassan, M.Q.; Jafferji, M.; Aqeilan, R.I.; Garzon, R.; Croce, C.M.; van Wijnen, A.J.; Stein, J.L.; Stein, G.S.; Lian, J.B. Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J. Biol. Chem. 2009, 284, 15676–15684. [Google Scholar] [CrossRef]
- Griffiths-Jones, S. miRBase: MicroRNA sequences and annotation. Curr. Protoc. Bioinform. 2010, 29, 12.9.1–12.9.10. [Google Scholar] [CrossRef]
- Zhang, P.P.; Wang, Y.C.; Cheng, C.; Zhang, F.; Ding, D.Z.; Chen, D.K. Runt-related transcription factor 2 influences cell adhesion-mediated drug resistance and cell proliferation in B-cell non-Hodgkin’s lymphoma and multiple myeloma. Leuk. Res. 2020, 92, 106340. [Google Scholar] [CrossRef]
- Hines, M.J.; Coffre, M.; Mudianto, T.; Panduro, M.; Wigton, E.J.; Tegla, C.; Osorio-Vasquez, V.; Kageyama, R.; Benhamou, D.; Perez, O.; et al. miR-29 sustains B cell survival and controls terminal differentiation via regulation of PI3K signaling. Cell Rep. 2020, 33, 108436. [Google Scholar] [CrossRef]
- Anastasiadou, E.; Boccellato, F.; Vincenti, S.; Rosato, P.; Bozzoni, I.; Frati, L.; Faggioni, A.; Presutti, C.; Trivedi, P. Epstein-Barr virus encoded LMP1 downregulates TCL1 oncogene through miR-29b. Oncogene 2010, 29, 1316–1328. [Google Scholar] [CrossRef]
- Teitell, M.; Damore, M.A.; Sulur, G.G.; Turner, D.E.; Stern, M.H.; Said, J.W.; Denny, C.T.; Wall, R. TCL1 oncogene expression in AIDS-related lymphomas and lymphoid tissues. Proc. Natl. Acad. Sci. USA 1999, 96, 9809–9814. [Google Scholar] [CrossRef]
- Said, J.W.; Hoyer, K.K.; French, S.W.; Rosenfelt, L.; Garcia-Lloret, M.; Koh, P.J.; Cheng, T.C.; Sulur, G.G.; Pinkus, G.S.; Kuehl, W.M.; et al. TCL1 oncogene expression in B cell subsets from lymphoid hyperplasia and distinct classes of B cell lymphoma. Lab. Investig. 2001, 81, 555–564. [Google Scholar] [CrossRef]
- Izumi, H.; Kosaka, N.; Shimizu, T.; Sekine, K.; Ochiya, T.; Takase, M. Bovine milk contains microRNA and messenger RNA that are stable under degradative conditions. J. Dairy Sci. 2012, 95, 4831–4841. [Google Scholar] [CrossRef]
- Wang, L.; Shi, Z.; Wang, X.; Mu, S.; Xu, X.; Shen, L.; Li, P. Protective effects of bovine milk exosomes against oxidative stress in IEC-6 cells. Eur. J. Nutr. 2021, 60, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Hatton, O.; Smith, M.M.; Alexander, M.; Mandell, M.; Sherman, C.; Stesney, M.W.; Hui, S.T.; Dohrn, G.; Medrano, J.; Ringwalt, K.; et al. Epstein-Barr virus latent membrane protein 1 regulates host B cell microRNA-155 and its target FOXO3a via PI3K p110α activation. Front. Microbiol. 2019, 10, 2692. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, I.; Li, Y.; Sharma, A.; Zhu, H.; Bodo, J.; Xu, W.; His, E.D.; Hill, B.T.; Almasan, A. Resistance to BTK inhibition by ibrutinib can be overcome by preventing FOXO3a nuclear export and PI3K/AKT activation in B-cell lymphoid malignancies. Cell Death Dis. 2019, 10, 924. [Google Scholar] [CrossRef] [PubMed]
- Campos, A.; Sharma, S.; Obermair, A.; Salomon, C. Extracellular vesicle-associated miRNAs and chemoresistance: A systematic review. Cancers 2021, 13, 4608. [Google Scholar] [CrossRef] [PubMed]
- Liao, T.L.; Hsieh, S.L.; Chen, Y.M.; Chen, H.H.; Liu, H.J.; Lee, H.C.; Chen, D.Y. Rituximab may cause increased hepatitis C virus viremia in rheumatoid arthritis patients through declining exosomal microRNA-155. Arthritis Rheumatol. 2018, 70, 1209–1219. [Google Scholar] [CrossRef] [PubMed]
- Rainy, N.; Zayoud, M.; Kloog, Y.; Rechavi, O.; Goldstein, I. Viral oncomiR spreading between B and T cells is employed by Kaposi’s sarcoma herpesvirus to induce non-cell-autonomous target gene regulation. Oncotarget 2016, 7, 41870–41884. [Google Scholar] [CrossRef]
- Zheng, Z.; Sun, R.; Zhao, H.J.; Fu, D.; Zhong, H.J.; Weng, X.Q.; Qu, B.; Zhao, Y.; Wang, L.; Zhao, W.L. MiR155 sensitized B-lymphoma cells to anti-PD-L1 antibody via PD-1/PD-L1-mediated lymphoma cell interaction with CD8+T cells. Mol. Cancer. 2019, 18, 54. [Google Scholar] [CrossRef]
- Poles, W.A.; Nishi, E.E.; de Oliveira, M.B.; Eugênio, A.I.P.; de Andrade, T.A.; Campos, A.H.F.M.; de Campos, R.R.; Vassallo, J.; Alves, A.C.; Scapulatempo Neto, C.; et al. Targeting the polarization of tumor-associated macrophages and modulating mir-155 expression might be a new approach to treat diffuse large B-cell lymphoma of the elderly. Cancer Immunol. Immunother. 2019, 68, 269–282. [Google Scholar] [CrossRef]
- Tingö, L.; Ahlberg, E.; Johansson, L.; Pedersen, S.A.; Chawla, K.; Sætrom, P.; Cione, E.; Simpson, M.R. Non-coding RNAs in human breast milk: A systematic review. Front. Immunol. 2021, 12, 725323. [Google Scholar] [CrossRef]
- Kirchner, B.; Pfaffl, M.W.; Dumpler, J.; Von Mutius, E.; Ege, M.J. MicroRNA in native and processed cow’s milk and its implication for the farm milk effect on asthma. J. Allergy Clin. Immunol. 2016, 137, 1893–1895. [Google Scholar] [CrossRef]
- Chutipongtanate, S.; Morrow, A.L.; Newburg, D.S. Human milk extracellular vesicles: A biological system with clinical implications. Cells 2022, 11, 2345. [Google Scholar] [CrossRef]
- Benmoussa, A.; Ly, S.; Shan, S.T.; Laugier, J.; Boilard, E.; Gilbert, C.; Provost, P. A subset of extracellular vesicles carries the bulk of microRNAs in commercial dairy cow’s milk. J. Extracell. Vesicles 2017, 6, 1401897. [Google Scholar] [CrossRef]
- Zhou, Q.; Li, M.; Wang, X.; Li, Q.; Wang, T.; Zhu, Q.; Zhou, X.; Wang, X.; Gao, X.; Li, X. Immune-related microRNAs are abundant in breast milk exosomes. Int. J. Biol. Sci. 2012, 8, 118–123. [Google Scholar] [CrossRef]
- Guo, M.M.; Zhang, K.; Zhang, J.H. Human breast milk-derived exosomal miR-148a-3p protects against necrotizing enterocolitis by regulating p53 and sirtuin 1. Inflammation 2022, 45, 1254–1268. [Google Scholar] [CrossRef]
- Gu, Y.; Li, M.; Wang, T.; Liang, Y.; Zhong, Z.; Wang, X.; Zhou, Q.; Chen, L.; Lang, Q.; He, Z.; et al. Lactation-related microRNA expression profiles of porcine breast milk exosomes. PLoS ONE 2012, 7, e43691. [Google Scholar] [CrossRef]
- Melnik, B.C.; Schmitz, G. MicroRNAs: Milk’s epigenetic regulators. Best. Pract. Res. Clin. Endocrinol. Metab. 2017, 31, 427–442. [Google Scholar] [CrossRef]
- Melnik, B.C. Milk disrupts p53 and DNMT1, the guardians of the genome: Implications for acne vulgaris and prostate cancer. Nutr. Metab. 2017, 14, 55. [Google Scholar] [CrossRef]
- Pappas, K.; Xu, J.; Zairis, S.; Resnick-Silverman, L.; Abate, F.; Steinbach, N.; Ozturk, S.; Saal, L.H.; Su, T.; Cheung, P.; et al. p53 maintains baseline expression of multiple tumor suppressor genes. Mol. Cancer Res. 2017, 15, 1051–1062. [Google Scholar] [CrossRef]
- Izquierdo, E.; Vorholt, D.; Blakemore, S.; Sackey, B.; Nolte, J.L.; Barbarino, V.; Schmitz, J.; Nickel, N.; Bachurski, D.; Lobastova, L.; et al. Extracellular vesicles and PD-L1 suppress macrophages, inducing therapy resistance in TP53-deficient B-cell malignancies. Blood 2022, 139, 3617–3629. [Google Scholar] [CrossRef]
- Qingjuan, L.; Xiaojuan, F.; Wei, Z.; Chao, W.; Pengpeng, K.; Hongbo, L.; Sanbing, Z.; Jun, H.; Min, Y.; Shuxia, L. miR-148a-3p overexpression contributes to glomerular cell proliferation by targeting PTEN in lupus nephritis. Am. J. Physiol. Cell. Physiol. 2016, 310, C470–C478. [Google Scholar] [CrossRef]
- Huang, F.; Zhao, J.L.; Wang, L.; Gao, C.C.; Liang, S.Q.; An, D.J.; Bai, J.; Chen, Y.; Han, H.; Qin, H.Y. miR-148a-3p mediates notch signaling to promote the differentiation and M1 activation of macrophages. Front. Immunol. 2017, 8, 1327. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Cai, M.; Zhu, J.; Xiao, W.; Liu, B.; Shi, Y.; Yang, X.; Liang, X.; Zheng, T.; Hu, S.; et al. miR-148a-3p promotes rabbit preadipocyte differentiation by targeting PTEN. In Vitro Cell Dev. Biol. Anim. 2018, 54, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Reif, S.; Elbaum Shiff, Y.; Golan-Gerstl, R. Milk-derived exosomes (MDEs) have a different biological effect on normal fetal colon epithelial cells compared to colon tumor cells in a miRNA-dependent manner. J. Transl. Med. 2019, 17, 325. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Zhang, P.; Gao, Y.; Fan, H.; Zhang, M.; Wu, J. Evaluation of AKT phosphorylation and PTEN loss and their correlation with the resistance of rituximab in DLBCL. Int. J. Clin. Exp. Pathol. 2015, 8, 14875–14884. [Google Scholar]
- Cui, W.; Ma, M.; Zheng, S.; Ma, Z.; Su, L.; Zhang, W. PIK3CA amplification and PTEN loss in diffused large B-cell lymphoma. Oncotarget 2017, 8, 66237–66247. [Google Scholar] [CrossRef]
- Wang, X.; Cao, X.; Sun, R.; Tang, C.; Tzankov, A.; Zhang, J.; Manyam, G.C.; Xiao, M.; Miao, Y.; Jabbar, K.; et al. Clinical significance of PTEN deletion, mutation, and loss of PTEN expression in de novo diffuse large B-cell lymphoma. Neoplasia 2018, 20, 574–593. [Google Scholar] [CrossRef]
- Ben Younes, K.; Doghri, R.; Mrad, K.; Bedhiafi, W.; Benammar-Elgaaied, A.; Sola, B.; Ben Aissa-Fennira, F. PTEN loss and cyclin A2 upregulation define a PI3K/AKT pathway activation in Helicobacter pylori-induced MALT and DLBCL gastric lymphoma with features of MALT. Appl. Immunohistochem. Mol. Morphol. 2021, 29, 56–61. [Google Scholar] [CrossRef]
- Leeman-Neill, R.J.; Bhagat, G. BCL6 as a therapeutic target for lymphoma. Expert Opin. Ther. Targets 2018, 22, 143–152. [Google Scholar] [CrossRef]
- Xiao, F.; Rui, K.; Shi, X.; Wu, H.; Cai, X.; Lui, K.O.; Lu, Q.; Ballestar, E.; Tian, J.; Zou, H.; et al. Epigenetic regulation of B cells and its role in autoimmune pathogenesis. Cell. Mol. Immunol. 2022, 19, 1215–1234. [Google Scholar] [CrossRef]
- Wigton, E.J.; Ansel, K.M. Noncoding RNAs in B cell responses. RNA Biol. 2021, 18, 633–639. [Google Scholar] [CrossRef]
- Melnik, B.C.; Schmitz, G. Milk’s role as an epigenetic regulator in health and disease. Diseases 2017, 5, 12. [Google Scholar] [CrossRef]
- Teng, G.; Hakimpour, P.; Landgraf, P.; Rice, A.; Tuschl, T.; Casellas, R.; Papavasiliou, F.N. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity 2008, 28, 621–629. [Google Scholar] [CrossRef]
- Borchert, G.M.; Holton, N.W.; Larson, E.D. Repression of human activation induced cytidine deaminase by miR-93 and miR-155. BMC Cancer 2011, 11, 347. [Google Scholar] [CrossRef]
- Frasca, D.; Diaz, A.; Romero, M.; Ferracci, F.; Blomberg, B.B. MicroRNAs miR-155 and miR-16 decrease AID and E47 in B Cells from elderly individuals. J. Immunol. 2015, 195, 2134–2140. [Google Scholar] [CrossRef]
- Recaldin, T.; Hobson, P.S.; Mann, E.H.; Ramadani, F.; Cousins, D.J.; Lavender, P.; Fear, D.J. miR-29b directly targets activation-induced cytidine deaminase in human B cells and can limit its inappropriate expression in naïve B cells. Mol. Immunol. 2018, 101, 419–428. [Google Scholar] [CrossRef]
- Alles, J.; Ludwig, N.; Rheinheimer, S.; Leidinger, P.; Grässer, F.A.; Keller, A.; Meese, E. MiR-148a impairs Ras/ERK signaling in B lymphocytes by targeting SOS proteins. Oncotarget 2017, 8, 56417–56427. [Google Scholar] [CrossRef]
- Yasuda, T.; Kometani, K.; Takahashi, N.; Imai, Y.; Aiba, Y.; Kurosaki, T. ERKs induce expression of the transcriptional repressor Blimp-1 and subsequent plasma cell differentiation. Sci. Signal. 2011, 4, ra25. [Google Scholar] [CrossRef]
- Allman, D.M.; Cancro, M.P. pERKing up the BLIMP in plasma cell differentiation. Sci. Signal. 2011, 4, pe21. [Google Scholar] [CrossRef]
- Minnich, M.; Tagoh, H.; Bönelt, P.; Axelsson, E.; Fischer, M.; Cebolla, B.; Tarakhovsky, A.; Nutt, S.L.; Jaritz, M.; Busslinger, M. Multifunctional role of the transcription factor Blimp-1 in coordinating plasma cell differentiation. Nat. Immunol. 2016, 17, 331–343. [Google Scholar] [CrossRef]
- Goodman, A.; Patel, S.P.; Kurzrock, R. PD-1-PD-L1 immune-checkpoint blockade in B-cell lymphomas. Nat. Rev. Clin. Oncol. 2017, 14, 203–220. [Google Scholar] [CrossRef]
- Vari, F.; Arpon, D.; Keane, C.; Hertzberg, M.S.; Talaulikar, D.; Jain, S.; Cui, Q.; Han, E.; Tobin, J.; Bird, R.; et al. Immune evasion via PD-1/PD-L1 on NK cells and monocyte/macrophages is more prominent in Hodgkin lymphoma than DLBCL. Blood 2018, 131, 1809–1819. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wu, L.; Tian, C.; Zhang, Y. PD-1-PD-L1 immune-checkpoint blockade in malignant lymphomas. Ann. Hematol. 2018, 97, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Xu-Monette, Z.Y.; Xiao, M.; Au, Q.; Padmanabhan, R.; Xu, B.; Hoe, N.; Rodríguez-Perales, S.; Torres-Ruiz, R.; Manyam, G.C.; Visco, C.; et al. Immune profiling and quantitative analysis decipher the clinical role of immune-checkpoint expression in the tumor immune microenvironment of DLBCL. Cancer Immunol. Res. 2019, 7, 644–657. [Google Scholar] [CrossRef] [PubMed]
- McCord, R.; Bolen, C.R.; Koeppen, H.; Kadel, E.E.; Oestergaard, M.Z.; Nielsen, T.; Sehn, L.H.; Venstrom, J.M. PD-L1 and tumor-associated macrophages in de novo DLBCL. Blood Adv. 2019, 3, 531–540. [Google Scholar] [CrossRef]
- Song, M.K.; Park, B.B.; Uhm, J. Understanding immune evasion and therapeutic targeting associated with PD-1/PD-L1 pathway in diffuse large B-cell lymphoma. Int. J. Mol. Sci. 2019, 20, 1326. [Google Scholar] [CrossRef]
- Xie, W.; Medeiros, L.J.; Li, S.; Yin, C.C.; Khoury, J.D.; Xu, J. PD-1/PD-L1 pathway and its blockade in patients with classic Hodgkin lymphoma and Non-Hodgkin large-cell lymphomas. Curr. Hematol. Malig. Rep. 2020, 15, 372–381. [Google Scholar] [CrossRef]
- Ashizawa, M.; Okayama, H.; Ishigame, T.; Thar Min, A.K.; Saito, K.; Ujiie, D.; Murakami, Y.; Kikuchi, T.; Nakayama, Y.; Noda, M.; et al. miRNA-148a-3p regulates immunosuppression in DNA mismatch repair-deficient colorectal cancer by targeting PD-L1. Mol. Cancer Res. 2019, 17, 1403–1413. [Google Scholar] [CrossRef]
- Wang, Q.; Lin, W.; Tang, X.; Li, S.; Guo, L.; Lin, Y.; Kwok, H.F. The roles of microRNAs in regulating the expression of PD-1/PD-L1 immune checkpoint. Int. J. Mol. Sci. 2017, 18, 2540. [Google Scholar] [CrossRef]
- Huang, C.; Melnick, A. Mechanisms of action of BCL6 during germinal center B cell development. Sci. China Life Sci. 2015, 58, 1226–1232. [Google Scholar] [CrossRef]
- Ye, B.H.; Lista, F.; Lo Coco, F.; Knowles, D.M.; Offit, K.; Chaganti, R.S.; Dalla-Favera, R. Alterations of a zinc finger-encoding gene, BCL-6, in diffuse large-cell lymphoma. Science 1993, 262, 747–750. [Google Scholar] [CrossRef]
- Basso, K.; Dalla-Favera, R. BCL6: Master regulator of the germinal center reaction and key oncogene in B cell lymphomagenesis. Adv. Immunol. 2010, 105, 193–210. [Google Scholar]
- van Esch, B.C.A.M.; Porbahaie, M.; Abbring, S.; Garssen, J.; Potaczek, D.P.; Savelkoul, H.F.J.; van Neerven, R.J.J. The impact of milk and its components on epigenetic programming of immune function in early life and beyond: Implications for allergy and asthma. Front. Immunol. 2020, 11, 2141. [Google Scholar] [CrossRef]
- Duy, C.; Yu, J.J.; Nahar, R.; Swaminathan, S.; Kweon, S.M.; Polo, J.M.; Valls, E.; Klemm, L.; Shojaee, S.; Cerchietti, L.; et al. BCL6 is critical for the development of a diverse primary B cell repertoire. J. Exp. Med. 2010, 207, 1209–1221. [Google Scholar] [CrossRef]
- Crotty, S.; Johnston, R.J.; Schoenberger, S.P. Effectors and memories: Bcl-6 and Blimp-1 in T and B lymphocyte differentiation. Nat. Immunol. 2010, 11, 114–120. [Google Scholar] [CrossRef]
- Atyeo, C.; Alter, G. The multifaceted roles of breast milk antibodies. Cell 2021, 184, 1486–1499. [Google Scholar] [CrossRef]
- Yu, J.C.; Khodadadi, H.; Malik, A.; Davidson, B.; Salles, É.D.S.L.; Bhatia, J.; Hale, V.L.; Baban, B. Innate immunity of neonates and infants. Front. Immunol. 2018, 9, 1759. [Google Scholar] [CrossRef]
- Yu, H.R.; Huang, L.H.; Li, S.C. Roles of microRNA in the immature immune system of neonates. Cancer Lett. 2018, 433, 99–106. [Google Scholar] [CrossRef]
- Pettersson, S.; Pobor, G.; Coutinho, A. Ontogenic development of B cell reactivities to cooperative cell signals: Dissociation between proliferation and antibody secretion. Eur. J. Immunol. 1982, 12, 653–658. [Google Scholar] [CrossRef]
- Melnik, B.C. Milk exosomal miRNAs: Potential drivers of AMPK-to-mTORC1 switching in β-cell de-differentiation of type 2 diabetes mellitus. Nutr. Metab. 2019, 16, 85. [Google Scholar] [CrossRef]
- Melnik, B.C.; Schmitz, G. Milk exosomal microRNAs: Postnatal promoters of β cell proliferation but potential inducers of β cell de-differentiation in adult life. Int. J. Mol. Sci. 2022, 23, 11503. [Google Scholar] [CrossRef]
- Dominguez, P.M.; Teater, M.; Chambwe, N.; Kormaksson, M.; Redmond, D.; Ishii, J.; Vuong, B.; Chaudhuri, J.; Melnick, A.; Vasanthakumar, A.; et al. DNA methylation dynamics of germinal center B cells are mediated by AID. Cell Rep. 2015, 12, 2086–2098. [Google Scholar] [CrossRef] [PubMed]
- Teater, M.; Dominguez, P.M.; Redmond, D.; Chen, Z.; Ennishi, D.; Scott, D.W.; Cimmino, L.; Ghione, P.; Chaudhuri, J.; Gascoyne, R.D.; et al. AICDA drives epigenetic heterogeneity and accelerates germinal center-derived lymphomagenesis. Nat. Commun. 2018, 9, 222. [Google Scholar] [CrossRef] [PubMed]
- You, H.; Xu-Monette, Z.Y.; Wei, L.; Nunns, H.; Nagy, M.L.; Bhagat, G.; Fang, X.; Zhu, F.; Visco, C.; Tzankov, A.; et al. Genomic complexity is associated with epigenetic regulator mutations and poor prognosis in diffuse large B-cell lymphoma. Oncoimmunology 2021, 10, 1928365. [Google Scholar] [CrossRef] [PubMed]
- Çakan, E.; Gunaydin, G. Activation induced cytidine deaminase: An old friend with new faces. Front. Immunol. 2022, 13, 965312. [Google Scholar] [CrossRef]
- Kozloski, G.A.; Lossos, I.S. LymphomiRs: MicroRNAs with regulatory roles in lymphomas. Curr. Opin. Hematol. 2015, 22, 362–368. [Google Scholar] [CrossRef]
- Aune, D.; Lau, R.; Chan, D.S.M.; Vieira, R.; Greenwood, D.C.; Kampman, E.; Norat, T. Dairy products and colorectal cancer risk: A systematic review and meta-analysis of cohort studies. Ann. Oncol. 2012, 23, 37–45. [Google Scholar] [CrossRef]
- Bakken, T.; Braaten, T.; Olsen, A.; Hjartåker, A.; Lund, E.; Skeie, G. Milk and risk of colorectal, colon and rectal cancer in the Norwegian Women and Cancer (NOWAC) Cohort Study. Br. J. Nutr. 2018, 119, 1274–1285. [Google Scholar] [CrossRef]
- Barrubés, L.; Babio, N.; Becerra-Tomás, N.; Rosique-Esteban, N.; Salas-Salvadó, J. Association between dairy product consumption and colorectal cancer risk in adults: A systematic review and meta-analysis of epidemiologic studies. Adv. Nutr. 2019, 10, S190–S211, Erratum in: Adv. Nutr. 2020, 11, 1055–1057. [Google Scholar] [CrossRef]
- Lumsden, A.L.; Mulugeta, A.; Hyppönen, E. Milk consumption and risk of twelve cancers: A large-scale observational and Mendelian randomisation study. Clin. Nutr. 2023, 42, 1–8. [Google Scholar] [CrossRef]
- Stremmel, W.; Weiskirchen, R.; Melnik, B.C. Milk exosomes prevent intestinal inflammation in a genetic mouse model of ulcerative colitis: A pilot experiment. Inflamm. Intest. Dis. 2020, 5, 117–123. [Google Scholar] [CrossRef]
- Reif, S.; Elbaum-Shiff, Y.; Koroukhov, N.; Shilo, I.; Musseri, M.; Golan-Gerstl, R. Cow and human milk-derived exosomes ameliorate colitis in DSS murine model. Nutrients 2020, 12, 2589. [Google Scholar] [CrossRef]
- Zhao, W.; Zheng, J.; Wei, G.; Yang, K.; Wang, G.; Sun, X. miR-148a inhibits cell proliferation and migration through targeting ErbB3 in colorectal cancer. Oncol. Lett. 2019, 18, 2530–2536. [Google Scholar] [CrossRef]
- Martino, E.; Balestrieri, A.; Mele, L.; Sardu, C.; Marfella, R.; D’Onofrio, N.; Campanile, G.; Balestrieri, M.L. Milk exosomal miR-27b worsen endoplasmic reticulum stress mediated colorectal cancer cell death. Nutrients 2022, 14, 5081. [Google Scholar] [CrossRef]
- Peng, X.; Chen, G.; Lv, B.; Lv, J. MicroRNA-148a/152 cluster restrains tumor stem cell phenotype of colon cancer via modulating CCT6A. Anticancer Drugs 2022, 33, e610–e621. [Google Scholar] [CrossRef]
- Melnik, B.C.; Stremmel, W.; Weiskirchen, R.; John, S.M.; Schmitz, G. Exosome-derived microRNAs of human milk and their effects on infant health and development. Biomolecules 2021, 11, 851. [Google Scholar] [CrossRef]
- Leroux, C.; Chervet, M.L.; German, J.B. Perspective: Milk microRNAs as important players in infant physiology and development. Adv. Nutr. 2021, 12, 1625–1635. [Google Scholar] [CrossRef]
- Melnik, B.C.; Schmitz, G. Exosomes of pasteurized milk: Potential pathogens of Western diseases. J. Transl. Med. 2019, 17, 3. [Google Scholar] [CrossRef]
- Melnik, B.C.; Weiskirchen, R.; Schmitz, G. Milk exosomal microRNAs: Friend or foe? A narrative review. ExRNA 2022, 4, 22. [Google Scholar] [CrossRef]
- Ghosh, S.; Iacucci, M. Diverse immune effects of bovine colostrum and benefits in human health and disease. Nutrients 2021, 13, 3798. [Google Scholar] [CrossRef]
- García-Martínez, J.; Pérez-Castillo, Í.M.; Salto, R.; López-Pedrosa, J.M.; Rueda, R.; Girón, M.D. Beneficial effects of bovine milk exosomes in metabolic interorgan cross-talk. Nutrients 2022, 14, 1442. [Google Scholar] [CrossRef]
- Cleveland Clinic. Health Essentials. A Healthy Diet for Diffuse Large B-Cell Lymphoma—Certain Foods Can Boost Your Well-Being During Treatment. 27 May 2022. Available online: https://health.clevelandclinic.org/diffuse-large-b-cell-lymphoma-diet/ (accessed on 15 March 2023).
- Christian, P.; Smith, E.R.; Lee, S.E.; Vargas, A.J.; Bremer, A.A.; Raiten, D.J. The need to study human milk as a biological system. Am. J. Clin. Nutr. 2021, 113, 1063–1072. [Google Scholar] [CrossRef] [PubMed]
- Wehbe, Z.; Kreydiyyeh, S. Cow’s milk may be delivering potentially harmful undetected cargoes to humans. Is it time to reconsider dairy recommendations? Nutr. Rev. 2022, 80, 874–888. [Google Scholar] [CrossRef] [PubMed]
- Klapper, W.; Kreuz, M.; Kohler, C.W.; Burkhardt, B.; Szczepanowski, M.; Salaverria, I.; Hummel, M.; Loeffler, M.; Pellissery, S.; Woessmann, W.; et al. Molecular mechanisms in Malignant Lymphomas Network Project of the Deutsche Krebshilfe. Patient age at diagnosis is associated with the molecular characteristics of diffuse large B-cell lymphoma. Blood 2012, 119, 1882–1887. [Google Scholar] [CrossRef] [PubMed]
- American Cancer Society. Non-Hodgkin Lymphoma Risk Factors. Available online: https://www.cancer.org/cancer/non-hodgkin-lymphoma/causes-risks-prevention/risk-factors.html (accessed on 5 March 2023).
- Epperla, N.; Vaughn, J.L.; Othus, M.; Hallack, A.; Costa, L.J. Recent survival trends in diffuse large B-cell lymphoma--Have we made any progress beyond rituximab? Cancer Med. 2020, 9, 5519–5525. [Google Scholar] [CrossRef]
- Shi, Y.; Han, Y.; Yang, J.; Liu, P.; He, X.; Zhang, C.; Zhou, S.; Zhou, L.; Qin, Y.; Song, Y.; et al. Clinical features and outcomes of diffuse large B-cell lymphoma based on nodal or extranodal primary sites of origin: Analysis of 1085 WHO classified cases in a single institution in China. Chin. J. Cancer Res. 2019, 31, 152–161. [Google Scholar] [CrossRef]
- Torfadottir, J.E.; Steingrimsdottir, L.; Mucci, L.; Aspelund, T.; Kasperzyk, J.L.; Olafsson, O.; Fall, K.; Tryggvadottir, L.; Harris, T.B.; Launer, L.; et al. Milk intake in early life and risk of advanced prostate cancer. Am. J. Epidemiol. 2012, 175, 144–153. [Google Scholar] [CrossRef]
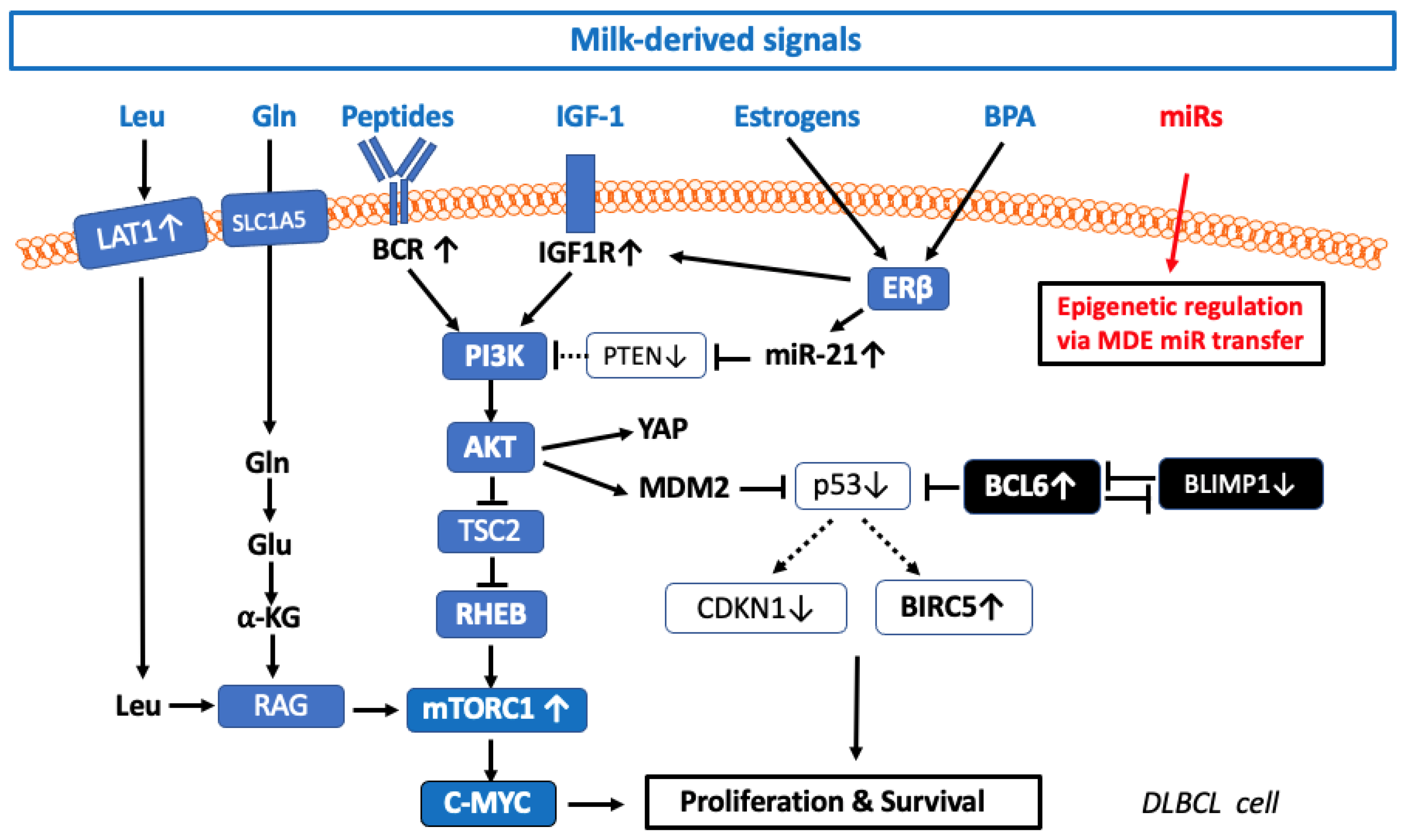
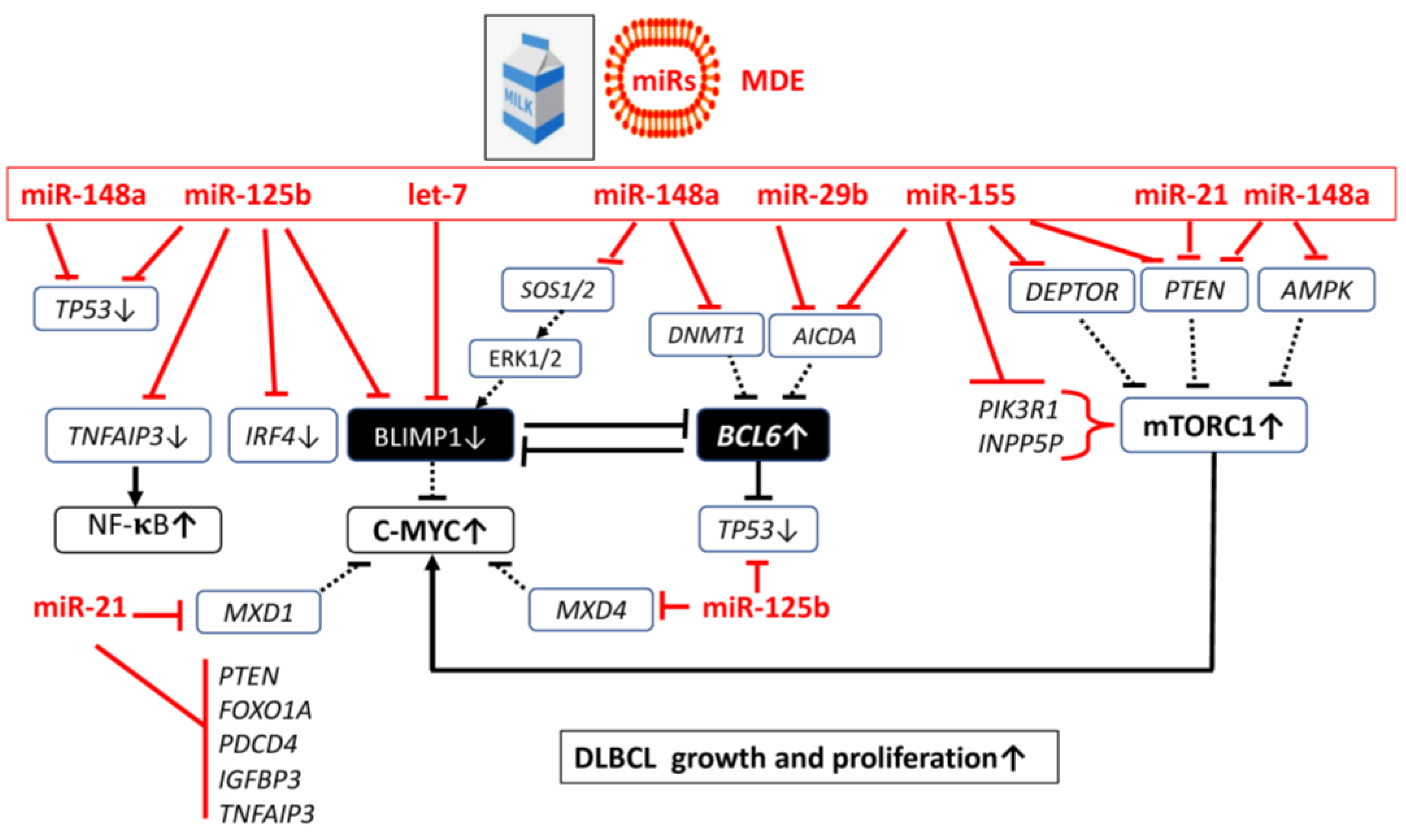
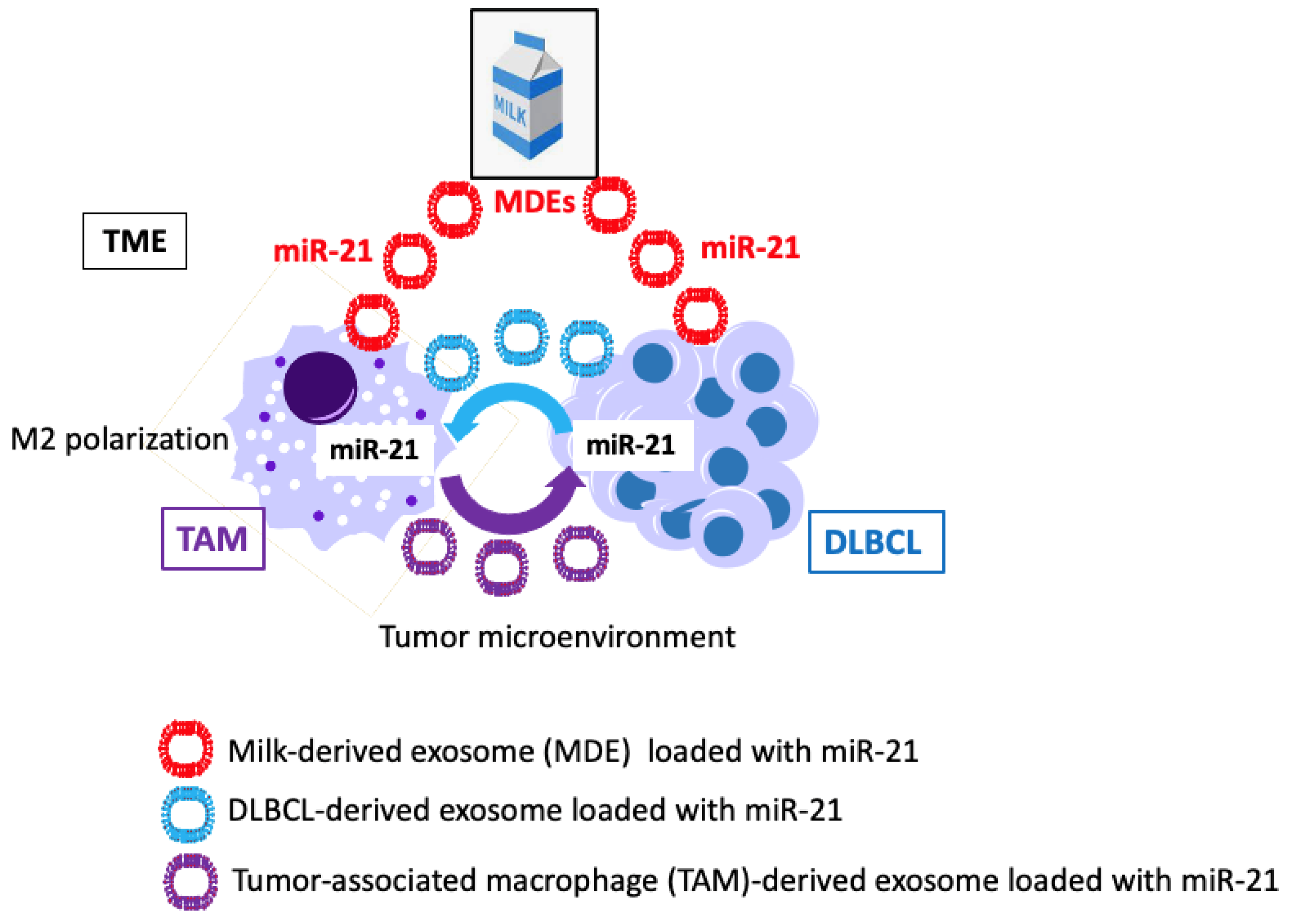

| Study Characteristics | Associated Risk | References |
|---|---|---|
| Meta-analysis 14 case-control studies and two cohort studies | Milk consumption is related to increased risk of non-Hodgkin lymphoma (NHL) RR = 1.49 (95% CI: 1.08–2.06); risk of NHL increased by 5% and 6% for each 200 g/day increment in total dairy product and milk consumption, respectively | [49] |
| An exposome-wide analysis based on the European Prospective Investigation into Cancer and Nutrition Study (n = 475,426) | Positive association between dairy intake and risk of B-cell lymphoma and DLBCL | [51] |
| Prospective China Kadoorie Biobank Study collecting ~0.5 million adults from 10 diverse (5 urban, 5 rural) areas across China during 2004–2008 | Increased lymphoma risk related to dairy consumption (mainly milk) for urban HR = 1.09 (95% CI: 0.89–1.34) and rural regions HR = 1.45 (95% CI: 1.07–1.96) | [50] |
| Compound | Cow’s Milk Intake | References | DLBCL | References |
|---|---|---|---|---|
| IGF-1 | Increased serum levels of IGF-1 activate mTORC1 | [67,68,69,70,71,72,73,74,75,76] | Increased IGF-1/IGF1R signaling | [61,62,63,64,65] |
| Leucine | Rich source of leucine (10 g leucine/100 g milk protein) activates mTORC1 | [92,93,94,95,96,97] | High expression of leucine transporter LAT1 associated with poor prognosis | [98,99] |
| Glutamine | Rich source of glutamine 9 g glutamine/100 g casein | [106,107] | Glutaminolysis activates TCA cycle and mTORC1 | [101,102,103] |
| Milk-derived peptides | B-cell epitopes stimulate BCR signaling | [147,148,149,150,151,152,153,154,155] | Persistent BCR activation augments mTORC1 signaling | [108,109,110,111,112,113,128,129,130,131,132] |
| Estrogens | Increased levels in milk of persistently pregnant dairy cows | [167,168,169,170,171,172,173,174] | High ERβ expression is related to poor prognosis | [157,158,159] |
| Bisphenol A (BPA) | Contamination of commercial milk, weak estrogenic activity increases IGF-1/mTORC1 | [201,202,203,204,205] | Lymphoma-promoting activities of BPA | [37,182,183,184,185,186] |
| BMMFs | Viral single-stranded DNA | [248,249] | Suspected to promote cancer induction early in life; Role in DLBCL not yet experimentally investigated | [250,256] |
| MiRs | DLBCL-Related miRs | References | Cow’s Milk- Derived miRs | References |
|---|---|---|---|---|
| let-7 | Over-expression of let-7 suppresses BLIMP1 (PRDM1) | [324,325] | Highly conserved in milk; component of MDEs | [411,427,428,429,430,431,432,433,434] |
| miR-125b | Increased serum levels of exosomal miR-125b-5p in DLBCL patients associated with shorter progression-free survival time and chemo-therapy resistance; targets TNFAIP3 (A20), the negative regulator of NF-κB; down-regulates the expression of interferon regulatory factor 4 (IRF4) and BLIMP1; targets TP53; reduces BLIMP1-mediated repression of pri-miR-21 | [287,288,326,328,329,330,342] | Resistant component of milk EVs and MDEs: Postprandial increase in plasma after consumption of commercial cow’s milk in humans; | [392,411,436,446] |
| miR-21 | Increased serum levels of patients with DLBCL miR-21; increased in exosome-enriched serum of patients with DLBCL; increased levels in DLBCL tumors; targets MAXD1 (MAX dimerization protein 1) promoting the formation of C-MYC-MAX heterodimers that activate C-MYC expression; targets Von Hippel Lindau mRNA (VHL); inversely correlated with the levels of FOXO1 and PTEN in DLBCL cell lines targets programmed cell death 4 (PDCD4); targets IGFBP3; drives M2 polarization of macrophages | [239,286,329,330,331,332,333,334,335,336,337,338,339,343,347,348,349,352,460,462,471,472,473,474] | Abundant signature miR of cow’s milk; component of MDEs; targets TNFAIP3, FOXO1, PTEN, BCL11B, and PDCD4; promotes M2 macrophage polarization | [238,267,353,354,427,430,446,447,448,449,450,451,452,453,454,455,456,457,473,474,475,476] |
| miR-29s | Transfection of U2932 DLBCL cells twith EBV-encoded latent membrane protein 1 (LMP1) exhibited significant upregulation of miR-29a,b and c; miR-29b enhances RUNX2 expression, which promotes proliferation of B-NHL cells; targets AID (AICDA) enhancing BCL6 expression | [496,498,538] | Component of MDEs; dose-dependent increase in plasma after intake of commercial cow’s milk; increasing RUNX2 expression in PBMCs | [409,411,448] |
| miR-155 | Higher levels of miR-155 are present in DLBCL with an ABC phenotype; highly expressed in activated B cells and proliferating lymphoblastoid cell lines; targets the PI3K inhibitor PIK3R1; activates BCR-PI3K-AKT-mTORC1 signaling at various checkpoints via targeting PTEN, DEPTOR, and SHIP1 (INPP5D); EBV upregulates miR-155. Higher miR-155 plasma levels are associated with short survival; higher levels of plasma exosomal miR-155 correlate with reduced R-CHOP response; targets AID (AICDA) enhancing BCL6 expression, the key suppressor of BLIMP1 | [28,240,241,299,358,359,360,361,362,363,364,365,366,371,372,377,378,535,536,537] | Immune-related miR of colostrum and cow’s milk; pretreatment of IEC-6 cells with MDEs increased intracellular levels of miR-155 | [429,504] |
| miR-148a | miR-148a-3p impairs RAS/ERK signaling in B cells by targeting SOS1/2 thereby reducing ERK-mediated expression of BLIMP1; miR-148a-3p targets DNMT1; enhances the expression of BCL6, the key suppressor of BLIMP1; elevated miR-148a-3p expression impaired B cell tolerance by promoting the survival of immature B cells | [28,235,299,380,526,534,539] | Signature miR of commercial cow’s milk; component of milk EVs and MDEs; targets DNMT1, TP53, PTEN, SOS1 and SOS2, thereby reduces ERK-mediated activation of BLIMP1; targets MITF, BACH2 and PDL1 | [384,427,447,511,515,517,526,527,539,551] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Melnik, B.C.; Stadler, R.; Weiskirchen, R.; Leitzmann, C.; Schmitz, G. Potential Pathogenic Impact of Cow’s Milk Consumption and Bovine Milk-Derived Exosomal MicroRNAs in Diffuse Large B-Cell Lymphoma. Int. J. Mol. Sci. 2023, 24, 6102. https://doi.org/10.3390/ijms24076102
Melnik BC, Stadler R, Weiskirchen R, Leitzmann C, Schmitz G. Potential Pathogenic Impact of Cow’s Milk Consumption and Bovine Milk-Derived Exosomal MicroRNAs in Diffuse Large B-Cell Lymphoma. International Journal of Molecular Sciences. 2023; 24(7):6102. https://doi.org/10.3390/ijms24076102
Chicago/Turabian StyleMelnik, Bodo C., Rudolf Stadler, Ralf Weiskirchen, Claus Leitzmann, and Gerd Schmitz. 2023. "Potential Pathogenic Impact of Cow’s Milk Consumption and Bovine Milk-Derived Exosomal MicroRNAs in Diffuse Large B-Cell Lymphoma" International Journal of Molecular Sciences 24, no. 7: 6102. https://doi.org/10.3390/ijms24076102
APA StyleMelnik, B. C., Stadler, R., Weiskirchen, R., Leitzmann, C., & Schmitz, G. (2023). Potential Pathogenic Impact of Cow’s Milk Consumption and Bovine Milk-Derived Exosomal MicroRNAs in Diffuse Large B-Cell Lymphoma. International Journal of Molecular Sciences, 24(7), 6102. https://doi.org/10.3390/ijms24076102








