Gut Microbiome Composition and Its Metabolites Are a Key Regulating Factor for Malignant Transformation, Metastasis and Antitumor Immunity
Abstract
1. Introduction
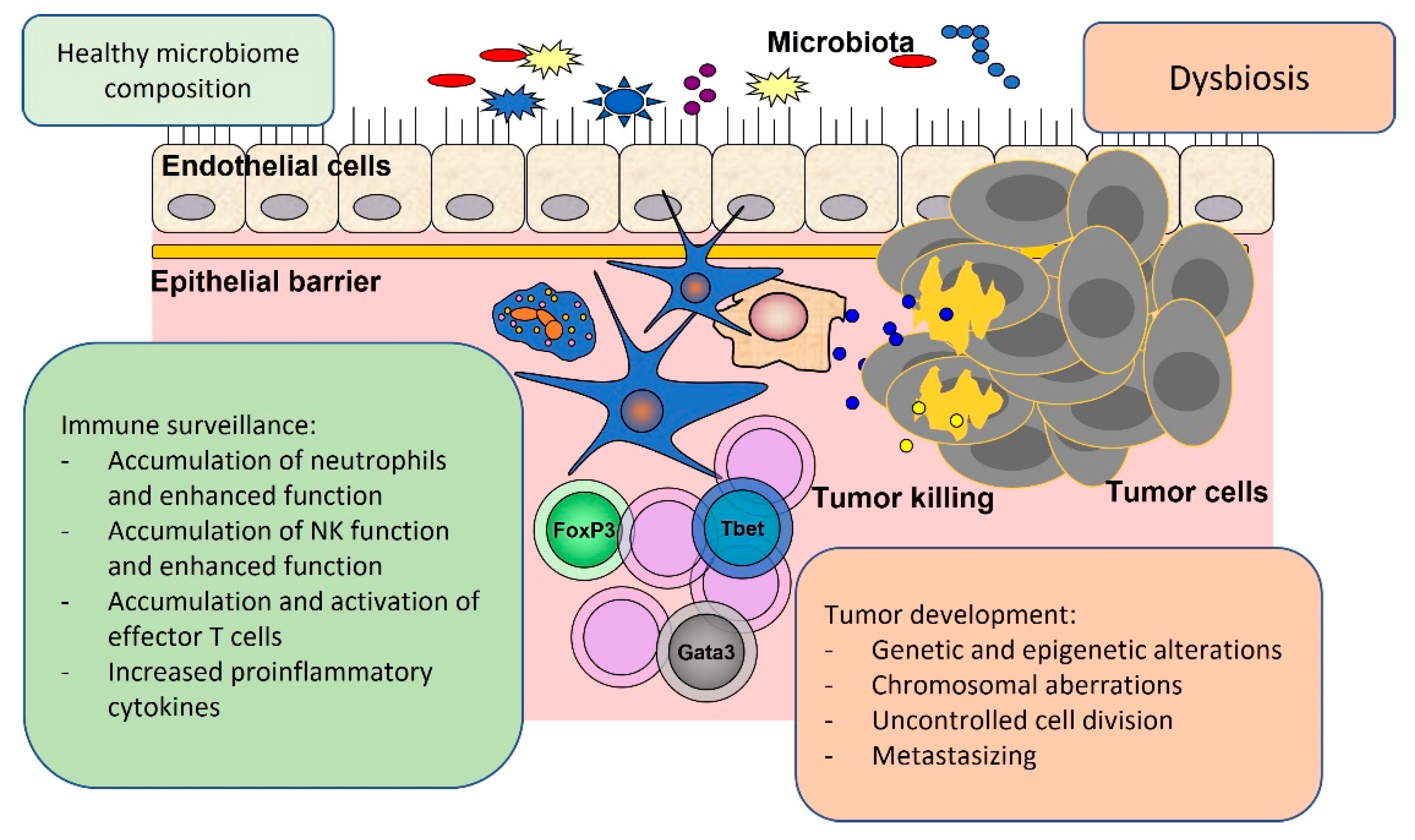
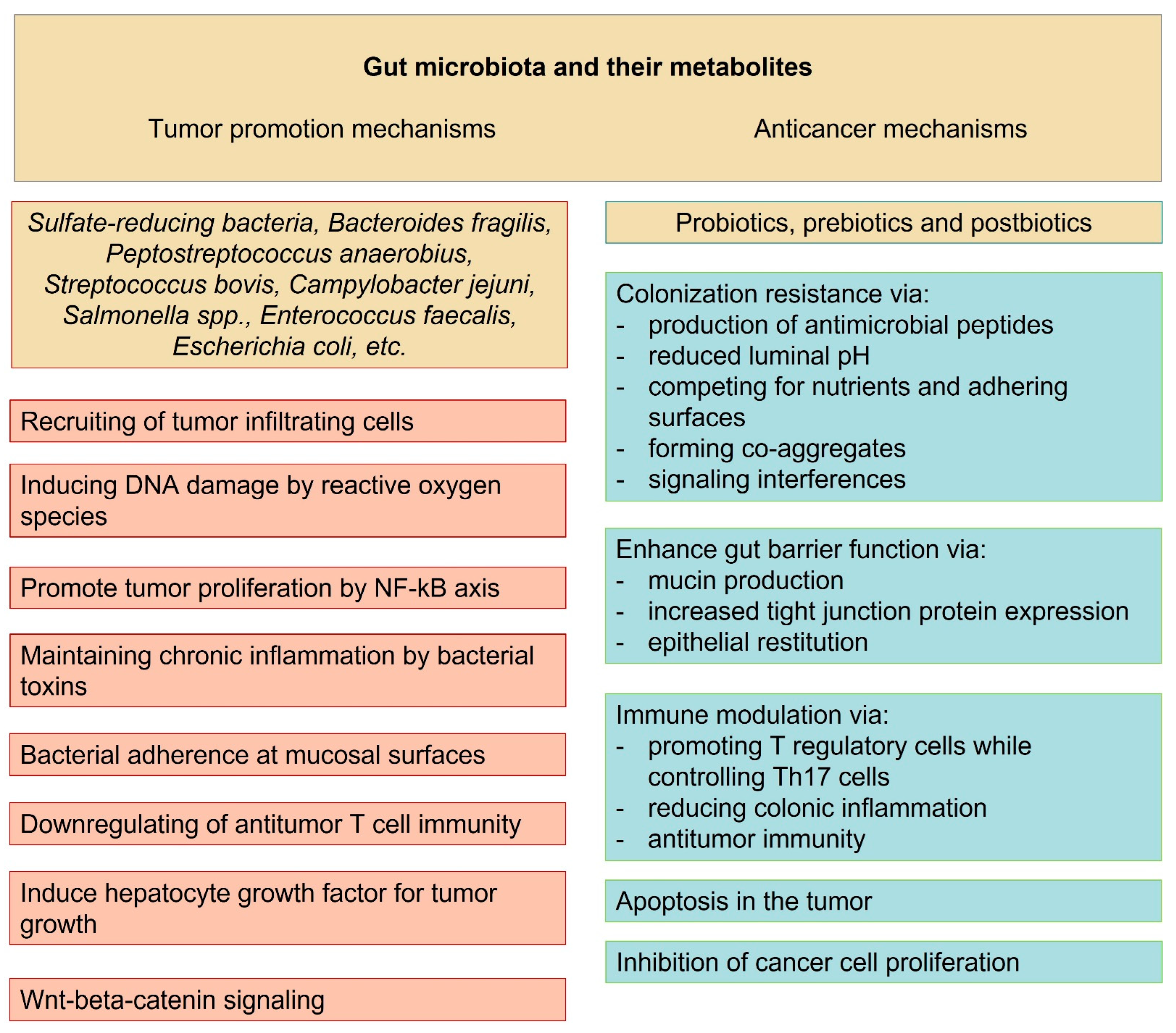
2. Local Oncogenic Effects of Gut Microbiota
3. Systemic Oncogenic Effects Related to Gut Microbiota
4. Tumor Suppressor Effects Related to Gut Microbiota
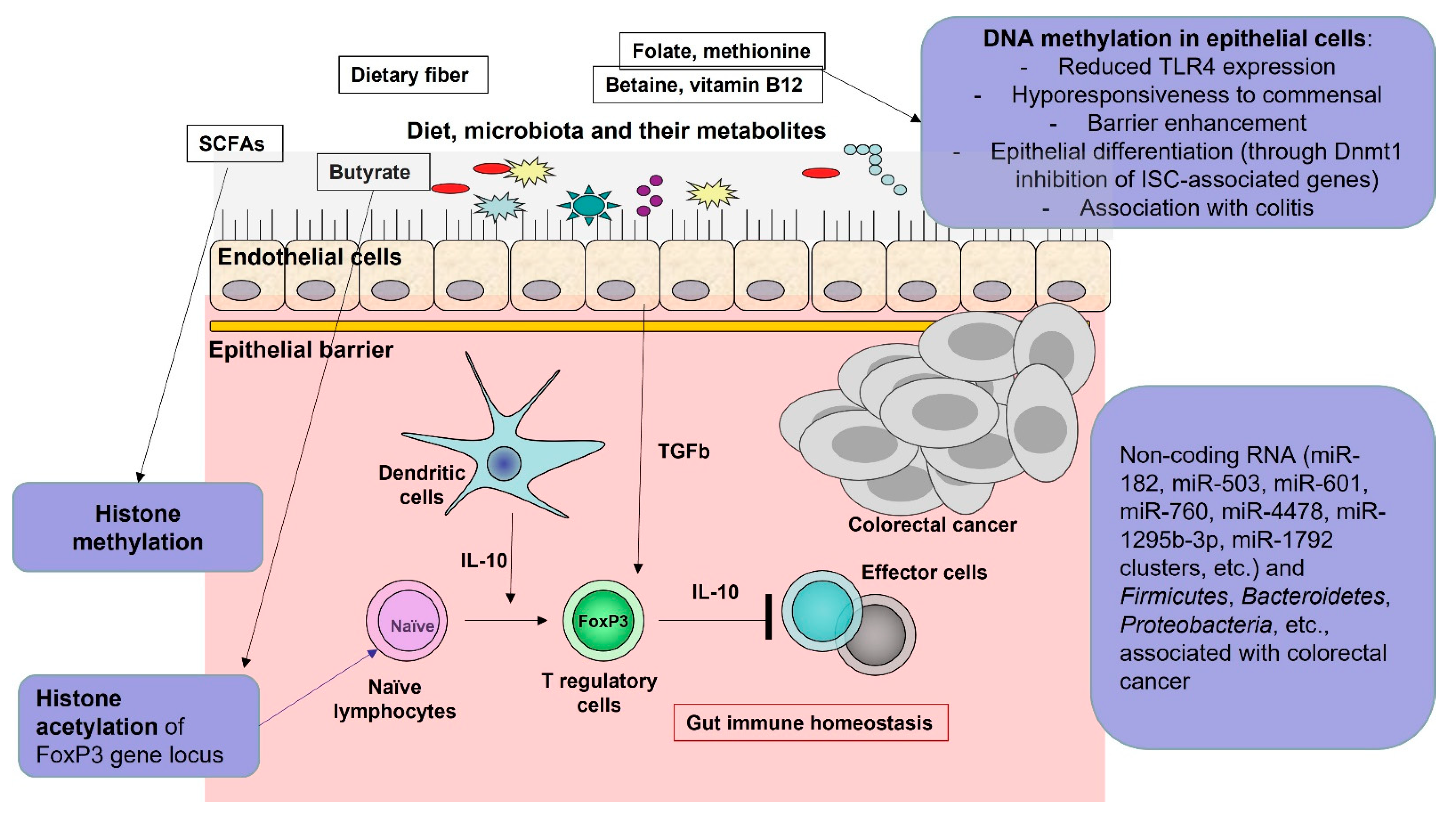
5. Role of the Microbiota-Produced Metabolites
6. Histone Deacetylases Pathways and Tumor Cell Apoptosis
7. Cancer Immunotherapy and Microbiome
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Thursby, E.; Juge, N. Introduction to the human gut microbiota. Biochem. J. 2017, 474, 1823–1836. [Google Scholar] [CrossRef] [PubMed]
- Klement, R.J.; Pazienza, V. Impact of Different Types of Diet on Gut Microbiota Profiles and Cancer Prevention and Treatment. Medicina 2019, 55, 84. [Google Scholar] [CrossRef]
- Grice, E.A.; Segre, J.A. The human microbiome: Our second genome. Annu. Rev. Genomics Hum. Genet. 2012, 13, 151–170. [Google Scholar] [CrossRef] [PubMed]
- Van Treuren, W.; Dodd, D. Microbial Contribution to the Human Metabolome: Implications for Health and Disease. Annu. Rev. Pathol. 2020, 15, 345–369. [Google Scholar] [CrossRef]
- Mair, R.D.; Sirich, T.L.; Plummer, N.S.; Meyer, T.W. Characteristics of Colon-Derived Uremic Solutes. Clin. J. Am. Soc. Nephrol. 2018, 13, 1398–1404. [Google Scholar] [CrossRef] [PubMed]
- Riazi-Rad, F.; Behrouzi, A.; Mazaheri, H.; Katebi, A.; Ajdary, S. Impact of gut microbiota on immune system. Acta Microbiol. Immunol. Hung. 2021, 68, 135–144. [Google Scholar] [CrossRef]
- Chen, W.; Liu, F.; Ling, Z.; Tong, X.; Xiang, C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS ONE 2012, 7, e39743. [Google Scholar] [CrossRef]
- Gao, Z.; Guo, B.; Gao, R.; Zhu, Q.; Qin, H. Microbiotammuneiss is associated with colorectal cancer. Front. Microbiol. 2015, 6, 20. [Google Scholar] [CrossRef]
- Xuan, C.; Shamonki, J.M.; Chung, A.; Dinome, M.L.; Chung, M.; Sieling, P.A.; Lee, D.J. Microbial dysbiosis is associated with human breast cancer. PLoS ONE 2014, 9, e83744. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, Y.; Li, S.; Peng, Z.; Liu, X.; Chen, J.; Zheng, X. Role of lung and gut microbiota on lung cancer pathogenesis. J. Cancer Res. Clin. Oncol. 2021, 147, 2177–2186. [Google Scholar] [CrossRef]
- Gasparyan, A.Y.; Ayvazyan, L.; Blackmore, H.; Kitas, G.D. Writing a narrative biomedical review: Considerations for authors, peer reviewers, and editors. Rheumatol. Int. 2011, 31, 1409–1417. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Ling, Z.; Li, L. The Intestinal Microbiota and Colorectal Cancer. Front. Immunol. 2020, 11, 615056. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.; Lee, C.G.; Jo, M.; Park, C.O.; Gwon, S.Y.; Hwang, S.; Yi, H.C.; Lee, S.Y.; Eom, Y.B.; Karim, B.; et al. Enterotoxigenic Bacteroides fragilis infection exacerbates tumorigenesis in AOM/DSS mouse model. Int. J. Med. Sci. 2020, 17, 145–152. [Google Scholar] [CrossRef]
- Velikova, T.V.; Miteva, L.; Stanilov, N.; Spassova, Z.; Stanilova, S.A. Interleukin-6 compared to the other Th17/Treg related cytokines in inflammatory bowel disease and colorectal cancer. World J. Gastroenterol. 2020, 26, 1912–1925. [Google Scholar] [CrossRef] [PubMed]
- Wernke, K.M.; Xue, M.; Tirla, A.; Kim, C.S.; Crawford, J.M.; Herzon, S.B. Structure and bioactivity of colibactin. Bioorg. Med. Chem. Lett. 2020, 30, 127280. [Google Scholar] [CrossRef]
- Dougherty, M.W.; Jobin, C. Shining a Light on Colibactin Biology. Toxins 2021, 13, 346. [Google Scholar] [CrossRef]
- Iftekhar, A.; Berger, H.; Bouznad, N.; Heuberger, J.; Boccellato, F.; Dobrindt, U.; Hermeking, H.; Sigal, M.; Meyer, T.F. Genomic aberrations after short-term exposure to colibactin-producing E. coli transform primary colon epithelial cells. Nat. Commun. 2021, 12, 1003. [Google Scholar] [CrossRef]
- Strakova, N.; Korena, K.; Karpiskova, R. Klebsiella pneumoniae producing bacterial toxin colibactin as a risk of colorectal cancer development—A systematic review. Toxicon 2021, 197, 126–135. [Google Scholar] [CrossRef]
- Eklöf, V.; Löfgren-Burström, A.; Zingmark, C.; Edin, S.; Larsson, P.; Karling, P.; Alexeyev, O.; Rutegård, J.; Wikberg, M.L.; Palmqvist, R. Cancer-associated fecal microbial markers in colorectal cancer detection. Int. J. Cancer 2017, 141, 2528–2536. [Google Scholar] [CrossRef]
- Jinadasa, R.N.; Bloom, S.E.; Weiss, R.S.; Duhamel, G.E. Cytolethal distending toxin: A conserved bacterial genotoxin that blocks cell cycle progression, leading to apoptosis of a broad range of mammalian cell lineages. Microbiology 2011, 157 Pt 7, 1851–1875. [Google Scholar] [CrossRef]
- Shenker, B.J.; Walker, L.M.; Zekavat, Z.; Ojcius, D.M.; Huang, P.R.; Boesze-Battaglia, K. Cytolethal distending toxin-induced release of interleukin-1β by human macrophages is dependent upon activation of glycogen synthase kinase 3β, spleen tyrosine kinase (Syk) and the noncanonical inflammasome. Cell. Microbiol. 2020, 22, e13194. [Google Scholar] [CrossRef] [PubMed]
- Boesze-Battaglia, K.; Dhingra, A.; Walker, L.M.; Zekavat, A.; Shenker, B.J. Internalization and Intoxication of Human Macrophages by the Active Subunit of the Aggregatibacter actinomycetemcomitans Cytolethal Distending Toxin Is Dependent Upon Cellugyrin (Synaptogyrin-2). Front. Immunol. 2020, 11, 1262. [Google Scholar] [CrossRef] [PubMed]
- Frisan, T. Bacterial genotoxins: The long journey to the nucleus of mammalian cells. Biochim. Biophys. Acta 2016, 1858, 567–575. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Gharaibeh, R.Z.; Newsome, R.C.; Pope, J.L.; Dougherty, M.W.; Tomkovich, S.; Pons, B.; Mirey, G.; Vignard, J.; Hendrixson, D.R.; et al. Campylobacter jejuni promotes colorectal tumorigenesis through the action of cytolethal distending toxin. Gut 2019, 68, 289–300. [Google Scholar] [CrossRef] [PubMed]
- Martin, O.C.B.; Bergonzini, A.; Amico, F.; Chen, P.; Shay, J.W.; Dupuy, J.; Svensson, M.; Masucci, M.G.; Frisan, T. Infection with genotoxin-producing Salmonella enterica synergises with loss of the tumour suppressor APC in promoting genomic instability via the PI3K pathway in colonic epithelial cells. Cell. Microbiol. 2019, 21, e13099. [Google Scholar] [CrossRef]
- Mughini-Gras, L.; Schaapveld, M.; Kramers, J.; Mooij, S.; Neefjes-Borst, E.A.; Pelt, W.V.; Neefjes, J. Increased colon cancer risk after severe Salmonella infection. PLoS ONE 2018, 13, e0189721. [Google Scholar] [CrossRef]
- Ye, X.; Wang, R.; Bhattacharya, R.; Boulbes, D.R.; Fan, F.; Xia, L.; Adoni, H.; Ajami, N.J.; Wong, M.C.; Smith, D.P.; et al. Fusobacterium Nucleatum Subspecies Animalis Influences Proinflammatory Cytokine Expression and Monocyte Activation in Human Colorectal Tumors. Cancer Prev. Res. 2017, 10, 398–409. [Google Scholar] [CrossRef]
- Sears, C.L.; Pardoll, D.M. Perspective: Alpha-bugs, their microbial partners, and the link to colon cancer. J. Infect. Dis. 2011, 203, 306–311. [Google Scholar] [CrossRef]
- Chen, L.; Wang, W.; Zhou, R.; Ng, S.C.; Li, J.; Huang, M.; Zhou, F.; Wang, X.; Shen, B.; A Kamm, M.; et al. Characteristics of fecal and mucosa-associated microbiota in Chinese patients with inflammatory bowel disease. Medicine 2014, 93, e51. [Google Scholar] [CrossRef]
- Saffarian, A.; Mulet, C.; Regnault, B.; Amiot, A.; Tran-Van-Nhieu, J.; Ravel, J.; Sobhani, I.; Sansonetti, P.J.; Pédron, T.; Barnich, N.; et al. Crypt- and Mucosa-Associated Core Microbiotas in Humans and Their Alteration in Colon Cancer Patients. MBio 2019, 10, 1315–1319. [Google Scholar] [CrossRef]
- Ahn, J.; Sinha, R.; Pei, Z.; Dominianni, C.; Wu, J.; Shi, J.; Goedert, J.J.; Hayes, R.B.; Yang, L. Human gut microbiome and risk for colorectal cancer. J. Natl. Cancer Inst. 2013, 105, 1907–1911. [Google Scholar] [CrossRef] [PubMed]
- Wirbel, J.; Pyl, P.T.; Kartal, E.; Zych, K.; Kashani, A.; Milanese, A.; Fleck, J.S.; Voigt, A.Y.; Palleja, A.; Ponnudurai, R.; et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat. Med. 2019, 25, 679–689. [Google Scholar] [CrossRef] [PubMed]
- Arima, K.; Zhong, R.; Ugai, T.; Zhao, M.; Haruki, K.; Akimoto, N.; Lau, M.C.; Okadome, K.; Mehta, R.S.; Väyrynen, J.P.; et al. Western-Style Diet, pks Island-Carrying Escherichia coli, and Colorectal Cancer: Analyses from Two Large Prospective Cohort Studies. Gastroenterology 2022, 163, 862–874. [Google Scholar] [CrossRef] [PubMed]
- Jian, X.; Zhu, Y.; Ouyang, J.; Wang, Y.; Lei, Q.; Xia, J.; Guan, Y.; Zhang, J.; Guo, J.; He, Y.; et al. Alterations of gut microbiome accelerate multiple myeloma progression by increasing the relative abundances of nitrogen-recycling bacteria. Microbiome 2020, 8, 74. [Google Scholar] [CrossRef]
- Ahmed, N.; Ghannoum, M.; Gallogly, M.; de Lima, M.; Malek, E. Influence of gut microbiome on multiple myeloma: Friend or foe? J. Immunother. Cancer 2020, 8, e000576. [Google Scholar] [CrossRef]
- Calcinotto, A.; Brevi, A.; Chesi, M.; Ferrarese, R.; Garcia Perez, L.; Grioni, M.; Kumar, S.; Garbitt, V.M.; Sharik, M.E.; Henderson, K.J.; et al. Microbiota-driven interleukin-17-producing cells and eosinophils synergize to accelerate multiple myeloma progression. Nat. Commun. 2018, 9, 4832. [Google Scholar] [CrossRef]
- Imbert, V.; Peyron, J.F. NF-κB in Hematological Malignancies. Biomedicines 2017, 5, 27. [Google Scholar] [CrossRef]
- Rajagopala, S.V.; Yooseph, S.; Harkins, D.M.; Moncera, K.J.; Zabokrtsky, K.B.; Torralba, M.G.; Tovchigrechko, A.; Highlander, S.K.; Pieper, R.; Sender, L.; et al. Gastrointestinal microbial populations can distinguish pediatric and adolescent Acute Lymphoblastic Leukemia (ALL) at the time of disease diagnosis. BMC Genom. 2016, 17, 635. [Google Scholar] [CrossRef]
- Yuan, L.; Wang, W.; Zhang, W.; Zhang, Y.; Wei, C.; Li, J.; Zhou, D. Gut Microbiota in Untreated Diffuse Large B Cell Lymphoma Patients. Front. Microbiol. 2021, 12, 646361. [Google Scholar] [CrossRef]
- Robles, A.I.; Traverso, G.; Zhang, M.; Roberts, N.J.; Khan, M.A.; Joseph, C.; Lauwers, G.Y.; Selaru, F.M.; Popoli, M.; Pittman, M.E.; et al. Whole-Exome Sequencing Analyses of Inflammatory Bowel Disease-Associated Colorectal Cancers. Gastroenterology 2016, 150, 931–943. [Google Scholar] [CrossRef]
- Greten, F.R.; Grivennikov, S.I. Inflammation and Cancer: Triggers, Mechanisms, and Consequences. Immunity 2019, 51, 27–41. [Google Scholar] [CrossRef] [PubMed]
- Andreeva, N.V.; Gabbasova, R.R.; Grivennikov, S.I. Microbiome in cancer progression and therapy. Curr. Opin. Microbiol. 2020, 56, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Aghamajidi, A.; Maleki Vareki, S. The Effect of the Gut Microbiota on Systemic and Antitumor Immunity and Response to Systemic Therapy against Cancer. Cancers 2022, 14, 3563. [Google Scholar] [CrossRef]
- Baruch, E.N.; Wang, J.; Wargo, J.A. Gut Microbiota and Antitumor Immunity: Potential Mechanisms for Clinical Effect. Cancer Immunol. Res. 2021, 9, 365–370. [Google Scholar] [CrossRef] [PubMed]
- Balachandran, V.P.; Łuksza, M.; Zhao, J.N.; Makarov, V.; Moral, J.A.; Remark, R.; Herbst, B.; Askan, G.; Bhanot, U.; Senbabaoglu, Y.; et al. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature 2017, 551, 512–516. [Google Scholar] [CrossRef] [PubMed]
- Su, L.F.; Kidd, B.A.; Han, A.; Kotzin, J.J.; Davis, M.M. Virus-specific CD4(+) memory-phenotype T cells are abundant in unexposed adults. Immunity 2013, 38, 373–383. [Google Scholar] [CrossRef]
- Velikova, T.; Krastev, B.; Lozenov, S.; Gencheva, R.; Peshevska-Sekulovska, M.; Nikolaev, G.; Peruhova, M. Antibiotic-Related Changes in Microbiome: The Hidden Villain behind Colorectal Carcinoma Immunotherapy Failure. Int. J. Mol. Sci. 2021, 22, 1754. [Google Scholar] [CrossRef]
- Vieira, R.S.; Castoldi, A.; Basso, P.J.; Hiyane, M.I.; Câmara, N.O.S.; Almeida, R.R. Butyrate Attenuates Lung Inflammation by Negatively Modulating Th9 Cells. Front. Immunol. 2019, 10, 67. [Google Scholar] [CrossRef]
- Matson, V.; Fessler, J.; Bao, R.; Chongsuwat, T.; Zha, Y.; Alegre, M.L.; Luke, J.J.; Gajewski, T.F. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 2018, 359, 104–108. [Google Scholar] [CrossRef]
- Jiang, L.; Xiao, X.; Ren, J.; Tang, Y.; Weng, H.; Yang, Q.; Wu, M.; Tang, W. Proteomic analysis of bladder cancer indicates Prx-I as a key molecule in BI-TK/GCV treatment system. PLoS ONE 2014, 9, e98764. [Google Scholar] [CrossRef]
- Yoon, Y.; Kim, G.; Jeon, B.N.; Fang, S.; Park, H. Bifidobacterium Strain-Specific Enhances the Efficacy of Cancer Therapeutics in Tumor-Bearing Mice. Cancers 2021, 13, 957. [Google Scholar] [CrossRef]
- Demin, N.N.; Karmanova, I.G.; Rubinskaya, N.L.; Khomutetskaya, O.E. Comparative neurochemical and physiological characteristics of catalepsy-type rest and sleep. Neurosci. Behav. Physiol. 1978, 9, 98–102. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Tinoco, R.; Elmen, L.; Segota, I.; Xian, Y.; Fujita, Y.; Sahu, A.; Zarecki, R.; Marie, K.; Feng, Y.; et al. Gut microbiota dependent antitumor immunity restricts melanoma growth in Rnf5(-/-) mice. Nat. Commun. 2019, 10, 1492. [Google Scholar] [CrossRef] [PubMed]
- Ghoneum, M.; Felo, N.; Agrawal, S.; Agrawal, A. A novel kefir product (PFT) activates dendritic cells to induce CD4+T and CD8+T cell responses in vitro. Int. J. Immunopathol. Pharmacol. 2015, 28, 488–496. [Google Scholar] [CrossRef] [PubMed]
- Jacouton, E.; Michel, M.L.; Torres-Maravilla, E.; Chain, F.; Langella, P.; Bermúdez-Humarán, L.G. Elucidating the Immune-Related Mechanisms by Which Probiotic Strain Lactobacillus casei BL23 Displays Anti-tumoral Properties. Front. Microbiol. 2019, 9, 3281. [Google Scholar] [CrossRef] [PubMed]
- Sugimura, N.; Li, Q.; Chu, E.S.H.; Lau, H.C.H.; Fong, W.; Liu, W.; Liang, C.; Nakatsu, G.; Su, A.C.Y.; Coker, O.O.; et al. Lactobacillus gallinarum modulates the gut microbiota and produces anti-cancer metabolites to protect against colorectal tumourigenesis. Gut 2022, 71, 2011–2021. [Google Scholar] [CrossRef] [PubMed]
- Hua, M.C.; Lin, T.Y.; Lai, M.W.; Kong, M.S.; Chang, H.J.; Chen, C.C. Probiotic Bio-Three induces Th1 and mmuneflammatory effects in PBMC and dendritic cells. World J. Gastroenterol. 2010, 16, 3529–3540. [Google Scholar] [CrossRef] [PubMed]
- Donohoe, D.R.; Holley, D.; Collins, L.B.; Montgomery, S.A.; Whitmore, A.C.; Hillhouse, A.; Curry, K.P.; Renner, S.W.; Greenwalt, A.; Ryan, E.P.; et al. A gnotobiotic mouse model demonstrates that dietary fiber protects against colorectal tumorigenesis in a microbiota- and butyrate-dependent manner. Cancer Discov. 2014, 4, 1387–1397. [Google Scholar] [CrossRef]
- Singh, N.; Gurav, A.; Sivaprakasam, S.; Brady, E.; Padia, R.; Shi, H.; Thangaraju, M.; Prasad, P.D.; Manicassamy, S.; Munn, D.H.; et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 2014, 40, 128–139. [Google Scholar] [CrossRef]
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly, Y.M.; Glickman, J.N.; Garrett, W.S. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013, 341, 569–573. [Google Scholar] [CrossRef]
- Salcedo, R.; Worschech, A.; Cardone, M.; Jones, Y.; Gyulai, Z.; Dai, R.M.; Wang, E.; Ma, W.; Haines, D.; O’hUigin, C.; et al. MyD88-mediated signaling prevents development of adenocarcinomas of the colon: Role of interleukin 18. J. Exp. Med. 2010, 207, 1625–1636. [Google Scholar] [CrossRef] [PubMed]
- Swanson, K.V.; Deng, M.; Ting, J.P. The NLRP3 inflammasome: Molecular activation and regulation to therapeutics. Nat. Rev. Immunol. 2019, 19, 477–489. [Google Scholar] [CrossRef] [PubMed]
- Geng, H.W.; Yin, F.Y.; Zhang, Z.F.; Gong, X.; Yang, Y. Butyrate Suppresses Glucose Metabolism of Colorectal Cancer Cells via GPR109a-AKT Signaling Pathway and Enhances Chemotherapy. Front. Mol. Biosci. 2021, 8, 634874. [Google Scholar] [CrossRef] [PubMed]
- Kespohl, M.; Vachharajani, N.; Luu, M.; Harb, H.; Pautz, S.; Wolff, S.; Sillner, N.; Walker, A.; Schmitt-Kopplin, P.; Boettger, T.; et al. The Microbial Metabolite Butyrate Induces Expression of Th1-Associated Factors in CD4+ T Cells. Front. Immunol. 2017, 8, 1036. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Chen, Y.; Zhang, S.; Dong, L. Gut microbiota-mediated immunomodulation in tumor. J. Exp. Clin. Cancer Res. 2021, 40, 221. [Google Scholar] [CrossRef]
- Ma, C.; Han, M.; Heinrich, B.; Fu, Q.; Zhang, Q.; Sandhu, M.; Agdashian, D.; Terabe, M.; Berzofsky, J.A.; Fako, V.; et al. Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science 2018, 360, eaan5931. [Google Scholar] [CrossRef]
- Zhang, W.; An, Y.; Qin, X.; Wu, X.; Wang, X.; Hou, H.; Song, X.; Liu, T.; Wang, B.; Huang, X.; et al. Gut Microbiota-Derived Metabolites in Colorectal Cancer: The Bad and the Challenges. Front. Oncol. 2021, 11, 739648. [Google Scholar] [CrossRef]
- Liu, T.; Song, X.; Khan, S.; Li, Y.; Guo, Z.; Li, C.; Wang, S.; Dong, W.; Liu, W.; Wang, B.; et al. The Gut Microbiota at the Intersection of Bile Acids and Intestinal Carcinogenesis: An Old Story, Yet Mesmerizing. Int. J. Cancer 2020, 146, 1780–1790. [Google Scholar] [CrossRef]
- Megna, B.W.; Carney, P.R.; Nukaya, M.; Geiger, P.; Kennedy, G.D. Indole-3-carbinol induces tumor cell death: Function follows form. J. Surg. Res. 2016, 204, 47–54. [Google Scholar] [CrossRef]
- Bae, S.; Ulrich, C.M.; Neuhouser, M.L.; Malysheva, O.; Bailey, L.B.; Xiao, L.; Brown, E.C.; Cushing-Haugen, K.L.; Zheng, Y.; Cheng, T.-Y.D.; et al. Plasma Choline Metabolites and Colorectal Cancer Risk in the Women’s Health Initiative Observational Study. Cancer Res. 2014, 74, 7442–7452. [Google Scholar] [CrossRef]
- Liu, X.; Liu, H.; Yuan, C.; Zhang, Y.; Wang, W.; Hu, S.; Liu, L.; Wang, Y. Preoperative Serum TMAO Level is a New Prognostic Marker for Colorectal Cancer. Biomark. Med. 2017, 11, 443–447. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Wang, Q.; Li, L. A Genome-Wide Systems Analysis Reveals Strong Link Between Colorectal Cancer and Trimethylamine N-Oxide (TMAO), a Gut Microbial Metabolite of Dietary Meat and Fat. BMC Genom. 2015, 16 (Suppl. S7), S4. [Google Scholar] [CrossRef] [PubMed]
- Yazici, C.; Wolf, P.G.; Kim, H.; Cross, T.L.; Vermillion, K.; Carroll, T.; Augustus, G.J.; Mutlu, E.; Tussing-Humphreys, L.; Braunschweig, C.; et al. Race-Dependent Association of Sulfidogenic Bacteria with Colorectal Cancer. Gut 2017, 66, 1983–1994. [Google Scholar] [CrossRef]
- Nguyen, L.H.; Ma, W.; Wang, D.D.; Cao, Y.; Mallick, H.; Gerbaba, T.K.; Lloyd-Price, J.; Abu-Ali, G.; Hall, A.B.; Sikavi, D.; et al. Association Between Sulfur-Metabolizing Bacterial Communities in Stool and Risk of Distal Colorectal Cancer in Men. Gastroenterology 2020, 158, 1313–1325. [Google Scholar] [CrossRef] [PubMed]
- Attene-Ramos, M.S.; Wagner, E.D.; Gaskins, H.R.; Plewa, M.J. Hydrogen Sulfide Induces Direct Radical-Associated DNA Damage. Mol. Cancer Res. 2007, 5, 455–459. [Google Scholar] [CrossRef] [PubMed]
- Bayerdörffer, E.; Mannes, G.A.; Richter, W.O.; Ochsenkühn, T.; Wiebecke, B.; Köpcke, W.; Paumgartner, G. Increased Serum Deoxycholic Acid Levels in Men with Colorectal Adenomas. Gastroenterology 1993, 104, 145–151. [Google Scholar] [CrossRef]
- Bayerdörffer, E.; Mannes, G.A.; Ochsenkühn, T.; Dirschedl, P.; Wiebecke, B.; Paumgartner, G. Unconjugated Secondary Bile Acids in the Serum of Patients with Colorectal Adenomas. Gut 1995, 36, 268–273. [Google Scholar] [CrossRef]
- Ocvirk, S.; Wilson, A.S.; Posma, J.M.; Li, J.V.; Koller, K.R.; Day, G.M.; Flanagan, C.A.; Otto, J.E.; Sacco, P.E.; Sacco, F.D.; et al. A Prospective Cohort Analysis of Gut Microbial Co-Metabolism in Alaska Native and Rural African People at High and Low Risk of Colorectal Cancer. Am. J. Clin. Nutr. 2020, 111, 406–419. [Google Scholar] [CrossRef]
- Liu, L.; Dong, W.; Wang, S.; Zhang, Y.; Liu, T.; Xie, R.; Wang, B.; Cao, H. Deoxycholic Acid Disrupts the Intestinal Mucosal Barrier and Promotes Intestinal Tumorigenesis. Food Funct. 2018, 9, 5588–5597. [Google Scholar] [CrossRef]
- Dong, W.; Liu, L.; Dou, Y.; Xu, M.; Liu, T.; Wang, S.; Zhang, Y.; Deng, B.; Wang, B.; Cao, H. Deoxycholic Acid Activates Epidermal Growth Factor Receptor and Promotes Intestinal Carcinogenesis by ADAM17-Dependent Ligand Release. J. Cell. Mol. Med. 2018, 22, 4263–4273. [Google Scholar] [CrossRef]
- Cao, H.; Xu, M.; Dong, W.; Deng, B.; Wang, S.; Zhang, Y.; Wang, S.; Luo, S.; Wang, W.; Qi, Y.; et al. Secondary Bile Acid-Induced Dysbiosis Promotes Intestinal Carcinogenesis. Int. J. Cancer 2017, 140, 2545–2556. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.Y.; Crawley, S.; Hokari, R.; Kwon, S.; Kim, Y.S. Bile Acid Regulates MUC2 Transcription in Colon Cancer Cells via Positive EGFR/PKC/Ras/ERK/CREB, PI3K/Akt/IkappaB/NF-kappaB and P38/MSK1/CREB Pathways and Negative JNK/c-Jun/AP-1 Pathway. Int. J. Oncol. 2010, 36, 941–953. [Google Scholar] [CrossRef] [PubMed]
- Fu, T.; Coulter, S.; Yoshihara, E.; Oh, T.G.; Fang, S.; Cayabyab, F.; Zhu, Q.; Zhang, T.; Leblanc, M.; Liu, S.; et al. FXR Regulates Intestinal Cancer Stem Cell Proliferation. Cell 2019, 176, 1098–1112.e18. [Google Scholar] [CrossRef] [PubMed]
- Jean-Louis, S.; Akare, S.; Ali, M.A.; Mash, E.A., Jr.; Meuillet, E.; Martinez, J.D. Deoxycholic Acid Induces Intracellular Signaling Through Membrane Perturbations. J. Biol. Chem. 2006, 281, 14948–14960. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Wang, P.P.; Zhao, J.; Green, R.; Sun, Z.; Roebothan, B.; Squires, J.; Buehler, S.; Dicks, E.; Zhao, J.; et al. Dietary N-Nitroso Compounds and Risk of Colorectal Cancer: A Case-Control Study in Newfoundland and Labrador and Ontario, Canada. Br. J. Nutr. 2014, 111, 1109–1117. [Google Scholar] [CrossRef]
- Hebels, D.G.; Briedé, J.J.; Khampang, R.; Kleinjans, J.C.; de Kok, T.M. Radical Mechanisms in Nitrosamine- and Nitrosamide-Induced Whole-Genome Gene Expression Modulations in Caco-2 Cells. Toxicol. Sci. 2010, 116, 194–205. [Google Scholar] [CrossRef]
- Hebels, D.G.; Jennen, D.G.; Kleinjans, J.C.; de Kok, T.M. Molecular Signatures of N-Nitroso Compounds in Caco-2 Cells: Implications for Colon Carcinogenesis. Toxicol. Sci. 2009, 108, 290–300. [Google Scholar] [CrossRef]
- Gottschalg, E.; Scott, G.B.; Burns, P.A.; Shuker, D.E. Potassium Diazoacetate-Induced P53 Mutations In Vitro in Relation to Formation of O6-Carboxymethyl- and O6-Methyl-2′-Deoxyguanosine DNA Adducts: Relevance for Gastrointestinal Cancer. Carcinogenesis 2007, 28, 356–362. [Google Scholar] [CrossRef]
- Lewin, M.H.; Bailey, N.; Bandaletova, T.; Bowman, R.; Cross, A.J.; Pollock, J.; Shuker, D.E.G.; Bingham, S.A. Red Meat Enhances the Colonic Formation of the DNA Adduct O6-Carboxymethyl Guanine: Implications for Colorectal Cancer Risk. Cancer Res. 2006, 66, 1859–1865. [Google Scholar] [CrossRef]
- Helmus, D.S.; Thompson, C.L.; Zelenskiy, S.; Tucker, T.C.; Li, L. Red Meat-Derived Heterocyclic Amines Increase Risk of Colon Cancer: A Population-Based Case-Control Study. Nutr. Cancer 2013, 65, 1141–1150. [Google Scholar] [CrossRef]
- Cross, A.J.; Ferrucci, L.M.; Risch, A.; Graubard, B.I.; Ward, M.H.; Park, Y.; Hollenbeck, A.R.; Schatzkin, A.; Sinha, R. A Large Prospective Study of Meat Consumption and Colorectal Cancer Risk: An Investigation of Potential Mechanisms Underlying This Association. Cancer Res. 2010, 70, 2406–2414. [Google Scholar] [CrossRef]
- Hasegawa, R.; Sano, M.; Tamano, S.; Imaida, K.; Shirai, T.; Nagao, M.; Sugimura, T.; Ito, N. Dose-Dependence of 2-Amino-1-Methyl-6-Phenylimidazo [4,5-B]-Pyridine (PhIP) Carcinogenicity in Rats. Carcinogenesis 1993, 14, 2553–2557. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Lin, X.; Li, Z.; Li, Q.; Bi, K. Quantitative Metabolomics for Investigating the Value of Polyamines in the Early Diagnosis and Therapy of Colorectal Cancer. Oncotarget 2017, 9, 4583–4592. [Google Scholar] [CrossRef] [PubMed]
- Manna, S.K.; Tanaka, N.; Krausz, K.W.; Haznadar, M.; Xue, X.; Matsubara, T.; Bowman, E.D.; Fearon, E.R.; Harris, C.C.; Shah, Y.M.; et al. Biomarkers of Coordinate Metabolic Reprogramming in Colorectal Tumors in Mice and Humans. Gastroenterology 2014, 146, 1313–1324. [Google Scholar] [CrossRef]
- Guo, Y.; Ye, Q.; Deng, P.; Cao, Y.; He, D.; Zhou, Z.; Wang, C.; Zaytseva, Y.Y.; Schwartz, C.E.; Lee, E.Y.; et al. Spermine Synthase and MYC Cooperate to Maintain Colorectal Cancer Cell Survival by Repressing Bim Expression. Nat. Commun. 2020, 11, 3243. [Google Scholar] [CrossRef] [PubMed]
- Clausen, M.R.; Mortensen, P.B. Fecal Ammonia in Patients with Adenomatous Polyps and Cancer of the Colon. Nutr. Cancer 1992, 18, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Visek, W.J. Diet and Cell Growth Modulation by Ammonia. Am. J. Clin. Nutr. 1978, 31, S216–S220. [Google Scholar] [CrossRef]
- Cheng, C.; Xie, Z.; Li, Y.; Wang, J.; Qin, C.; Zhang, Y. PTBP1 Knockdown Overcomes the Resistance to Vincristine and Oxaliplatin in Drug-Resistant Colon Cancer Cells Through Regulation of Glycolysis. BioMed Pharm. 2018, 108, 194–200. [Google Scholar] [CrossRef]
- Haas, R.; Smith, J.; Rocher-Ros, V.; Nadkarni, S.; Montero-Melendez, T.; D’Acquisto, F.; Bland, E.J.; Bombardieri, M.; Pitzalis, C.; Perretti, M.; et al. Lactate Regulates Metabolic and Pro-Inflammatory Circuits in Control of T Cell Migration and Effector Functions. PloS Biol. 2015, 13, e1002202. [Google Scholar] [CrossRef]
- Gao, R.; Gao, Z.; Huang, L.; Qin, H. Gut microbiota and colorectal cancer. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 757–769. [Google Scholar] [CrossRef]
- Chen, F.; Dai, X.; Zhou, C.; Li, K.-X.; Zhang, Y.-J.; Lou, X.-Y.; Zhu, Y.-M.; Sun, Y.-L.; Peng, B.-X.; Cui, W. Integrated analysis of the faecal metagenome and serum metabolome reveals the role of gut microbiome-associated metabolites in the detection of colorectal cancer and adenoma. Gut 2022, 71, 1315–1325. [Google Scholar] [CrossRef] [PubMed]
- Woo, V.; Alenghat, T. Epigenetic regulation by gut microbiota. Gut Microbes. 2022, 14, 2022407. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Li, Y.; Stoll, M.L.; Tollefsbol, T.O. The Epigenetic Connection Between the Gut Microbiome in Obesity and Diabetes. Front. Genet. 2020, 10, 1329. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wang, C.Z.; Wan, J.Y.; Yao, H.; Yuan, C.S. Dissecting the Interplay Mechanism between Epigenetics and Gut Microbiota: Health Maintenance and Disease Prevention. Int. J. Mol. Sci. 2021, 22, 6933. [Google Scholar] [CrossRef] [PubMed]
- Peruhova, M.; Peshevska-Sekulovska, M.; Krastev, B.; Panayotova, G.; Georgieva, V.; Konakchieva, R.; Nikolaev, G.; Velikova, T.V. What could microRNA expression tell us more about colorectal serrated pathway carcinogenesis? World J. Gastroenterol. 2020, 26, 6556–6571. [Google Scholar] [CrossRef] [PubMed]
- Ropero, S.; Esteller, M. The role of histone deacetylases (HDACs) in human cancer. Mol. Oncol. 2007, 1, 19–25. [Google Scholar] [CrossRef]
- Boffa, L.C.; Lupton, J.R.; Mariani, M.R.; Ceppi, M.; Newmark, H.L.; Scalmati, A.; Lipkin, M. Modulation of colonic epithelial cell proliferation, histone acetylation, and luminal short chain fatty acids by variation of dietary fiber (wheat bran) in rats. Cancer Res. 1992, 52, 5906–5912. [Google Scholar]
- Miro-Blanch, J.; Yanes, O. Epigenetic Regulation at the Interplay Between Gut Microbiota and Host Metabolism. Front. Genet. 2019, 10, 638. [Google Scholar] [CrossRef]
- Yang, W.; Yu, T.; Huang, X.; Bilotta, A.J.; Xu, L.; Lu, Y.; Sun, J.; Pan, F.; Zhou, J.; Zhang, W.; et al. Intestinal microbiota-derived short-chain fatty acids regulation of immune cell IL-22 production and gut immunity. Nat. Commun. 2020, 11, 4457. [Google Scholar] [CrossRef]
- Fellows, R.; Denizot, J.; Stellato, C.; Cuomo, A.; Jain, P.; Stoyanova, E.; Balázsi, S.; Hajnády, Z.; Liebert, A.; Kazakevych, J.; et al. Microbiota derived short chain fatty acids promote histone crotonylation in the colon through histone deacetylases. Nat. Commun. 2018, 9, 105. [Google Scholar] [CrossRef]
- Kaiko, G.E.; Ryu, S.H.; Koues, O.I.; Collins, P.L.; Solnica-Krezel, L.; Pearce, E.J.; Pearce, E.L.; Oltz, E.M.; Stappenbeck, T.S. The Colonic Crypt Protects Stem Cells from Microbiota-Derived Metabolites. Cell 2016, 165, 1708–1720. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.E.; Hashimoto-Hill, S.; Woo, V.; Eshleman, E.M.; Whitt, J.; Engleman, L.; Karns, R.; Denson, L.A.; Haslam, D.B.; Alenghat, T. Microbiota-derived metabolite promotes HDAC3 activity in the gut. Nature 2020, 586, 108–112. [Google Scholar] [CrossRef]
- Turgeon, N.; Gagné, J.M.; Blais, M.; Gendron, F.P.; Boudreau, F.; Asselin, C. The acetylome regulators Hdac1 and Hdac2 differently modulate intestinal epithelial cell dependent homeostatic responses in experimental colitis. Am. J. Physiol. Gastrointest. Liver Physiol. 2014, 306, G594–G605. [Google Scholar] [CrossRef]
- Gonneaud, A.; Turgeon, N.; Boudreau, F.; Perreault, N.; Rivard, N.; Asselin, C. Distinct Roles for Intestinal Epithelial Cell-Specific Hdac1 and Hdac2 in the Regulation of Murine Intestinal Homeostasis. J. Cell. Physiol. 2016, 231, 436–448. [Google Scholar] [CrossRef] [PubMed]
- Zimberlin, C.D.; Lancini, C.; Sno, R.; Rosekrans, S.L.; McLean, C.M.; Vlaming, H.; van den Brink, G.R.; Bots, M.; Medema, J.P.; Dannenberg, J.H. HDAC1 and HDAC2 collectively regulate intestinal stem cell homeostasis. FASEB J. 2015, 29, 2070–2080. [Google Scholar] [CrossRef]
- Liu, S.; Chang, W.; Jin, Y.; Feng, C.; Wu, S.; He, J.; Xu, T. The function of histone acetylation in cervical cancer development. Biosci. Rep. 2019, 39, BSR20190527. [Google Scholar] [CrossRef] [PubMed]
- Taur, Y.; Jenq, R.R.; Perales, M.A.; Littmann, E.R.; Morjaria, S.; Ling, L.; No, D.; Gobourne, A.; Viale, A.; Dahi, P.B.; et al. The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood 2014, 124, 1174–1182. [Google Scholar] [CrossRef]
- Bhatt, A.P.; Redinbo, M.R.; Bultman, S.J. The role of the microbiome in cancer development and therapy. CA Cancer J. Clin. 2017, 67, 326–344. [Google Scholar] [CrossRef]
- Peruhova, M.; Peshevska-Sekulovska, M.; Velikova, T. Interactions between human microbiome, liver diseases, and immunosuppression after liver transplant. World J. Immunol. 2021, 11, 11–16. [Google Scholar] [CrossRef]
- Nakov, R.; Velikova, T. Chemical Metabolism of Xenobiotics by Gut Microbiota. Curr. Drug. Metab. 2020, 21, 260–269. [Google Scholar] [CrossRef]
- Velikova, T.; Nakov, R.; Ianiro, G. 2.10—Medication and Health Risks Associated with Neglected Side Effects on Gut Microbiota. In Comprehensive Gut Microbiota; Elsevier: Amsterdam, The Netherlands, 2022; Volume 112, p. 124. [Google Scholar] [CrossRef]
- Wallace, B.D.; Wang, H.; Lane, K.T.; Scott, J.E.; Orans, J.; Koo, J.S.; Venkatesh, M.; Jobin, C.; Yeh, L.-A.; Mani, S.; et al. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science 2010, 330, 831–835. [Google Scholar] [CrossRef] [PubMed]
- Zitvogel, L.; Galluzzi, L.; Viaud, S.; Vétizou, M.; Daillère, R.; Merad, M.; Kroemer, G. Cancer and the gut microbiota: An unexpected link. Sci. Transl. Med. 2015, 7, 271ps1. [Google Scholar] [CrossRef] [PubMed]
- Raza, M.H.; Gul, K.; Arshad, A.; Riaz, N.; Waheed, U.; Rauf, A.; Aldakheel, F.; Alduraywish, S.; Rehman, M.U.; Abdullah, M.; et al. Microbiota in cancer development and treatment. J. Cancer Res. Clin. Oncol. 2019, 145, 49–63. [Google Scholar] [CrossRef] [PubMed]
- Hanus, M.; Parada-Venegas, D.; Landskron, G.; Wielandt, A.M.; Hurtado, C.; Alvarez, K.; Hermoso, M.A.; López-Köstner, F.; De la Fuente, M. Immune System, Microbiota, and Microbial Metabolites: The Unresolved Triad in Colorectal Cancer Microenvironment. Front. Immunol. 2021, 12, 612826. [Google Scholar] [CrossRef] [PubMed]
- Alhinai, E.A.; Walton, G.E.; Commane, D.M. The Role of the Gut Microbiota in Colorectal Cancer Causation. Int. J. Mol. Sci. 2019, 20, 5295. [Google Scholar] [CrossRef]
- Liu, Y.; Baba, Y.; Ishimoto, T.; Gu, X.; Zhang, J.; Nomoto, D.; Okadome, K.; Baba, H.; Qiu, P. Gut microbiome in gastrointestinal cancer: A friend or foe? Int. J. Biol. Sci. 2022, 18, 4101–4117. [Google Scholar] [CrossRef]
- Elkrief, A.; Derosa, L.; Kroemer, G.; Zitvogel, L.; Routy, B. The negative impact of antibiotics on outcomes in cancer patients treated with immunotherapy: A new independent prognostic factor? Ann. Oncol. 2019, 30, 1572–1579. [Google Scholar] [CrossRef]
- Yi, M.; Jiao, D.; Qin, S.; Chu, Q.; Li, A.; Wu, K. Manipulating Gut Microbiota Composition to Enhance the Therapeutic Effect of Cancer Immunotherapy. Integr. Cancer Ther. 2019, 18, 1534735419876351. [Google Scholar] [CrossRef]
- Perillo, F.; Amoroso, C.; Strati, F.; Giuffrè, M.R.; Díaz-Basabe, A.; Lattanzi, G.; Facciotti, F. Gut Microbiota Manipulation as a Tool for Colorectal Cancer Management: Recent Advances in Its Use for Therapeutic Purposes. Int. J. Mol. Sci. 2020, 21, 5389. [Google Scholar] [CrossRef]
- Fong, W.; Li, Q.; Yu, J. Gut microbiota modulation: A novel strategy for prevention and treatment of colorectal cancer. Oncogene 2020, 39, 4925–4943. [Google Scholar] [CrossRef]
- Kim, J.; Lee, H.K. Potential Role of the Gut Microbiome in Colorectal Cancer Progression. Front. Immunol. 2022, 12, 807648. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.; Ye, X.; Yang, X.; Jiang, R.; Qian, C.; Wang, X. The cross-talk between intestinal bacterial microbiota and immune cells in colorectal cancer progression. Clin. Transl. Oncol. 2022, 25, 620–632. [Google Scholar] [CrossRef] [PubMed]
| Microbiota-Derived Metabolite | Source | Effects | References |
|---|---|---|---|
| SCFA | Anaerobes | Antitumor effect, reducing inflammation | [58,59,60,61,62,63,64] |
| Secondary bile acids | B. fragilis, Bacteroides vulgatus, Clostridium perfringens, Eubacterium, Lactobacillus and Bifidobacterium | Contribute to CRC progression | [65,66,68] |
| Indoles | Gram-positive anaerobe (i.e., P. anaerobius) | Tumor prevention | [69,70] |
| TMAO | Gram-negative, Gram-positive, anaerobe (i.e., E. coli, Clostridium and Desulfovibrios) | Positive association with CRC risk, new prognostic marker | [71,72] |
| H2S | Gram-negative anaerobes (i.e., F. nucleatum and Desulfovibrios) | Potential environmental risk factors for CRC | [73,74,75] |
| DCA | Gram-positive, Gram-negative anaerobes (i.e., Desulfovibrios and Clostridium) | Positive associations with colorectal adenomas and CRC; contributes to CRC development and carcinogenesis promotion | [76,77,78,79,80,81,82,83,84] |
| NOCs | Facultative and anaerobes | Positive association with CRC risk | [85,86,87,88,89] |
| HCAs | Bacteroides, Lactobacilli | Positive association with CRC risk; Bacteroides convert HCA to carcinogens and Lactobacilli reduce their mutagenic effect | [90,91,92] |
| Polyamines | Gram-negative anaerobes (i.e., B. fragilis and F. nucleatum) | Positive association with CRC | [93,94,95] |
| Ammonia | Gram-negative anaerobes, clostridia, enterobacteria and Bacillus spp. Gram-positive non-sporing anaerobes, streptococci | Contribute to CRC development and promote neoplastic transformation | [96,97] |
| Lactate | Lactobacillus, Leuconostoc, Pediococcus, Lactococcus and Streptococcus | Promote CRC | [98,99] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lozenov, S.; Krastev, B.; Nikolaev, G.; Peshevska-Sekulovska, M.; Peruhova, M.; Velikova, T. Gut Microbiome Composition and Its Metabolites Are a Key Regulating Factor for Malignant Transformation, Metastasis and Antitumor Immunity. Int. J. Mol. Sci. 2023, 24, 5978. https://doi.org/10.3390/ijms24065978
Lozenov S, Krastev B, Nikolaev G, Peshevska-Sekulovska M, Peruhova M, Velikova T. Gut Microbiome Composition and Its Metabolites Are a Key Regulating Factor for Malignant Transformation, Metastasis and Antitumor Immunity. International Journal of Molecular Sciences. 2023; 24(6):5978. https://doi.org/10.3390/ijms24065978
Chicago/Turabian StyleLozenov, Stefan, Boris Krastev, Georgi Nikolaev, Monika Peshevska-Sekulovska, Milena Peruhova, and Tsvetelina Velikova. 2023. "Gut Microbiome Composition and Its Metabolites Are a Key Regulating Factor for Malignant Transformation, Metastasis and Antitumor Immunity" International Journal of Molecular Sciences 24, no. 6: 5978. https://doi.org/10.3390/ijms24065978
APA StyleLozenov, S., Krastev, B., Nikolaev, G., Peshevska-Sekulovska, M., Peruhova, M., & Velikova, T. (2023). Gut Microbiome Composition and Its Metabolites Are a Key Regulating Factor for Malignant Transformation, Metastasis and Antitumor Immunity. International Journal of Molecular Sciences, 24(6), 5978. https://doi.org/10.3390/ijms24065978









