Co-Expression Network Analysis Identifies Molecular Determinants of Loneliness Associated with Neuropsychiatric and Neurodegenerative Diseases
Abstract
1. Introduction
2. Results
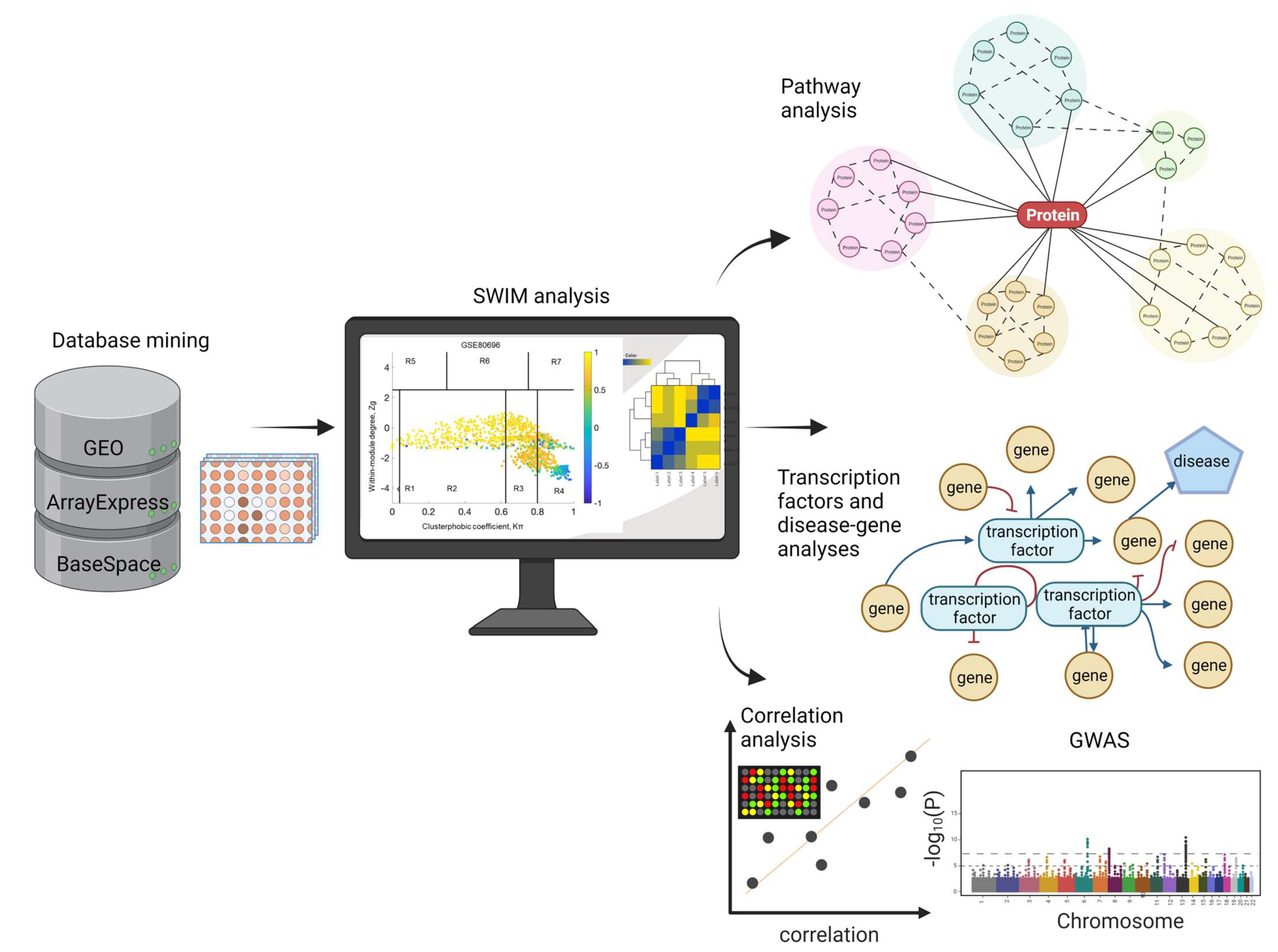
2.1. Database Mining and Study Selection
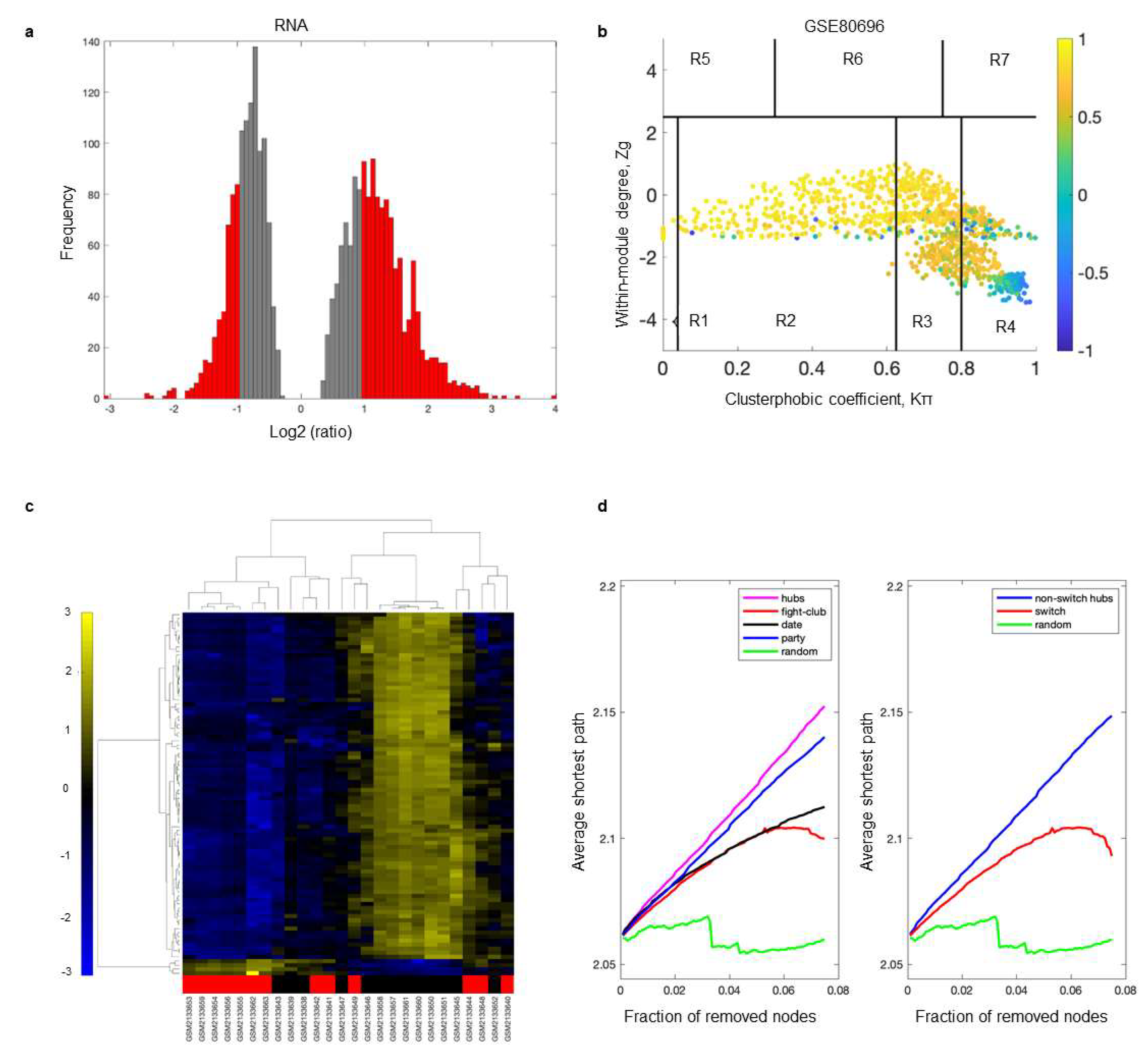
2.2. Identification of Switch Genes Associated with Loneliness
2.3. Biological and Functional Analysis of Switch Genes
2.4. Gene–Disease Association Analysis
2.5. Gene–Transcription Factor Network Analysis
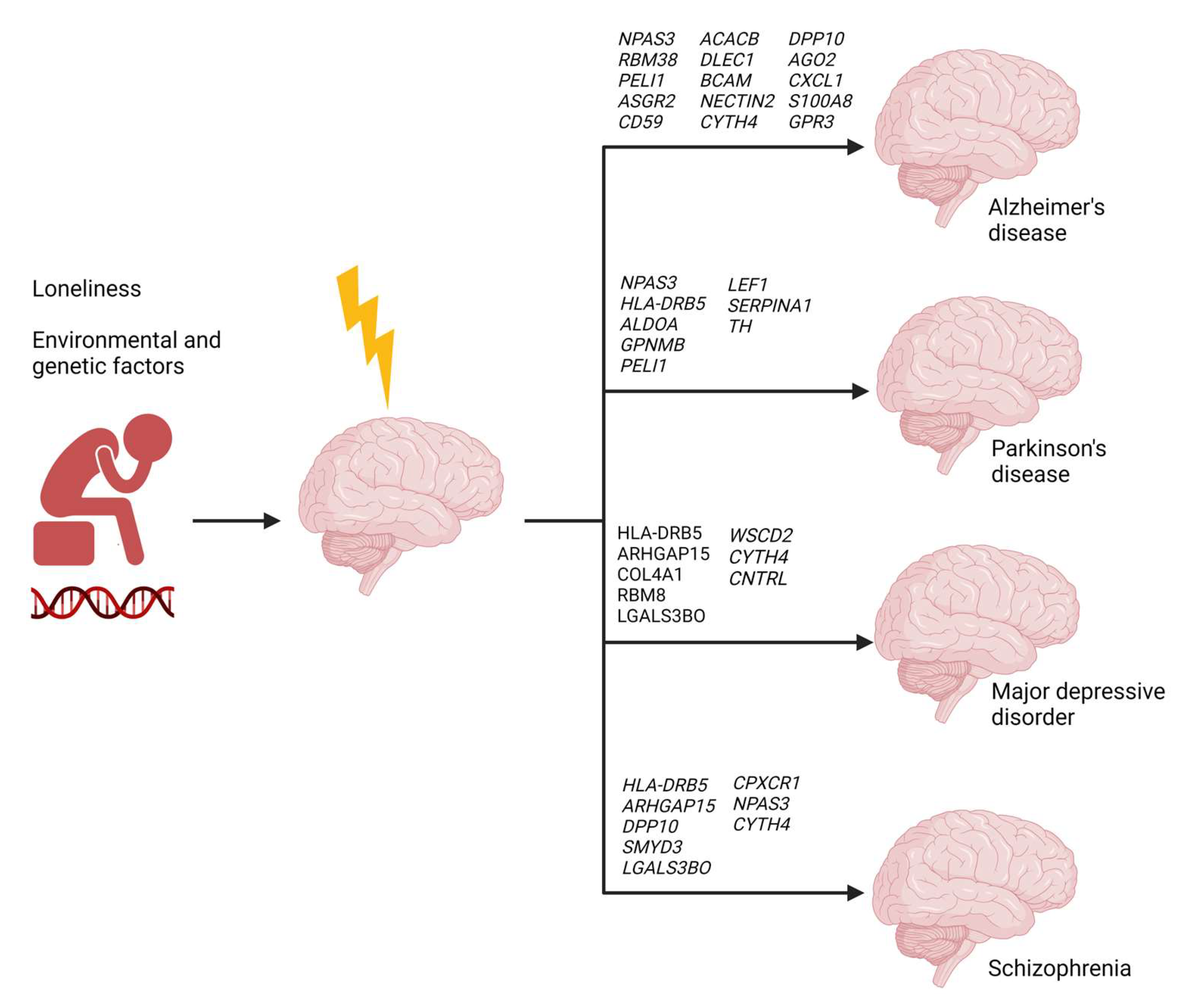
2.6. Loneliness-Related Switch Genes Associated with Neuropsychiatric and Neurodegenerative Diseases
2.7. Comparative Gene Correlation Analysis between Loneliness and Neuropsychiatric and Neurodegenerative Diseases
3. Discussion
4. Materials and Methods
4.1. Microarray Dataset Selection
4.2. Identification of Switch Genes
4.3. Functional Analysis of Switch Genes
4.4. Transcription Factor Analysis
4.5. Gene Expression and Correlation Analyses
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Canli, T.; Yu, L.; Yu, X.; Zhao, H.; Fleischman, D.; Wilson, R.S.; De Jager, P.L.; Bennett, D.A. Loneliness 5 years ante-mortem is associated with disease-related differential gene expression in postmortem dorsolateral prefrontal cortex. Transl. Psychiatry 2018, 8, 2. [Google Scholar] [CrossRef]
- Beutel, M.E.; Klein, E.M.; Brähler, E.; Reiner, I.; Jünger, C.; Michal, M.; Wiltink, J.; Wild, P.S.; Münzel, T.; Lackner, K.J.; et al. Loneliness in the general population: Prevalence, determinants and relations to mental health. BMC Psychiatry 2017, 17, 97. [Google Scholar] [CrossRef]
- Kuiper, J.S.; Zuidersma, M.; Oude Voshaar, R.C.; Zuidema, S.U.; van den Heuvel, E.R.; Stolk, R.P.; Smidt, N. Social relationships and risk of dementia: A systematic review and meta-analysis of longitudinal cohort studies. Ageing Res. Rev. 2015, 22, 39–57. [Google Scholar] [CrossRef] [PubMed]
- Lam, J.A.; Murray, E.R.; Yu, K.E.; Ramsey, M.; Nguyen, T.T.; Mishra, J.; Martis, B.; Thomas, M.L.; Lee, E.E. Neurobiology of loneliness: A systematic review. Neuropsychopharmacology 2021, 46, 1873–1887. [Google Scholar] [CrossRef] [PubMed]
- Cole, S.W.; Hawkley, L.C.; Arevalo, J.M.; Sung, C.Y.; Rose, R.M.; Cacioppo, J.T. Social regulation of gene expression in human leukocytes. Genome Biol. 2007, 8, R189. [Google Scholar] [CrossRef] [PubMed]
- Cole, S.W.; Hawkley, L.C.; Arevalo, J.M.G.; Cacioppo, J.T. Transcript origin analysis identifies antigen-presenting cells as primary targets of socially regulated gene expression in leukocytes. Proc. Natl. Acad. Sci. USA 2011, 108, 3080–3085. [Google Scholar] [CrossRef] [PubMed]
- Canli, T.; Wen, R.; Wang, X.; Mikhailik, A.; Yu, L.; Fleischman, D.; Wilson, R.S.; Bennett, D.A. Differential transcriptome expression in human nucleus accumbens as a function of loneliness. Mol. Psychiatry 2016, 22, 1069–1078. [Google Scholar] [CrossRef]
- Rilling, J.K.; Gutman, D.A.; Zeh, T.R.; Pagnoni, G.; Berns, G.S.; Kilts, C.D. A Neural Basis for Social Cooperation. Neuron 2002, 35, 395–405. [Google Scholar] [CrossRef]
- Davey, C.G.; Allen, N.B.; Harrison, B.J.; Dwyer, D.B.; Yücel, M. Being liked activates primary reward and midline self-related brain regions. Hum. Brain Mapp. 2009, 31, 660–668. [Google Scholar] [CrossRef]
- Paci, P.; Colombo, T.; Fiscon, G.; Gurtner, A.; Pavesi, G.; Farina, L. SWIM: A computational tool to unveiling crucial nodes in complex biological networks. Sci. Rep. 2017, 7, 44797. [Google Scholar] [CrossRef]
- Fiscon, G.; Conte, F.; Farina, L.; Paci, P. Network-Based Approaches to Explore Complex Biological Systems towards Network Medicine. Genes 2018, 9, 437. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Horvath, S. A General Framework for Weighted Gene Co-Expression Network Analysis. Stat. Appl. Genet. Mol. Biol. 2005, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Paci, P.; Fiscon, G.; Conte, F.; Licursi, V.; Morrow, J.; Hersh, C.; Cho, M.; Castaldi, P.; Glass, K.; Silverman, E.K.; et al. Integrated transcriptomic correlation network analysis identifies COPD molecular determinants. Sci. Rep. 2020, 10, 3361. [Google Scholar] [CrossRef] [PubMed]
- Potashkin, J.A.; Bottero, V.; Santiago, J.A.; Quinn, J.P. Computational identification of key genes that may regulate gene expression reprogramming in Alzheimer’s patients. PLoS ONE 2019, 14, e0222921. [Google Scholar] [CrossRef] [PubMed]
- Potashkin, J.A.; Bottero, V.; Santiago, J.A.; Quinn, J.P. Bioinformatic Analysis Reveals Phosphodiesterase 4D-Interacting Protein as a Key Frontal Cortex Dementia Switch Gene. Int. J. Mol. Sci. 2020, 21, 3787. [Google Scholar] [CrossRef]
- Bottero, V.; Powers, D.; Yalamanchi, A.; Quinn, J.; Potashkin, J. Key Disease Mechanisms Linked to Alzheimer’s Disease in the Entorhinal Cortex. Int. J. Mol. Sci. 2021, 22, 3915. [Google Scholar] [CrossRef]
- Santiago, J.; Quinn, J.; Potashkin, J. Network Analysis Identifies Sex-Specific Gene Expression Changes in Blood of Amyotrophic Lateral Sclerosis Patients. Int. J. Mol. Sci. 2021, 22, 7150. [Google Scholar] [CrossRef]
- Bottero, V.; Santiago, J.A.; Quinn, J.P.; Potashkin, J.A. Key Disease Mechanisms Linked to Amyotrophic Lateral Sclerosis in Spinal Cord Motor Neurons. Front. Mol. Neurosci. 2022, 15, 825031. [Google Scholar] [CrossRef]
- Santiago, J.A.; Quinn, J.P.; Potashkin, J.A. Physical Activity Rewires the Human Brain against Neurodegeneration. Int. J. Mol. Sci. 2022, 23, 6223. [Google Scholar] [CrossRef]
- Fiscon, G.; Conte, F.; Licursi, V.; Nasi, S.; Paci, P. Computational identification of specific genes for glioblastoma stem-like cells identity. Sci. Rep. 2018, 8, 7769. [Google Scholar] [CrossRef]
- Fiscon, G.; Conte, F.; Paci, P. SWIM tool application to expression data of glioblastoma stem-like cell lines, corresponding primary tumors and conventional glioma cell lines. BMC Bioinform. 2018, 19, 436. [Google Scholar] [CrossRef] [PubMed]
- Long, J.; Ray, B.; Lahiri, D.K. MicroRNA-339-5p Down-regulates Protein Expression of β-Site Amyloid Precursor Protein-Cleaving Enzyme 1 (BACE1) in Human Primary Brain Cultures and Is Reduced in Brain Tissue Specimens of Alzheimer Disease Subjects. J. Biol. Chem. 2014, 289, 5184–5198. [Google Scholar] [CrossRef]
- Pircs, K.; Petri, R.; Madsen, S.; Brattås, P.L.; Vuono, R.; Ottosson, D.R.; St-Amour, I.; Hersbach, B.; Matusiak-Brückner, M.; Lundh, S.H.; et al. Huntingtin Aggregation Impairs Autophagy, Leading to Argonaute-2 Accumulation and Global MicroRNA Dysregulation. Cell Rep. 2018, 24, 1397–1406. [Google Scholar] [CrossRef] [PubMed]
- Patel, D.; Zhang, X.; Farrell, J.; Lunetta, K.; Farrer, L. Set-Based Rare Variant Expression Quantitative Trait Loci in Blood and Brain from Alzheimer Disease Study Participants. Genes 2021, 12, 419. [Google Scholar] [CrossRef]
- Hampel, H.; Caraci, F.; Cuello, A.C.; Caruso, G.; Nisticò, R.; Corbo, M.; Baldacci, F.; Toschi, N.; Garaci, F.; Chiesa, P.A.; et al. A Path Toward Precision Medicine for Neuroinflammatory Mechanisms in Alzheimer’s Disease. Front. Immunol. 2020, 11, 456. [Google Scholar] [CrossRef]
- Eyre, H.A.; Eskin, A.; Nelson, S.F.; St Cyr, N.M.; Siddarth, P.; Baune, B.T.; Lavretsky, H. Genomic predictors of remission to antidepressant treatment in geriatric depression using genome-wide expression analyses: A pilot study. Int. J. Geriatr. Psychiatry 2015, 31, 510–517. [Google Scholar] [CrossRef] [PubMed]
- Gusev, F.E.; Reshetov, D.A.; Mitchell, A.C.; Andreeva, T.V.; Dincer, A.; Grigorenko, A.P.; Fedonin, G.; Halene, T.; Aliseychik, M.; Filippova, E.; et al. Chromatin profiling of cortical neurons identifies individual epigenetic signatures in schizophrenia. Transl. Psychiatry 2019, 9, 256. [Google Scholar] [CrossRef]
- Berger, M.; Cooter, M.; Roesler, A.S.; Chung, S.; Park, J.; Modliszewski, J.L.; VanDusen, K.W.; Thompson, J.W.; Moseley, A.; Devinney, M.J.; et al. APOE4 Copy Number-Dependent Proteomic Changes in the Cerebrospinal Fluid. J. Alzheimer’s Dis. 2021, 79, 511–530. [Google Scholar] [CrossRef] [PubMed]
- Smyth, L.C.D.; Murray, H.C.; Hill, M.; van Leeuwen, E.; Highet, B.; Magon, N.J.; Osanlouy, M.; Mathiesen, S.N.; Mockett, B.; Singh-Bains, M.K.; et al. Neutrophil-vascular interactions drive myeloperoxidase accumulation in the brain in Alzheimer’s disease. Acta Neuropathol. Commun. 2022, 10, 38. [Google Scholar] [CrossRef]
- Katzeff, J.S.; Bright, F.; Lo, K.; Kril, J.J.; Connolly, A.; Crossett, B.; Ittner, L.M.; Kassiou, M.; Loy, C.T.; Hodges, J.R.; et al. Altered serum protein levels in frontotemporal dementia and amyotrophic lateral sclerosis indicate calcium and immunity dysregulation. Sci. Rep. 2020, 10, 13741. [Google Scholar] [CrossRef]
- Lodeiro, M.; Puerta, E.; Ismail, M.-A.; Rodriguez-Rodriguez, P.; Rönnbäck, A.; Codita, A.; Parrado-Fernandez, C.; Maioli, S.; Gil-Bea, F.; Merino-Serrais, P.; et al. Aggregation of the Inflammatory S100A8 Precedes Aβ Plaque Formation in Transgenic APP Mice: Positive Feedback for S100A8 and Aβ Productions. J. Gerontol. Ser. A 2016, 72, 319–328. [Google Scholar] [CrossRef]
- Rajkumar, A.P.; Bidkhori, G.; Shoaie, S.; Clarke, E.; Morrin, H.; Hye, A.; Williams, G.; Ballard, C.; Francis, P.T.; Aarsland, D. Postmortem Cortical Transcriptomics of Lewy Body Dementia Reveal Mitochondrial Dysfunction and Lack of Neuroinflammation. Am. J. Geriatr. Psychiatry 2019, 28, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Perez-Nievas, B.G.; Johnson, L.; Beltran-Lobo, P.; Hughes, M.M.; Gammallieri, L.; Tarsitano, F.; Myszczynska, M.A.; Vazquez-Villasenor, I.; Jimenez-Sanchez, M.; Troakes, C.; et al. Astrocytic C–X–C motif chemokine ligand-1 mediates β-amyloid-induced synaptotoxicity. J. Neuroinflamm. 2021, 18, 306. [Google Scholar] [CrossRef]
- Rezazadeh, M.; Gharesouran, J.; Movafagh, A.; Taheri, M.; Darvish, H.; Emamalizadeh, B.; Shahmohammadibeni, N.; Khorshid, H.R.K.; Behmanesh, M.; Sahraian, M.A.; et al. Dominant and Protective Role of the CYTH4 Primate-Specific GTTT-Repeat Longer Alleles Against Neurodegeneration. J. Mol. Neurosci. 2015, 56, 593–596. [Google Scholar] [CrossRef]
- Khademi, E.; Alehabib, E.; Shandiz, E.E.; Ahmadifard, A.; Andarva, M.; Jamshidi, J.; Rahimi-Aliabadi, S.; Pouriran, R.; Nejad, F.R.; Mansoori, N.; et al. Support for “Disease-Only” Genotypes and Excess of Homozygosity at the CYTH4 Primate-Specific GTTT-Repeat in Schizophrenia. Genet. Test. Mol. Biomarkers 2017, 21, 485–490. [Google Scholar] [CrossRef] [PubMed]
- Dai, D.; Yuan, J.; Wang, Y.; Xu, J.; Mao, C.; Xiao, Y. Peli1 controls the survival of dopaminergic neurons through modulating microglia-mediated neuroinflammation. Sci. Rep. 2019, 9, 8034. [Google Scholar] [CrossRef]
- Young, J.; Gallagher, E.; Koska, K.; Guetta-Baranes, T.; Morgan, K.; Thomas, A.; Brookes, K.J. Genome-wide association findings from the brains for dementia research cohort. Neurobiol. Aging 2021, 107, 159–167. [Google Scholar] [CrossRef]
- Yin, P.; Xue, Y.; Wang, T.; Zhong, D.; Li, G. The Therapeutic Targets of Fingolimod (FTY720) Are Involved in Pathological Processes in the Frontal Cortex of Alzheimer’s Disease Patients: A Network Pharmacology Study. Front. Aging Neurosci. 2021, 13, 609679. [Google Scholar] [CrossRef]
- Rossi, J.J.; Rosenfeld, J.A.; Chan, K.M.; Streff, H.; Nankivell, V.; Peet, D.J.; Whitelaw, M.L.; Bersten, D.C. Molecular characterisation of rare loss-of-function NPAS3 and NPAS4 variants identified in individuals with neurodevelopmental disorders. Sci. Rep. 2021, 11, 6602. [Google Scholar] [CrossRef] [PubMed]
- Bradshaw, N.J.; Korth, C. Protein misassembly and aggregation as potential convergence points for non-genetic causes of chronic mental illness. Mol. Psychiatry 2018, 24, 936–951. [Google Scholar] [CrossRef]
- Michaelson, J.J.; Shin, M.-K.; Koh, J.-Y.; Brueggeman, L.; Zhang, A.; Katzman, A.; McDaniel, L.; Fang, M.; Pufall, M.; Pieper, A. Neuronal PAS Domain Proteins 1 and 3 Are Master Regulators of Neuropsychiatric Risk Genes. Biol. Psychiatry 2017, 82, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Nucifora, L.G.; Wu, Y.C.; Lee, B.J.; Sha, L.; Margolis, R.L.; Ross, C.A.; Sawa, A.; Nucifora, F.C., Jr. A Mutation in NPAS3 That Segregates with Schizophrenia in a Small Family Leads to Protein Aggregation. Complex Psychiatry 2016, 2, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Arbez, N.; Nucifora, L.G.; Sell, G.L.; Delisi, L.E.; Ross, C.A.; Margolis, R.L.; Nucifora, F.C., Jr. A mutation in NPAS3 segregates with mental illness in a small family. Mol. Psychiatry 2013, 19, 7–8. [Google Scholar] [CrossRef]
- Sha, L.; MacIntyre, L.; Machell, J.A.; Kelly, M.P.; Porteous, D.J.; Brandon, N.J.; Muir, W.J.; Blackwood, D.H.; Watson, D.G.; Clapcote, S.J.; et al. Transcriptional regulation of neurodevelopmental and metabolic pathways by NPAS3. Mol. Psychiatry 2011, 17, 267–279. [Google Scholar] [CrossRef]
- Diaz-Ortiz, M.E.; Seo, Y.; Posavi, M.; Cordon, M.C.; Clark, E.; Jain, N.; Charan, R.; Gallagher, M.D.; Unger, T.L.; Amari, N.; et al. GPNMB confers risk for Parkinson’s disease through interaction with α-synuclein. Science 2022, 377, eabk0637. [Google Scholar] [CrossRef]
- Haavik, J.; Toska, K. Tyrosine hydroxylase and Parkinson’s disease. Mol. Neurobiol. 1998, 16, 285–309. [Google Scholar] [CrossRef] [PubMed]
- Tabrez, S.; Jabir, N.R.; Shakil, S.; Greig, N.H.; Alam, Q.; Abuzenadah, A.M.; Damanhouri, G.A.; Kamal, M.A. A Synopsis on the Role of Tyrosine Hydroxylase in Parkinson’s Disease. CNS Neurol. Disord. Drug Targets 2012, 11, 395–409. [Google Scholar] [CrossRef]
- Yang, L.-B.; Li, R.; Meri, S.; Rogers, J.; Shen, Y. Deficiency of Complement Defense Protein CD59 May Contribute to Neurodegeneration in Alzheimer’s Disease. J. Neurosci. 2000, 20, 7505–7509. [Google Scholar] [CrossRef]
- Goetzl, E.J.; Schwartz, J.B.; Abner, E.L.; Jicha, G.A.; Kapogiannis, D. High complement levels in astrocyte-derived exosomes of Alzheimer disease. Ann. Neurol. 2018, 83, 544–552. [Google Scholar] [CrossRef]
- Siitonen, M.; Börjesson-Hanson, A.; Pöyhönen, M.; Ora, A.; Pasanen, P.; Bras, J.; Kern, S.; Kern, J.; Andersen, O.; Stanescu, H.; et al. Multi-infarct dementia of Swedish type is caused by a 3’UTR mutation of COL4A1. Brain 2017, 140, e29. [Google Scholar] [CrossRef]
- Hamilton, A.; Vasefi, M.; Vander Tuin, C.; McQuaid, R.J.; Anisman, H.; Ferguson, S.S. Chronic Pharmacological mGluR5 Inhibition Prevents Cognitive Impairment and Reduces Pathogenesis in an Alzheimer Disease Mouse Model. Cell Rep. 2016, 15, 1859–1865. [Google Scholar] [CrossRef] [PubMed]
- Abd-Elrahman, K.S.; Hamilton, A.; Hutchinson, S.R.; Liu, F.; Russell, R.C.; Ferguson, S.S.G. mGluR5 antagonism increases autophagy and prevents disease progression in the zQ175 mouse model of Huntington’s disease. Sci. Signal. 2017, 10, 1–11. [Google Scholar] [CrossRef]
- Abd-Elrahman, K.S.; Hamilton, A.; Vasefi, M.; Ferguson, S.S.G. Autophagy is increased following either pharmacological or genetic silencing of mGluR5 signaling in Alzheimer’s disease mouse models. Mol. Brain 2018, 11, 19. [Google Scholar] [CrossRef] [PubMed]
- Seipold, L.; Altmeppen, H.; Koudelka, T.; Tholey, A.; Kasparek, P.; Sedlacek, R.; Schweizer, M.; Bär, J.; Mikhaylova, M.; Glatzel, M.; et al. In vivo regulation of the A disintegrin and metalloproteinase 10 (ADAM10) by the tetraspanin 15. Cell. Mol. Life Sci. 2018, 75, 3251–3267. [Google Scholar] [CrossRef]
- Shigemizu, D.; Asanomi, Y.; Akiyama, S.; Mitsumori, R.; Niida, S.; Ozaki, K. Whole-genome sequencing reveals novel ethnicity-specific rare variants associated with Alzheimer’s disease. Mol. Psychiatry 2022, 27, 2554–2562. [Google Scholar] [CrossRef]
- Pascual-Morena, C.; Cavero-Redondo, I.; Reina-Gutiérrez, S.; Saz-Lara, A.; López-Gil, J.F.; Martínez-Vizcaíno, V. Prevalence of Neuropsychiatric Disorders in Duchenne and Becker Muscular Dystrophies: A Systematic Review and Meta-analysis. Arch. Phys. Med. Rehabil. 2022, 103, 2444–2453. [Google Scholar] [CrossRef]
- Nouri, P.; Götz, S.; Rauser, B.; Irmler, M.; Peng, C.; Trümbach, D.; Kempny, C.; Lechermeier, C.G.; Bryniok, A.; Dlugos, A.; et al. Dose-Dependent and Subset-Specific Regulation of Midbrain Dopaminergic Neuron Differentiation by LEF1-Mediated WNT1/b-Catenin Signaling. Front. Cell Dev. Biol. 2020, 8, 587778. [Google Scholar] [CrossRef]
- Huang, Y.; Skwarek-Maruszewska, A.; Horré, K.; Vandewyer, E.; Wolfs, L.; Snellinx, A.; Saito, T.; Radaelli, E.; Corthout, N.; Colombelli, J.; et al. Loss of GPR3 reduces the amyloid plaque burden and improves memory in Alzheimer’s disease mouse models. Sci. Transl. Med. 2015, 7, 309ra164. [Google Scholar] [CrossRef] [PubMed]
- Barbagallo, C.; Di Martino, M.; Grasso, M.; Salluzzo, M.; Scionti, F.; Cosentino, F.; Caruso, G.; Barbagallo, D.; Di Pietro, C.; Ferri, R.; et al. Uncharacterized RNAs in Plasma of Alzheimer’s Patients Are Associated with Cognitive Impairment and Show a Potential Diagnostic Power. Int. J. Mol. Sci. 2020, 21, 7644. [Google Scholar] [CrossRef] [PubMed]
- Bezerra, G.A.; Dobrovetsky, E.; Seitova, A.; Fedosyuk, S.; Dhe-Paganon, S.; Gruber, K. Structure of human dipeptidyl peptidase 10 (DPPY): A modulator of neuronal Kv4 channels. Sci. Rep. 2015, 5, 8769. [Google Scholar] [CrossRef]
- Djurovic, S.; Gustafsson, O.; Mattingsdal, M.; Athanasiu, L.; Bjella, T.; Tesli, M.; Agartz, I.; Lorentzen, S.; Melle, I.; Morken, G. A genome-wide association study of bipolar disorder in Norwegian individuals, followed by replication in Icelandic sample. J. Affect. Disord. 2010, 126, 312–316. [Google Scholar] [CrossRef]
- Mitchell, A.C.; Bharadwaj, R.; Whittle, C.; Krueger, W.; Mirnics, K.; Hurd, Y.; Rasmussen, T.; Akbarian, S. The Genome in Three Dimensions: A New Frontier in Human Brain Research. Biol. Psychiatry 2013, 75, 961–969. [Google Scholar] [CrossRef]
- Strickland, S.L.; Reddy, J.S.; Allen, M.; N’Songo, A.; Burgess, J.D.; Corda, M.M.; Ballard, T.; Wang, X.; Carrasquillo, M.M.; Biernacka, J.M.; et al. MAPT haplotype–stratified GWAS reveals differential association for AD risk variants. Alzheimer’s Dement. 2020, 16, 983–1002. [Google Scholar] [CrossRef]
- Seki, T.; Kanagawa, M.; Kobayashi, K.; Kowa, H.; Yahata, N.; Maruyama, K.; Iwata, N.; Inoue, H.; Toda, T. Galectin 3–binding protein suppresses amyloid-β production by modulating β-cleavage of amyloid precursor protein. J. Biol. Chem. 2020, 295, 3678–3691. [Google Scholar] [CrossRef] [PubMed]
- Halbgebauer, S.; Nagl, M.; Klafki, H.; Haußmann, U.; Steinacker, P.; Oeckl, P.; Kassubek, J.; Pinkhardt, E.; Ludolph, A.C.; Soininen, H.; et al. Modified serpinA1 as risk marker for Parkinson’s disease dementia: Analysis of baseline data. Sci. Rep. 2016, 6, 26145. [Google Scholar] [CrossRef] [PubMed]
- Abu-Rumeileh, S.; Halbgebauer, S.; Steinacker, P.; Anderl-Straub, S.; Polischi, B.; Ludolph, A.C.; Capellari, S.; Parchi, P.; Otto, M. CSF SerpinA1 in Creutzfeldt–Jakob disease and frontotemporal lobar degeneration. Ann. Clin. Transl. Neurol. 2020, 7, 191–199. [Google Scholar] [CrossRef]
- Barba, L.; Halbgebauer, S.; Paoletti, F.P.; Bellomo, G.; Abu-Rumeileh, S.; Steinacker, P.; Massa, F.; Parnetti, L.; Otto, M. Specific Cerebrospinal Fluid SerpinA1 Isoform Pattern in Alzheimer’s Disease. Int. J. Mol. Sci. 2022, 23, 6922. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Wei, J.; Du, Y.; Chen, P.; Liu, X.; Liu, H.; Alzheimer’s Disease Neuroimaging Initiative. Improved cognitive impairments by silencing DMP1 via enhancing the proliferation of neural progenitor cell in Alzheimer-like mice. Aging Cell 2022, 21, e13601. [Google Scholar] [CrossRef]
- Hiew, L.-F.; Poon, C.-H.; You, H.-Z.; Lim, L.-W. TGF-β/Smad Signalling in Neurogenesis: Implications for Neuropsychiatric Diseases. Cells 2021, 10, 1382. [Google Scholar] [CrossRef] [PubMed]
- Tesseur, I.; Zou, K.; Esposito, L.; Bard, F.; Berber, E.; Van Can, J.; Lin, A.H.; Crews, L.; Tremblay, P.; Mathews, P.; et al. Deficiency in neuronal TGF-β signaling promotes neurodegeneration and Alzheimer’s pathology. J. Clin. Investig. 2006, 116, 3060–3069. [Google Scholar] [CrossRef]
- Dumitriu, A.; Latourelle, J.; Hadzi, T.C.; Pankratz, N.; Garza, D.; Miller, J.P.; Vance, J.; Foroud, T.; Beach, T.G.; Myers, R. Gene Expression Profiles in Parkinson Disease Prefrontal Cortex Implicate FOXO1 and Genes under Its Transcriptional Regulation. PLoS Genet. 2012, 8, e1002794. [Google Scholar] [CrossRef]
- Pardeshi, R.; Bolshette, N.; Gadhave, K.; Ahire, A.; Ahmed, S.; Cassano, T.; Gupta, V.B.; Lahkar, M. Insulin signaling: An opportunistic target to minify the risk of Alzheimer’s disease. Psychoneuroendocrinology 2017, 83, 159–171. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Dong, H.H. FoxO integration of insulin signaling with glucose and lipid metabolism. J. Endocrinol. 2017, 233, R67–R79. [Google Scholar] [CrossRef] [PubMed]
- Santiago, J.A.; Bottero, V.; Potashkin, J.A. Transcriptomic and Network Analysis Highlight the Association of Diabetes at Different Stages of Alzheimer’s Disease. Front. Neurosci. 2019, 13, 1273. [Google Scholar] [CrossRef] [PubMed]
- Srikanth, V.; Maczurek, A.; Phan, T.; Steele, M.; Westcott, B.; Juskiw, D.; Münch, G. Advanced glycation endproducts and their receptor RAGE in Alzheimer’s disease. Neurobiol. Aging 2011, 32, 763–777. [Google Scholar] [CrossRef]
- Guo, Q.; Zhu, X.; Wei, R.; Zhao, L.; Zhang, Z.; Yin, X.; Zhang, Y.; Chu, C.; Wang, B.; Li, X. miR-130b-3p regulates M1 macrophage polarization via targeting IRF1. J. Cell. Physiol. 2020, 236, 2008–2022. [Google Scholar] [CrossRef]
- Chu, Y.-B.; Li, J.; Jia, P.; Cui, J.; Zhang, R.; Kang, X.; Lv, M.; Zhang, S. Irf1- and Egr1-activated transcription plays a key role in macrophage polarization: A multiomics sequencing study with partial validation. Int. Immunopharmacol. 2021, 99, 108072. [Google Scholar] [CrossRef]
- Gao, T.; Jernigan, J.; Raza, S.A.; Dammer, E.B.; Xiao, H.; Seyfried, N.T.; Levey, A.I.; Rangaraju, S. Transcriptional regulation of homeostatic and disease-associated-microglial genes by IRF1, LXRβ, and CEBPα. Glia 2019, 67, 1958–1975. [Google Scholar] [CrossRef]
- Ponnusamy, M.; Wang, S.; Yuksel, M.; Hansen, M.T.; Blazier, D.M.; McMillan, J.D.; Zhang, X.; Dammer, E.B.; Collier, L.; Thinakaran, G. Loss of forebrain BIN1 attenuates hippocampal pathology and neuroinflammation in a tauopathy model. Brain 2022. [Google Scholar] [CrossRef]
- Sudwarts, A.; Ramesha, S.; Gao, T.; Ponnusamy, M.; Wang, S.; Hansen, M.; Kozlova, A.; Bitarafan, S.; Kumar, P.; Beaulieu-Abdelahad, D.; et al. BIN1 is a key regulator of proinflammatory and neurodegeneration-related activation in microglia. Mol. Neurodegener. 2022, 17, 33. [Google Scholar] [CrossRef]
- Vied, C.M.; Freudenberg, F.; Wang, Y.; Raposo, A.A.S.F.; Feng, D.; Nowakowski, R.S. A multi-resource data integration approach: Identification of candidate genes regulating cell proliferation during neocortical development. Front. Neurosci. 2014, 8, 257. [Google Scholar] [CrossRef]
- Knepper, J.L.; James, A.C.; Ming, J.E. TGIF, a gene associated with human brain defects, regulates neuronal development. Dev. Dyn. 2006, 235, 1482–1490. [Google Scholar] [CrossRef]
- Cui, A.; Fan, H.; Zhang, Y.; Zhang, Y.; Niu, D.; Liu, S.; Liu, Q.; Ma, W.; Shen, Z.; Shen, L.; et al. Dexamethasone-induced Krüppel-like factor 9 expression promotes hepatic gluconeogenesis and hyperglycemia. J. Clin. Investig. 2019, 129, 2266–2278. [Google Scholar] [CrossRef]
- Piccinin, E.; Sardanelli, A.; Seibel, P.; Moschetta, A.; Cocco, T.; Villani, G. PGC-1s in the Spotlight with Parkinson’s Disease. Int. J. Mol. Sci. 2021, 22, 3487. [Google Scholar] [CrossRef] [PubMed]
- Zheng, B.; Liao, Z.; Locascio, J.J.; Lesniak, K.A.; Roderick, S.S.; Watt, M.L.; Eklund, A.C.; Zhang-James, Y.; Kim, P.D.; Hauser, M.A.; et al. PGC-1α, A Potential Therapeutic Target for Early Intervention in Parkinson’s Disease. Sci. Transl. Med. 2010, 2, 52ra73. [Google Scholar] [CrossRef]
- Liu, E.Y.; Cali, C.P.; Lee, E.B. RNA metabolism in neurodegenerative disease. Dis. Model. Mech. 2017, 10, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Nussbacher, J.K.; Tabet, R.; Yeo, G.W.; Lagier-Tourenne, C. Disruption of RNA Metabolism in Neurological Diseases and Emerging Therapeutic Interventions. Neuron 2019, 102, 294–320. [Google Scholar] [CrossRef]
- Santiago, J.A.; Potashkin, J.A. Network-based metaanalysis identifies HNF4A and PTBP1 as longitudinally dynamic biomarkers for Parkinson’s disease. Proc. Natl. Acad. Sci. USA 2015, 112, 2257–2262. [Google Scholar] [CrossRef]
- Vinogradov, A.E.; Anatskaya, O.V. Growth of Biological Complexity from Prokaryotes to Hominids Reflected in the Human Genome. Int. J. Mol. Sci. 2021, 22, 11640. [Google Scholar] [CrossRef] [PubMed]
- Martin, L.A.; Neighbors, H.W.; Griffith, D.M. The experience of symptoms of depression in men vs women: Analysis of the National Comorbidity Survey Replication. JAMA Psychiatry 2013, 70, 1100–1106. [Google Scholar] [CrossRef]
- Santiago, J.A.; Quinn, J.P.; Potashkin, J.A. Sex-specific transcriptional rewiring in the brain of Alzheimer’s disease patients. Front Aging Neurosci. 2022, 14, 1009368. [Google Scholar] [CrossRef] [PubMed]
- Naj, A.C.; Beecham, G.W.; Martin, E.R.; Gallins, P.J.; Powell, E.H.; Konidari, I.; Whitehead, P.L.; Cai, G.; Haroutunian, V.; Scott, W.K.; et al. Dementia Revealed: Novel Chromosome 6 Locus for Late-Onset Alzheimer Disease Provides Genetic Evidence for Folate-Pathway Abnormalities. PLoS Genet. 2010, 6, e1001130. [Google Scholar] [CrossRef]
- Logue, M.; Schu, M.; Vardarajan, B.N.; Buros, J.; Green, R.C.; Go, R.C.; Griffith, P.; Obisesan, T.O.; Shatz, R.; Borenstein, A.; et al. A Comprehensive Genetic Association Study of Alzheimer Disease in African Americans. Arch. Neurol. 2011, 68, 1569–1579. [Google Scholar] [CrossRef]
- Hu, X.; Pickering, E.; Liu, Y.C.; Hall, S.; Fournier, H.; Katz, E.; Dechairo, B.; John, S.; Van Eerdewegh, P.; Soares, H.; et al. Meta-Analysis for Genome-Wide Association Study Identifies Multiple Variants at the BIN1 Locus Associated with Late-Onset Alzheimer’s Disease. PLoS ONE 2011, 6, e16616. [Google Scholar] [CrossRef] [PubMed]
- Wijsman, E.M.; Pankratz, N.D.; Choi, Y.; Rothstein, J.H.; Faber, K.M.; Cheng, R.; Lee, J.H.; Bird, T.D.; Bennett, D.A.; Diaz-Arrastia, R.; et al. Genome-Wide Association of Familial Late-Onset Alzheimer’s Disease Replicates BIN1 and CLU and Nominates CUGBP2 in Interaction with APOE. PLoS Genet. 2011, 7, e1001308. [Google Scholar] [CrossRef] [PubMed]
- Hollingworth, P.; Harold, D.; Sims, R.; Gerrish, A.; Lambert, J.-C.; Carrasquillo, M.M.; Abraham, R.; Hamshere, M.L.; Pahwa, J.S.; Moskvina, V.; et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat. Genet. 2011, 43, 429–435. [Google Scholar] [CrossRef]
- Lambert, J.-C.; Heath, S.; Even, G.; Campion, D.; Sleegers, K.; Hiltunen, M.; Combarros, O.; Zelenika, D.; Bullido, M.J.; Tavernier, B.; et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat. Genet. 2009, 41, 1094–1099. [Google Scholar] [CrossRef]
- Antúnez, C.; Boada, M.; González-Pérez, A.; Gayán, J.; Ramírez-Lorca, R.; Marin, J.; Hernandez, I.; Moreno-Rey, C.; Morón, F.J.; López-Arrieta, J.; et al. The membrane-spanning 4-domains, subfamily A (MS4A) gene cluster contains a common variant associated with Alzheimer’s disease. Genome Med. 2011, 3, 33. [Google Scholar] [CrossRef] [PubMed]
- Harold, D.; Abraham, R.; Hollingworth, P.; Sims, R.; Gerrish, A.; Hamshere, M.L.; Pahwa, J.S.; Moskvina, V.; Dowzell, K.; Williams, A.; et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat. Genet. 2009, 41, 1088–1093. [Google Scholar] [CrossRef]
- Martinelli-Boneschi, F.; Giacalone, G.; Magnani, G.; Biella, G.; Coppi, E.; Santangelo, R.; Brambilla, P.; Esposito, F.; Lupoli, S.; Clerici, F.; et al. Pharmacogenomics in Alzheimer’s disease: A genome-wide association study of response to cholinesterase inhibitors. Neurobiol. Aging 2013, 34, 1711.e7–1711.e13. [Google Scholar] [CrossRef]
- Pankratz, N.; Beecham, G.W.; DeStefano, A.L.; Dawson, T.M.; Doheny, K.F.; Factor, S.A.; Hamza, T.H.; Hung, A.Y.; Hyman, B.T.; Ivinson, A.J.; et al. Meta-analysis of Parkinson’s Disease: Identification of a novel locus, RIT2. Ann. Neurol. 2012, 71, 370–384. [Google Scholar] [CrossRef] [PubMed]
- Nalls, M.A.; Pankratz, N.; Lill, C.M.; Do, C.B.; Hernandez, D.G.; Saad, M.; DeStefano, A.L.; Kara, E.; Bras, J.; Sharma, M.; et al. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat. Genet. 2014, 46, 989–993. [Google Scholar] [CrossRef] [PubMed]
- Chang, D.; Nalls, M.A.; Hallgrímsdóttir, I.B.; Hunkapiller, J.; Van Der Brug, M.; Cai, F.; International Parkinson’s Disease Genomics Consortium; 23andMe Research Team; Kerchner, G.A.; Ayalon, G.; et al. A meta-analysis of genome-wide association studies identifies 17 new Parkinson’s disease risk loci. Nat. Genet. 2017, 49, 1511–1516. [Google Scholar] [CrossRef] [PubMed]
- Simón-Sánchez, J.; Schulte, C.; Bras, J.M.; Sharma, M.; Gibbs, J.R.; Berg, D.; Paisan-Ruiz, C.; Lichtner, P.; Scholz, S.W.; Hernandez, D.G.; et al. Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat. Genet. 2009, 41, 1308–1312. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.L.; Pearce, E.; Ajnakina, O.; Johnson, S.; Lewis, G.; Mann, F.; Pitman, A.; Solmi, F.; Sommerlad, A.; Steptoe, A.; et al. The association between loneliness and depressive symptoms among adults aged 50 years and older: A 12-year population-based cohort study. Lancet Psychiatry 2020, 8, 48–57. [Google Scholar] [CrossRef]
- Shyn, S.I.; Shi, J.; Kraft, J.B.; Potash, J.B.; Knowles, J.A.; Weissman, M.M.; Garriock, H.A.; Yokoyama, J.S.; McGrath, P.J.; Peters, E.J.; et al. Novel loci for major depression identified by genome-wide association study of Sequenced Treatment Alternatives to Relieve Depression and meta-analysis of three studies. Mol. Psychiatry 2009, 16, 202–215. [Google Scholar] [CrossRef]
- Wray, N.R.; Pergadia, M.L.; Blackwood, D.H.R.; Penninx, B.W.J.H.; Gordon, S.D.; Nyholt, D.R.; Ripke, S.; MacIntyre, D.J.; A McGhee, K.; Maclean, A.W.; et al. Genome-wide association study of major depressive disorder: New results, meta-analysis, and lessons learned. Mol. Psychiatry 2010, 17, 36–48. [Google Scholar] [CrossRef]
- Garriock, H.A.; Kraft, J.B.; Shyn, S.I.; Peters, E.J.; Yokoyama, J.S.; Jenkins, G.D.; Reinalda, M.S.; Slager, S.L.; McGrath, P.J.; Hamilton, S.P. A Genomewide Association Study of Citalopram Response in Major Depressive Disorder. Biol. Psychiatry 2010, 67, 133–138. [Google Scholar] [CrossRef]
- Ikeda, M.; Aleksic, B.; Kinoshita, Y.; Okochi, T.; Kawashima, K.; Kushima, I.; Ito, Y.; Nakamura, Y.; Kishi, T.; Okumura, T.; et al. Genome-Wide Association Study of Schizophrenia in a Japanese Population. Biol. Psychiatry 2011, 69, 472–478. [Google Scholar] [CrossRef]
- O’Donovan, M.C.; Craddock, N.; Norton, N.; Williams, H.; Peirce, T.; Moskvina, V.; Nikolov, I.; Hamshere, M.; Carroll, L.; Georgieva, L.; et al. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat. Genet. 2008, 40, 1053–1055. [Google Scholar] [CrossRef]
- Shi, J.; Levinson, D.F.; Duan, J.; Sanders, A.R.; Zheng, Y.; Pe’Er, I.; Dudbridge, F.; Holmans, P.A.; Whittemore, A.S.; Mowry, B.J.; et al. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature 2009, 460, 753–757. [Google Scholar] [CrossRef] [PubMed]
- The Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium. Genome-wide association study identifies five new schizophrenia loci. Nat. Genet. 2011, 43, 969–976. [Google Scholar] [CrossRef] [PubMed]
- Contreras, A.; Valiente, C.; Vázquez, C.; Trucharte, A.; Peinado, V.; Varese, F.; Bentall, R.P. The network structure of paranoia dimensions and its mental health correlates in the general population: The core role of loneliness. Schizophr. Res. 2022, 246, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Deng, W.; Liu, X.; Li, M.; Chen, Z.; He, Z.; Wang, Y.; Wang, Q.; Hu, X.; Collier, D.A.; et al. A genome-wide association study for quantitative traits in schizophrenia in China. Genes Brain Behav. 2011, 10, 734–739. [Google Scholar] [CrossRef]
- Steen, O.D.; Ori, A.P.S.; Wardenaar, K.J.; van Loo, H.M. Loneliness associates strongly with anxiety and depression during the COVID pandemic, especially in men and younger adults. Sci. Rep. 2022, 12, 9517. [Google Scholar] [CrossRef]
- Czeisler, M.É.; Lane, R.I.; Petrosky, E.; Wiley, J.F.; Christensen, A.; Njai, R.; Weaver, M.D.; Robbins, R.; Facer-Childs, E.R.; Barger, L.K.; et al. Mental Health, Substance Use, and Suicidal Ideation During the COVID-19 Pandemic—United States, June 24–30, 2020. MMWR Morb. Mortal. Wkly. Rep. 2020, 69, 1049–1057. [Google Scholar] [CrossRef]
- Chételat, G.; Lutz, A.; Arenaza-Urquijo, E.; Collette, F.; Klimecki, O.; Marchant, N. Why could meditation practice help promote mental health and well-being in aging? Alzheimer’s Res. Ther. 2018, 10, 57. [Google Scholar] [CrossRef]
- Lazar, S.W.; Kerr, C.E.; Wasserman, R.H.; Gray, J.R.; Greve, D.N.; Treadway, M.T.; McGarvey, M.; Quinn, B.T.; Dusek, J.A.; Benson, H.; et al. Meditation experience is associated with increased cortical thickness. Neuroreport 2005, 16, 1893–1897. [Google Scholar] [CrossRef]
- Memon, A.A.; Coleman, J.J.; Amara, A.W. Effects of exercise on sleep in neurodegenerative disease. Neurobiol. Dis. 2020, 140, 104859. [Google Scholar] [CrossRef]
- Mamalaki, E.; Ntanasi, E.; Hatzimanolis, A.; Basta, M.; Kosmidis, M.H.; Dardiotis, E.; Hadjigeorgiou, G.M.; Sakka, P.; Scarmeas, N.; Yannakoulia, M. The Association of Adherence to the Mediterranean Diet with Depression in Older Adults Longitudinally Taking into Account Cognitive Status: Results from the HELIAD Study. Nutrients 2023, 15, 359. [Google Scholar] [CrossRef]
- Zhou, G.; Soufan, O.; Ewald, J.; Hancock, R.E.W.; Basu, N.; Xia, J. NetworkAnalyst 3.0: A visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019, 47, W234–W241. [Google Scholar] [CrossRef] [PubMed]
- Kupershmidt, I.; Su, Q.J.; Grewal, A.; Sundaresh, S.; Halperin, I.; Flynn, J.; Shekar, M.; Wang, H.; Park, J.; Cui, W.; et al. Ontology-Based Meta-Analysis of Global Collections of High-Throughput Public Data. PLoS ONE 2010, 5, e13066. [Google Scholar] [CrossRef] [PubMed]


| Switch Gene | Name | Dataset (High vs. Low Loneliness) | Neurodegenerative Diseases | Neuropsychiatric Diseases | References |
|---|---|---|---|---|---|
| AGO2 | Argonaute 2 | Males | Silencing of AGO2 enhances the expression of APP-cleaving enzyme (BACE1) in vitro. AGO2 accumulation leads to dysregulation of miRNAs and impairment of autophagy in Huntington’s disease. | [22,23] | |
| HLA-DRB5 | Major histocompatibility complex, class II, DR beta 5 | Males | HLA-DRB5 variants are linked to neuroinflammation and AD. | HLA-DRB5 is associated with remission to antidepressant treatment and epigenetic alterations in schizophrenia. | [24,25,26,27] |
| ALDOA | Aldolase, fructose-bisphosphate A | Males | Increased APOE4 copy number is associated with increased ALDOA levels in CSF of AD patients. | [28] | |
| S100A8 | S100 calcium-binding protein A8 | Males | Myeloperoxidase is accumulated in S100A8(+) neutrophils in the AD human brain. S100A8 is increased in the serum of FTD patients. Aggregation of S100A8 precedes Aβ formation in mice. | [29,30,31] | |
| CTSG | Cathepsin G | Males | Dysregulated expression of CTSG is associated with Lewy body dementia. | [32] | |
| CXCL1 | C-X-C motif chemokine ligand 1 | Males | Upregulated CXCL1 mediates Aβ toxicity in the human AD brain. | [33] | |
| CYTH4 | Cytohesin 4 | Males | Expression levels of CYTH4 alleles are increased in AD patients. | CYTH4 variants are associated with schizophrenia in primates and bipolar disorder in humans. | [34,35] |
| PELI1 | Pellino E3 ubiquitin-protein ligase 1 | Males | PELI1 is involved in microglial activation and dopaminergic cell death in PD. | [36] | |
| FPR1 | Formyl peptide receptor 1 | Males | FPR1 is associated with dementia risk. | [37,38] | |
| NPAS3 | Males | NPAS3 variants are implicated in schizophrenia and intellectual disability. | [39,40,41,42,43,44] | ||
| GPNMB | Glycoprotein nonmetastatic melanoma protein B | All | GPNMB confers risk for PD. It is increased in the plasma of PD patients and associated with disease severity. GPNMB is a switch gene in the frontal cortex of FTD patients. | [15,45] | |
| TH | Tyrosine hydroxylase | All | A deficiency of the TH enzyme is a critical feature in PD. | [46,47] | |
| CD59 | Complement defense 59 | All | CD59 deficits are observed in AD patients’ frontal cortex, hippocampus, and plasma. | [48,49] | |
| COL4A1 | Collagen type IV alpha 1 chain | All | COL4A1 mutation is associated with multi-infarct dementia. | [50] | |
| ZBTB16 | Zinc finger and BTB domain-containing 16 | All | ZBTB16 is linked to increased autophagy in Huntington’s and AD mice models. | [51,52,53] | |
| TSPAN15 | Tetraspanin 15 | All | TSPAN15 is increased in human and animal models of AD. | [54] | |
| DMD | Dystrophin | All | DMD is a crucial hub gene associated with AD risk variants. | There is a higher prevalence of neuropsychiatric diseases among patients with Duchene and Becker muscular dystrophies. | [55,56] |
| LEF1 | Lymphoid enhancer-binding factor 1 | All | LEF1 is involved in the differentiation of midbrain dopaminergic neurons. | [57] | |
| GPR3 | G protein-coupled receptor 3 | All | Loss of GPR3 reduced Aβ formation and improved memory in AD mouse models. Elevated GPR3 expression is correlated with AD progression in humans. | [58] | |
| UBE2V1 | Ubiquitin-conjugating enzyme E2 V1 | All | UBE2V1 is downregulated in the blood of AD patients. | [59] | |
| DPP10 | Dipeptidyl peptidase like 10 | All | DPP10 malfunctioning is associated with AD and FTD. | DPP10 variants are associated with bipolar disorder and schizophrenia | [60,61,62] |
| NECTIN2 | Nectin cell adhesion molecule 2 | All | NECTIN2 variants are associated with AD risk. | [63] | |
| LGALS3BP | Galectin-3-binding protein | All | Increased secretion of GAL3BP suppressed Aβ production in a cellular model of AD. | [64] | |
| CDKN1A | Cyclin-dependent kinase inhibitor 1A | All | CDKN1A is increased in the blood of AD patients. | [64] | |
| SERPINA1 | Serpin family A member 1 | All | SERPINA1 is a risk marker for PD dementia. SERPINA1 is upregulated in CSF of Creutzfeldt-Jakob disease and FTD patients. SERPINA1 isoforms were differentially expressed in CSF of AD and LBD patients. | [65,66,67] | |
| DMP1 | Dentin matrix acidic phosphoprotein 1 | All | Silencing of DMP1 improved cognitive impairment and enhanced the proliferation of neural progenitor cells in AD mice. | [68] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santiago, J.A.; Quinn, J.P.; Potashkin, J.A. Co-Expression Network Analysis Identifies Molecular Determinants of Loneliness Associated with Neuropsychiatric and Neurodegenerative Diseases. Int. J. Mol. Sci. 2023, 24, 5909. https://doi.org/10.3390/ijms24065909
Santiago JA, Quinn JP, Potashkin JA. Co-Expression Network Analysis Identifies Molecular Determinants of Loneliness Associated with Neuropsychiatric and Neurodegenerative Diseases. International Journal of Molecular Sciences. 2023; 24(6):5909. https://doi.org/10.3390/ijms24065909
Chicago/Turabian StyleSantiago, Jose A., James P. Quinn, and Judith A. Potashkin. 2023. "Co-Expression Network Analysis Identifies Molecular Determinants of Loneliness Associated with Neuropsychiatric and Neurodegenerative Diseases" International Journal of Molecular Sciences 24, no. 6: 5909. https://doi.org/10.3390/ijms24065909
APA StyleSantiago, J. A., Quinn, J. P., & Potashkin, J. A. (2023). Co-Expression Network Analysis Identifies Molecular Determinants of Loneliness Associated with Neuropsychiatric and Neurodegenerative Diseases. International Journal of Molecular Sciences, 24(6), 5909. https://doi.org/10.3390/ijms24065909






