Abstract
The whitefly, Bemisia tabaci MED (Hemiptera: Aleyrodidae), is an omnivorous agricultural pest, which causes huge economic losses to agriculture and is highly resistant to many pesticides. The overexpression of cytochrome P450 may play an important role in host adaptation and insecticide resistance in B. tabaci MED. Therefore, the present study systematically analyzed the cytochrome P450 gene family at the genome-wide level to understand its function in B. tabaci MED. Our analysis identified 58 cytochrome P450 genes in B. tabaci MED, among which 24 were novel. Phylogenetic analysis revealed broad functional and species-specific diversification in B. tabaci MED P450, suggesting the role of multiple P450 genes in detoxifying. Reverse transcription-real time quantitative PCR (RT-qPCR) showed that CYP4CS2, CYP4CS5, CYP4CS6, CYP4CS8, CYP6DW4, CYP6DW5, CYP6DW6, CYP6DZ8, and CYP6EN1 genes increased significantly after two days of exposure to imidacloprid. Interestingly, all nine genes belonged to the CYP4 and CYP6 families. A decrease in the expression of five genes (CYP6DW4, CYP6DW5, CYP6DW6, CYP6DZ8, and CYP4CS6) via RNA interference (RNAi) resulted in a significant increase in the mortalities of whiteflies when exposed to imidacloprid. These results indicate that the overexpression of the P450 genes may play an essential role in imidacloprid tolerance of B. tabaci MED. Thus, the present study provides basic information on P450 genes in B. tabaci MED, which will further help elucidate the insecticide resistance mechanism in the agricultural pest whitefly.
1. Significance
The whitefly, Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae), is the main pest worldwide that is resistant to many insecticides. Therefore, it is necessary to develop new and effective compounds against B. tabaci. We sequenced the whole genome of B. tabaci to detect cytochrome P450 protein, and knocked out five genes by RNA interference, which led to an increase in mortality after imidacloprid exposure, and confirmed their role in insecticide resistance. These findings provide a theoretical basis for further studying the specific function of cytochrome P450 protein and clarifying the protein–compound interaction to develop effective pesticides.
2. Introduction
The whitefly, B. tabaci, is one of the most destructive polyphagous phloem-feeders worldwide [1]. B. tabaci is a cryptic species complex including the species “MEAM1” and “MED” (also known as biotype B and biotype Q, respectively), acquiring a globally invasive insect status and are widely distributed throughout China, Southern United States, and Africa [2,3]. B. tabaci do direct damage by feeding on plant sap and indirect damage by transmitting plant viruses to vegetable crops [4,5,6]. Because of its ability to transmit begomoviruses and criniviruses very efficiently into many vegetable crops [7,8,9,10], farmers respond with calendar application of insecticides, often with insecticides sharing the same mode of action, resulting in resistance development in exposed populations [11,12].
Cytochrome P450 contributes to growth, development, nutrition, and detoxification of xenobiotics, and is used to metabolize a variety of endogenous and exogenous compounds, forming a polygenic enzyme superfamily [13,14]. Cytochrome P450 is the main detoxification enzyme system involved in the metabolism of various pesticides and other exogenous and endogenous compounds in insects, and participates in the synthesis and metabolism of various endogenous substances, such as ecdysone, juvenile hormone, and fatty acids [15]. In addition, P450 can enhance the detoxification activity by overexpressing and accumulating detoxification proteins or changing these proteins’ structure [16,17]. In the P450 supergene family, CYP4 and CYP6 family genes are mainly related to pests’ insecticide resistance [18]. Studies have also proven that P450 plays a key role in insecticide resistance due to its genetic diversity, broad substrate specificity, and catalytic versatility [19]. However, no systematic study has been conducted on the cytochrome P450 gene family at the genome-wide level in B. tabaci MED.
As an alternative to organophosphates and pyrethroids, neonicotinoid insecticides have been used to control whiteflies in fields and greenhouses; however, the extensive use has led to the rapid development of neonicotinoid resistance [20,21]. B. tabaci MED has a high level of resistance to neonicotinoid insecticides, which may contribute to genetic cluster changes [22].
Neonicotinoid insecticides were introduced in 1991, with imidacloprid being the first product in the market. Imidacloprid targeted nicotinic acetylcholine receptor (nAChR) receptors in the insect nervous system [23]. Because of its broad-spectrum action, high efficiency, low toxicity, and low residue, it was widely adopted in many cultivations throughout the world, including China. Furthermore, imidacloprid had multiple modes of action, such as contact killing, stomach toxicity, and systemic absorption, resulting in the slow evolution of resistance in some cropping systems [24]. However, because of its genetic ability to better metabolize insecticides and selection pressure imposed by the calendar application of imidacloprid, populations of B. tabaci MED have become extremely tolerant to the commercial products of this insecticide [25]. Studies have demonstrated that the overexpression of CYP6CM1 in B. tabaci leads to imidacloprid resistance [26]. However, only a few P450s have been published in B. tabaci compared with other insects, such as Bombyx mori (84) [27], Leptinotarsa decemlineata (96) [18], Rhynchophorus ferrugineus (77) [28], and Drosophila melanogaster (85) [29]. Therefore, to better understand the resistance mechanism in B. tabaci MED, there is a need to identify more P450 genes of B. tabaci MED.
The present study aimed to identify the cytochrome P450 gene family in B. tabaci MED and assess their roles in insecticide resistance. We conducted a genome-wide analysis of P450 sequences in the complete genome of B. tabaci MED and investigated their roles in insecticide resistance using expression analysis, RNAi approach, and a bioassay.
3. Results
3.1. Genome-Wide Identification of the P450 Gene Superfamily in B. tabaci
We obtained about 105.92 Gb of clean data by sequencing the B. tabaci library on the PacBio platform. The total sequencing depth was about 166.16 x, the read N50 was 23.42 Kb, and the average read length was 14.74 Kb (Table 1). After searching by BlastP and BlastN, 24 genes encoding P450 protein were identified in the MED genome of B. tabaci MED. The predicted proteins encoded by the 24 BtP450 genes were initially classified using NCBI CDD analysis into four families, including CYP4, CYP6, CYP9, CYP301-318, and CYP18a (Table 2).

Table 1.
Summary of the Bemisia tabaci MED genome assembly.

Table 2.
Numbers of genes in P450 clans and families identified in Bemisia tabaci.
Further, in order to evaluate the phylogenetic relationship between each family of cytochrome P450 proteins in B. tabaci MED, all P450 proteins were compared with MEGA7 to generate a phylogenetic tree. As shown in Figure 1, the P450 proteins from the same family clustered together. The topology of the phylogenetic tree reconstructed by the ML method and PhyML was roughly in agreement with the NJ method, demonstrating our results’ reliability. The CYP4 and CYP6 families of B. tabaci MED were the largest families (Figure 1).
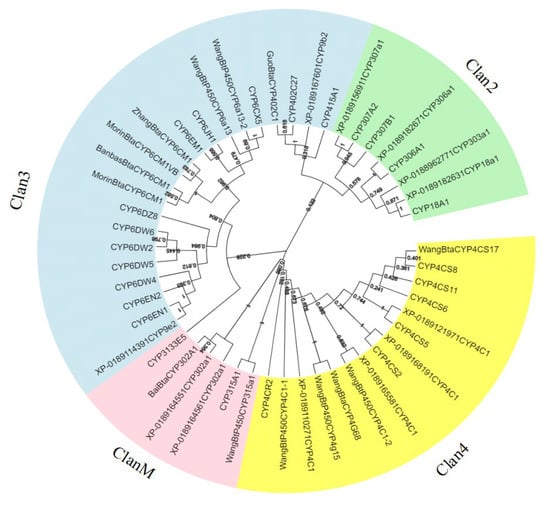
Figure 1.
Phylogenetic relationships of P450 families from Bemisia tabaci MED. The unrooted phylogentic tree was constructed using MEGA7 by the neighbor-joining method. The bootstrap test was performed with 1000 replicates.
3.2. Phylogenetic and Protein Structure Analyses of P450s in B. tabaci MED
Then, in order to explore the structural diversity of B. tabaci MED P450 superfamily, MEME was used to analyze the conserved motifs. As shown in Figure 2, the motifs of P450 proteins in the same family are also extremely similar, and these motifs are highly conserved among B. tabaci MED P450 proteins. Meanwhile, a few P450 proteins from sister branches also shared a common motif composition (Figure 2). This phenomenon is probably associated with the gene structure and phylogenetic relationships.
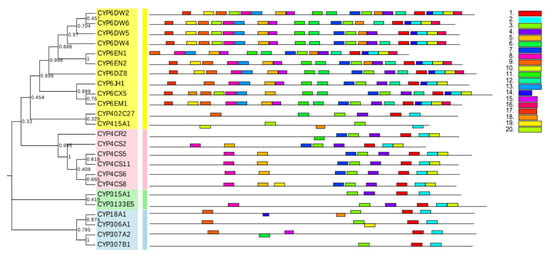
Figure 2.
Phylogenetic relationships and protein motif analysis of Bemisia tabaci MED CYP. The unrooted phylogenetic tree was constructed using MEGA 7 by the neighbor-joining method and the bootstrap test was performed with 1000 replicates. The colored shadow marks the different CYP families. All motifs were identified by MEME database with the complete amino acid sequences of CYPs. Lengths of motifs for each CYP protein are exhibited proportionally.
3.3. Differential Expression of BtP450 Genes under Imidacloprid Treatment
After B. tabaci MED adults were fed 50 ppm imidacloprid, we quantified the P450 protein-encoding 24 non-redundant genes (Figure 3). Among these 24 genes, CYP4CS2, CYP4CS5, CYP4CS6, CYP4CS8, CYP6DW4, CYP6DW5, CYP6DW6, CYP6DZ8, and CYP6EN1 were found to be significantly upregulated. Then, to examine the involvement of BtaP450 in the whitefly imidacloprid stress response, the upregulated and downregulated genes were selected as candidates for RNA interference. Further qPCR analysis showed that the expression of significantly upregulated genes in B. tabaci MED was suppressed considerably after two days of dsRNA feeding (Figure 3).
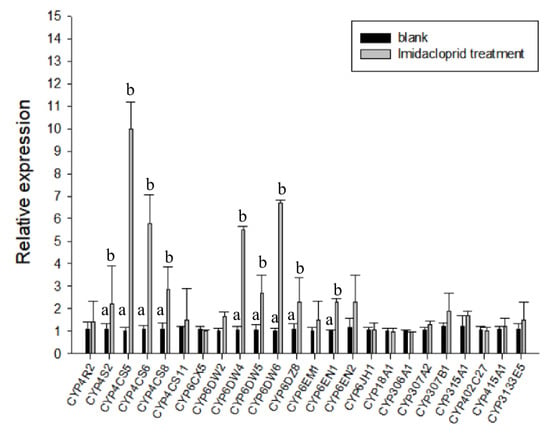
Figure 3.
Expression of different genes in Bemisia tabaci MED fed imidacloprid (50 ppm). Quantitative real-time PCR (RT-qPCR) analysis of the expression profile of the BtP450 gene superfamily in B. tabaci MED. Gene names are shown on the x-axis and expression levels are shown on the y-axis. Different letters on the column indicate that the expression levels of different genes are significantly different (p < 0.05).
3.4. Effect of BtP450 RNAi on Imidacloprid Tolerance of B. tabaci MED
To further investigate the role of BtP450 in the B. tabaci MED insecticide stress response, five genes (CYP6DW4, CYP6DW5, CYP6DW6, CYP6DZ8, and CYP4CS6) significantly upregulated after imidacloprid treatment were selected as candidates for RNAi. The expression of these five genes of the CYP4 and CYP6 family was suppressed considerably after two days of dsRNA feeding, as shown by qPCR (Figure 4).
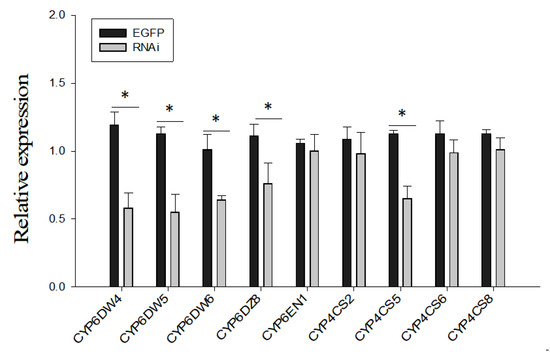
Figure 4.
The mRNA level of different P450 in laboratory Bemisia tabaci MED after feeding dsRNA. The mRNA levels of P450 were assessed by RT-qPCR after 48 h dsRNA treatment. Values are means ± SE of three biological replicates. The asterisk (*) indicates a significant difference (p < 0.05).
After feeding dsRNA for two days, we performed biological assays with 100 ppm of imidacloprid (Figure 5). RNAi of CYP6DW4, CYP6DW5, CYP6DW6, CYP6DZ8, and CYP4CS6 significantly increased the susceptibility of B. tabaci MED to imidacloprid. After knocking out these genes, the mortality rate for whiteflies exposed to dsRNA was significantly higher in comparison to whiteflies exposed to dsEGFP.
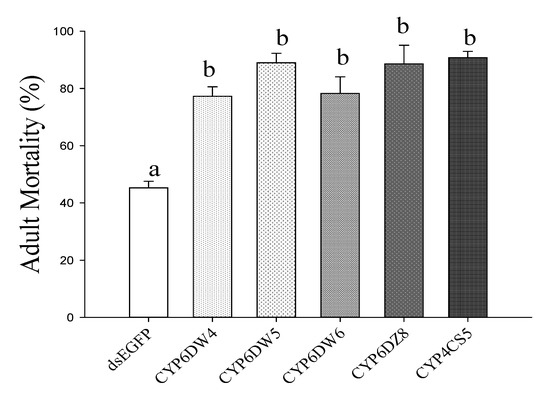
Figure 5.
24 h mortality of Bemisia tabaci MED fed imidacloprid (100 ppm) after five P450 genes silencing. Means for different letters were significantly different (p < 0.05).
4. Discussion
In 2003, B. tabaci MED was identified as an alien species in China, which seriously impacted agroecology [30]. With the rapid development of genome sequencing technology, whole genome sequencing helped researchers study the gene families. The genome sequencing of B. tabaci MED has also been completed, which provides a good foundation for understanding the evolutionary relationship of each gene family of B. tabaci MED. This study used the transcriptome data of B. tabaci MED to identify the P450 gene family and analyze the location and evolutionary relationship of the various P450 genes in the genome. The results detected 24 P450 gene sequences in the B. tabaci MED genome, which were divided into four clans, including Clan 2, Clan 3, Clan 4, and Clan M. Compared with the published P450 genes, the P450 genes of Clan 3 and Clan 4 in the B. tabaci genome showed an apparent gene expansion. Previous studies have shown that the P450 gene families: CYP 4, 6, and 9 are associated with pesticide resistance and host adaptation. For example, in B. tabaci, the CYP6CM1 gene is related to imidacloprid resistance [26]. In the brown planthopper, the CYP6ER1 gene is associated with imidacloprid resistance [31]. Similarly, the bumblebee CYP9Q3 efficiently metabolizes thiacloprid [32,33], and CYP9Q6 metabolizes thiacloprid and acetamiprid [34]. The P450 CYP4, 6, and 9 families are Clan 3 and Clan 4 genes in B. tabaci MED; therefore, it is very likely that these clans of genes might be related to the insecticide resistance in B. tabaci MED as well.
Glutathione S-transferase, esterase, and P450 are generally regarded as detoxification enzymes and help ward off and catabolize toxins, including pesticides. Studies have demonstrated point mutations in pesticide target genes can lead to an increase in gene expression and diversification of coding sequences with the variable outcome for insecticide metabolism in insects [35]. For instance, in B. tabaci MED, the increase in susceptibility has been primarily associated with the mutations in the P450 monooxygenase rather than the nicotinic acetylcholine receptor, which is the target site of imidacloprid action [36]. The P450-specific inhibitor piperonyl butanol significantly reduced imidacloprid resistance levels, demonstrating the important role of P450s in insecticide resistance [37]. Similarly, P450-mediated effects of insecticide resistance have been reported in aphids [38]. In addition, specific P450 genes have been associated with neonicotinoid resistance. For example, Drosophila CYP6G1 confers imidacloprid resistance [39,40]. Puinean proposed that CYP6Y3 confers neonicotinoid resistance to Myzus persicae [41]. Puinean significantly reduced imidacloprid resistance levels using the P450s-specific inhibitor piperonyl butanol, which demonstrated an important role for P450s in insecticide resistance [37]. It is reported that members of CYP3 and CYP4 families participate in the metabolism of pesticides because of long-term use of pesticides [19]. Elevated expression levels of CYP6CM1 and CYP4C64 were associated with imidacloprid resistance in B. tabaci [25,26], whereas overexpression of CYP6ER1 and CYP6AY1 was closely associated with imidacloprid resistance in N. lugens [42]. In red palm weevil (RPW), CYP345J1 and CYP6NR1 overexpression metabolized pesticide molecules more efficiently [28], thereby enhancing tolerance of RPW in palm fields, both CYP345 and CYP6 belong to a single gene clan of P450.
This study identified nine highly expressed P450 genes after imidacloprid treatment in MED. Subsequent follow-up studies using RNAi and qPCR revealed that feeding MED with dsRNA corresponding to these genes resulted in a significant reduction in the expression of five genes (belonging to CYP4 and CYP6 families). Furthermore, the known down of these genes resulted in a significant increase in mortality of B. tabaci MED exposed to imidacloprid signifying these genes had a direct role in imidacloprid metabolism in B. tabaci MED. Similarly, previous studies with other coleopterans, such as Tribolium castaneum and Leptinotarsa decemlineata, have also reported that the role of CYP4 173 and CYP6 family genes in the metabolism of xenobiotics compounds [18,43,44]. Taken together, data from this study and previous studies suggest that the xenobiotic metabolism role of these genes might be conserved in insects. However, further studies are warranted to fully comprehend the role these genes might play in insecticide metabolism and insecticide resistance development in insect pests.
The current study provides a genomic database of B. tabaci MED P450 and a comprehensive picture of the B. tabaci MED P450 family based on gene identification and imidacloprid-induced expression profiling. We systematically analyzed 24 B. tabaci MED P450 genes, which provide a basis for further molecular and functional characterization of P450. The phylogenetic tree divided the B. tabaci MED P450 into four clans, of which clan 2 is dominant. Our research provides a solid foundation for future insecticide design. However, to guide the rational development of pesticide-related compounds and improve the accuracy and efficiency of computational-based drug construction, a deeper understanding of the interaction of imidacloprid-related heterocyclic compounds with P450s is required. Our study reveals various roles of B. tabaci MED P450 and provides a reference for better research on the functions of B. tabaci MED P450.
5. Materials and Methods
5.1. Insect Sampling and Rearing
The B. tabaci MED Lingshui population was collected from Lingshui City, Hainan Province, in January 2017. Since the collection, all populations have not been exposed to pesticides, and they are raised under laboratory conditions, without any further selection pressure, and each generation is about every 25 days. These insects were reared on the common tobacco variety NC89 for a long time in insect-proof cages (temperature 27 ± 1 °C, relative humidity 60 ± 5%, and light cycle 16L:8D).
5.2. P450 Gene Classification and Phylogenetic Analysis
The experimental process is carried out according to the standard protocol provided by Pacbio, and the library construction includes six steps: (1) use g-TUBE to interrupt the DNA sample; (2) repair the damaged DNA sample; (3) terminal repair of DNA; (4) connect the dumbbell-shaped connector; (5) exonuclease digestion of nucleic acid; and (6) screening the target fragment with BluePippin to obtain the sequencing library. The present study used three strategies to predict the gene structure: de novo prediction, homologous species-based prediction, and Unigene-based prediction. De novo prediction was carried out using Genscan [45], Augustus (v2.4) [46], GlimmerHMM (v3.0.4) [47], GeneID (v1.4), and SNAP (v2006-07-28) [48] programs. The homologous species-based prediction was obtained by running GeMoMa (v1.3.1) [49,50], Hisat (v2.0.4) [51]. and Stringtie (v1.2.3) [52] for reference transcript-based assembly, followed by gene prediction using TransDecoder (v2.0) and GeneMarkS -T (v5.1) [53]. Meanwhile, PASA (v2.0.2) [54] was used to predict Unigene sequences based on transcriptome data without reference assembly. Finally, EVM (v1.1.1) was used to integrate the prediction results obtained by the above three methods and modified with PASA. The predicted gene sequences were compared with NR [55], KOG [56], GO [57], KEGG [58], TrEMBL [59], and other functional databases by BLAST (v2.2.31) [60] with an e-value of 1e-5. Then, the KEGG pathway annotation analysis and KOG function annotation analysis of genes were carried out. The B. tabaci samples were obtained from Lingshui, Hainan, in 2017 and identified as MED using the mtCOI gene [61,62]. Genes annotated as cytochrome P450 based on the genome annotation library were selected as candidate genes.
In order to analyze the phylogenetic tree, P450 amino acid sequences of various insects were downloaded from NCBI, and then merged with 24 P450 amino acid sequences of B. tabaci. The phylogenetic tree was analyzed by MEGA7 NJ method and corrected for 1000 times. Then the conserved motifs of 24 P450 sequences were analyzed by MEME, and the screened P450 genes were classified according to the phylogenetic tree, and further named according to the molecular weight, and then handed over to Dr. David Nielsen (CYP Nomenclature Committee) for gene naming.
5.3. Imidacloprid Treatment
Then, to express P450 under imidacloprid stress, whiteflies were treated with imidacloprid at a concentration of 50 ppm for two days, using a blank control and triplicates per treatment; approximately 100 adults were maintained per replicate.
5.4. RNA Extraction, cDNA Synthesis, and P450 Cloning
The RNA samples were extracted from 100 adults using TRIzol reagent (Thermo Fisher, Waltham, MA, USA), and the first-strand cDNA was prepared with the PrimeScript RT Kit (Takara, Dalian, China), following the manufacturer’s protocol. The cDNA was used as a template for full-sequence cloning and real-time PCR (qPCR). Use (Takara) thermal cycle program for PCR amplification: 94 °C for 5 min (pre-denaturation), 94 °C for 1 min (deformation), 55 °C for 30 s, 72 °C for 40 s, then 40 cycles, and finally 72 °C for 72 °C 10 min. The amplicon was checked by agarose gel electrophoresis (1%) and recycled with SteadyPure Agarose Gel DNA Purification Kit (Accurate Biotechnology, Changsha, China) to ensure the accuracy of the operation. The relative expression levels of the P450 genes were calculated using the 2−ΔΔCT method [63]. The primers used in this study are shown in Supplementary Materials.
5.5. Double-Stranded RNA (dsRNA) Synthesis, RNAi, and Real-Time PCR (qPCR)
Using primers containing T7 RNA polymerase promoter, cytochrome P450 and enhanced green fluorescent protein (EGFP) genes were amplified by PCR. The PCR products were purified by Accurate Biology, and dsRNA was synthesized by using T7 RiboMAX Express RNAi kit (Vazyme, Nanjing, China) according to the manufacturer’s instructions. The concentration of dsRNA was determined by nano-drop spectrophotometer, and its integrity was verified by agarose gel electrophoresis (1.5%).
The RNAi assay was performed by feeding dsRNA to B. tabaci MED adults in a feeding chamber (5 cm × 10 cm) according to a previously reported method [64]. After 48 h of feeding on artificial diet with dsRNA, the bioassay was conducted, using four biological replicates per treatment. Mortality was recorded after 24 h of feeding. The differences in the relative expression levels and mortality were assessed following Student’s t-test and analysis of variance (ANOVA) using SPSS (v.21) (IBM-SPSS, Armonk, NY, USA), and considered significant at p < 0.05.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms24065899/s1.
Funding
This research was funded by the National Natural Science Foundation of China (31872030), the Taishan Scholar Foundation of Shandong Province (tstp20221135), the First-Class Grassland Science Discipline Program in Shandong Province (1619002).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data generated or analysed during this study are included in this published article.
Acknowledgments
The authors are grateful to David. R. Nelson (University of Tennessee) for the P450 nomenclature. We are grateful to the editor and reviewers for their numerous perceptive and constructive comments, which helped us to improve our manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Barinaga, M. Is devastating whitefly invader really a new species? Science 1993, 259, 30. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; De Barro, P.; Zhao, H.; Wang, J.; Nardi, F.; Liu, S.S. An extensive field survey combined with a phylogenetic analysis reveals rapid and widespread invasion of two alien whiteflies in China. PLoS ONE 2011, 6, e16061. [Google Scholar] [CrossRef] [PubMed]
- Saurabh, G.; Michael, S.C.; Bhabesh, D.; Timothy, C.; Alvin, M.S.; Andre, S.; William, E.S.; Rajagopalbabu, S. Low Genetic Variability in Bemisia tabaci MEAM1 Populations within Farmscapes of Georgia, USA. Insect 2020, 11, 834. [Google Scholar]
- De Barro, P.J.; Dinsdale, A. Bemisia tabaci: A Statement of Species Status. Annu. Rev. Entomol. 2011, 56, 1–19. [Google Scholar] [CrossRef] [PubMed]
- De Barro, P.J.; Bourne, A.; Khan, S.A.; Brancatini, V.A.L. Host plant and biotype density interactions—Their role in the establishment of the invasive B biotype of Bemisia tabaci. Biol. Invasions 2006, 8, 287–294. [Google Scholar] [CrossRef]
- Guo, C.L.; Zhu, Y.Z.; Zhang, Y.J. Invasion biology and management of alien whitefly (Bemisia tabaci) in China. J. Integr. Pest Manag. 2021, 12, 1–16. [Google Scholar] [CrossRef]
- Gautam, S.; Mugerwa, H.; Buck, J.W.; Dutta, B.; Coolong, T.; Adkins, S.; Srinivasan, R. Differential Transmission of Old and New World Begomoviruses by Middle East-Asia Minor 1 (MEAM1) and Mediterranean (MED) Cryptic Species of Bemisia tabaci. Viruses 2022, 14, 1104. [Google Scholar] [CrossRef]
- Habibu, M.; Saurabh, G.; Michael, A.C.; Bhabesh, D.; Judith, K.B.; Scott, A.; Rajagopalbabu, S. Differential Transcriptional Responses in Two Old World Bemisia tabaci Cryptic Species Post Acquisition of Old and New World Begomoviruses. Cells 2022, 11, 2060. [Google Scholar]
- Pan, H.P.; Chu, D.; Yan, W.Q.; Su, Q.; Liu, B.M.; Wang, S.L.; Wu, Q.J.; Xie, W.; Jiao, X.G.; Li, R.M.; et al. Rapid spread of tomato yellow leaf curl virus in China is aided differentially by two invasive whiteflies. PLoS ONE 2012, 7, e34817. [Google Scholar] [CrossRef]
- Wendy, G.; Saurabh, G.; Bhabesh, D.; Rajagopalbabu, S. Whitefly-Mediated Transmission and Subsequent Acquisition of Highly Similar and Naturally Occurring Tomato Yellow Leaf Curl Virus Variants. Phytopathology 2022, 112, 720–728. [Google Scholar]
- Rami Murad, G.; Emmanouil, R.; Ralf, N.; Isaac, I. Insecticide resistance and its management in Bemisia tabaci species. J. Pest Sci. 2020, 93, 893–910. [Google Scholar]
- Li, M.J.; Yang, S.W.; Chen, G.H.; Dou, W.J.; Shang, H.P.; Zhang, X.M. Density and seasonal dynamics of Bemisia tabaci and its predators in different agricultural landscapes in South China. Front. Plant Sci. 2022, 13, 928634. [Google Scholar] [CrossRef] [PubMed]
- Feyereisen, R.; Andersen, J.; Carino, F.; Cohen, M.; Koener, J. Cytochrome P450 in the house fly: Structure, catalytic activity and regulation of expression of CYP6A1 in an insecticide-resistant strain. Pestic. Sci. 1995, 43, 233–239. [Google Scholar] [CrossRef]
- Feyereisen, R. Insect CYP genes and P450 enzymes. Insect Mol. Biol. Biochem. 2011, 8, 236–316. [Google Scholar]
- Iga, M.; Kataoka, H. Recent Studies on Insect Hormone Metabolic Pathways Mediated by Cytochrome P450 Enzymes. Biol. Pharm. Bull. 2012, 35, 838–843. [Google Scholar] [CrossRef]
- Kim, I.Y.; Choi, B.; Park, W.R.; Kim, Y.J.; Kim, B.E.; Mun, S.; Kim, D.Y. Nuclear receptor HR96 up-regulates cytochrome P450 for insecticide detoxification in Tribolium castaneum. Pest. Manag. Sci. 2022, 78, 230–239. [Google Scholar] [CrossRef]
- Fotoukkiaii, S.M.; Wybouw, N.; Kurlovs, A.H.; Leeuwen, T.V. High-resolution genetic mapping reveals cis-regulatory and copy number variation in loci associated with cytochrome P450-mediated detoxification in a generalist arthropod pest. PLoS Genet. 2021, 17, e1009422. [Google Scholar] [CrossRef]
- Zhu, F.; Moural, T.W.; Nelson, D.R.; Palli, S.R. A specialist herbivore pest adaptation to xenobiotics through up-regulation of multiple cytochrome P450s. Sci. Rep. 2016, 6, 20421. [Google Scholar] [CrossRef]
- Liu, N.; Li, M.; Gong, Y.; Liu, F.; Li, T. Cytochrome P450s-their expression, regulation, and role in insecticide resistance. Pestic. Biochem. Physiol. 2015, 120, 77–81. [Google Scholar] [CrossRef]
- Chen, J.C.; Wang, Z.H.; Cao, L.J.; Gong, Y.J.; Hoffmann, A.A.; Wei, S.J. Toxicity of seven insecticides to different developmental stages of the whitefly Bemisia tabaci MED (Hemiptera: Aleyrodidae) in multiple field populations of China. Ecotoxicology 2018, 27, 742–751. [Google Scholar] [CrossRef]
- Daborn, P.; Boundy, S.; Yen, J.; Pittendrigh, B. DDT resistance in Drosophila correlates with Cyp6g1 over-expression and confers cross-resistance to the neonicotinoid imidacloprid. Mol. Genet. Genom. 2001, 266, 556–563. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.; Kim, S.; Lee, S.H.; Lee, J.H. Insecticide resistance trait may contribute to genetic cluster change in Bemisia tabaci MED (Hemiptera: Aleyrodidae) as a potential driving force. Pest Manag. Sci. 2021, 77, 3581–3587. [Google Scholar] [CrossRef] [PubMed]
- Buckingham, S.; Lapied, B.; Corronc, H.; Sattelle, F. Imidacloprid actions on insect neuronal acetylcholine receptors. J. Exp. Biol. 1997, 200, 2685–2692. [Google Scholar] [CrossRef] [PubMed]
- Tomizama, M.; Casida, J.E. Neonicotinoid insecticide toxicology: Mechanisms of selective action. Annu. Rev. Pharmacol. Toxicol. 2005, 45, 247–268. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Xie, W.; Wang, S.L.; Wu, Q.J.; Pan, H.P.; Li, R.M.; Yang, N.N.; Liu, B.M.; Xu, B.Y.; Zhou, X.M.; et al. Two cytochrome P450 genes are involved in imidacloprid resistance in field populations of the whitefly, Bemisia tabaci, in China. Pestic. Biochem. Physiol. 2013, 107, 343–350. [Google Scholar] [CrossRef]
- Karunker, I.; Benting, J.; Lueke, B.; Ponge, T.; Nauen, R.; Roditakis, E.; Vontas, J.; Gorman, K.; Denholm, I.; Morin, S. Over-expression of cytochrome P450 CYP6CM1 is associated with high resistance to imidacloprid in the B and Q biotypes of Bemisia tabaci (Hemiptera: Aleyrodidae). Insect Biochem. Mol. Biol. 2008, 38, 634–644. [Google Scholar] [CrossRef]
- Ai, J.W.; Zhu, Y.; Duan, J.; Yu, Q.Y.; Xiang, Z.H. Genome-wide analysis of cytochrome P450 monooxygenase genes in the silkworm. Bombyx Mori. 2011, 48, 42–50. [Google Scholar]
- Binu, A.; Jibin, J.; Mahmoud, M.; Arnab, P. Global transcriptome profiling and functional analysis reveal that tissue-specific constitutive overexpression of cytochrome P450s confers tolerance to imidacloprid in palm weevils in date palm fields. BMC Genom. 2019, 20, 440. [Google Scholar]
- Nelson, D.R. The cytochrome p450 homepage. Hum. Genom. 2009, 4, 1. [Google Scholar] [CrossRef]
- Pan, H.P.; Ge, D.Q.; Wang, S.L.; Wu, Q.J.; Xu, B.Y.; Xie, W.; Zhang, Y.J. Replacement of B biotype Bemisia tabaci by Q biotype B. tabaci in some areas of Beijing and Hebei. Plant Prot. 2010, 36, 40–44. [Google Scholar]
- Zimmer, C.T.; Garrood, W.T.; Singh, K.S.; Randall, E.; Lueke, B.; Gutbrod, O.; Matthiesen, S.; Kohler, M.; Nauen, R.; Davies, T.G.E.; et al. Neofunctionalization of duplicated P450 genes drives the evolution of insecticide resistance in the brown planthopper. Curr. Biol. 2018, 28, 268–274.e5. [Google Scholar] [CrossRef] [PubMed]
- Manjon, C.; Troczka, B.J.; Zaworra, M.; Beadle, K.; Randall, E.; Hertlein, G.; Singh, K.S.; Zimmer, C.T.; Homem, R.A. Unravelling the molecular determinants of bee sensitivity to neonicotinoid insecticides. Curr. Biol. 2018, 28, 1137–1143.e5. [Google Scholar] [CrossRef]
- Beadle, K.; Singh, K.S.; Troczka, B.J.; Randall, E.; Zaworra, M.; Zimmer, C.T.; Hayward, A.; Reid, R.L. Genomic insights into neonico-tinoid sensitivity in the solitary bee Osmia bicornis. PLoS Genet. 2019, 15, e1007903. [Google Scholar] [CrossRef]
- Troczka, B.J.; Homem, R.A.; Reid, R.; Beadle, K.; Kohler, M.; Zaworra, M.; Field, L.M. Identification and Functional Characterisation of a Novel N-cyanoamidine Neonicotinoid Metabolising Cytochrome P450, CYP9Q6, From the Buff-Tailed Bumblebee Bombus Terrestris. Insect Biochem. Mol. Biol. 2019, 111, 103171. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Schuler MABerenbaum, M.R. Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu. Rev. Entomol. 2007, 52, 231–253. [Google Scholar] [CrossRef] [PubMed]
- Rauch, N.; Nauen, R. Identification of biochemical markers linked to neonicotinoid cross resistance in Bemisia tabaci (Hemiptera: Aleyrodidae). Arch. Insect. Biochem. 2003, 54, 165–176. [Google Scholar] [CrossRef]
- Puinean, A.M.; Denholm, I.; Millar, N.S.; Nauen, R.; Williamson, M.S. Characterisation of imidacloprid resistance mechanisms in the brown planthopper, Nilaparvata lugens Stal (Hemiptera: Delphacidae). Pestic. Biochem. Phys. 2010, 97, 129–132. [Google Scholar] [CrossRef]
- Le, G. Xenobiotic response in Drosophila melanogaster: Sex dependence of P450 and GST gene induction. Insect. Biochem. Mol. Biol. 2006, 36, 674–682. [Google Scholar]
- Daborn, P.J.; Yen, J.L.; Bogwitz, M.R.; Le Goff, G.; Feil, E.; Jeffers, S.; Tijet, N.; Perry, T.; Heckel, D.; Batterham, P.; et al. A single p450 allele associated with insecticide resistance in Drosophila. Science 2002, 297, 2253–2256. [Google Scholar] [CrossRef]
- Philippou, D.; Field, L.; Moores, G. Metabolic enzyme(s) confer imidacloprid resistance in a clone of Myzus persicae (Sulzer) (Hemiptera: Aphididae) from Greece. Pest Manag. Sci. 2010, 66, 390–395. [Google Scholar] [CrossRef]
- Puinean, A.M.; Foster, P.; Oliphant, L.; Denholm, I.; Field, L.M.; Millar, N.S.; Williamson, M.S.; Bass, C. Amplification of a cytochrome P450 gene is asso-ciated with resistance to neonicotinoid insecticides in the aphid Myzus persicae. PLoS Genet. 2010, 6, e1000999. [Google Scholar] [CrossRef] [PubMed]
- Bass, C.; Carvalho, R.; Oliphant, L.; Puinean, A.; Field, L.G.; Gorman, K. Overexpression of a cytochrome P450 monooxygenase, CYP6ER1, is associated with resistance to imidacloprid in the brown planthopper, Nilaparvata lugens. Insect Mol. Biol. 2011, 20, 763–773. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Moural, T.W.; Shah, K.; Palli, S.R. Integrated analysis of cytochrome P450 gene superfamily in the red flour beetle, Tribolium castaneum. BMC Genom. 2013, 14, 1. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Zhang, K.; Wei, L.; Wei, G.; Xiong, W.; Lu, Y.; Zhang, Y.; Gao, A.; Li, B. Insecticidal activity of artemisia vulgaris essential oil and transcriptome analysis of Tribolium castaneum in response to oil exposure. Front. Genet. 2020, 11, 589. [Google Scholar] [CrossRef] [PubMed]
- Burge, C.; Karlin, S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 1997, 268, 78–94. [Google Scholar] [CrossRef]
- Stanke, M.; Waack, S. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics 2003, 19, ii215–ii225. [Google Scholar] [CrossRef] [PubMed]
- Majoros, W.H.; Pertea, M.; Salzberg, S.L. TigrScan and GlimmerHMM: Two open source ab initio eukaryotic gene-finders. Bioinformatics 2004, 20, 2878–2879. [Google Scholar] [CrossRef]
- Korf, I. Gene finding in novel genomes. BMC Bioinform. 2004, 5, 59. [Google Scholar] [CrossRef]
- Keilwagen, J.; Wenk, M.; Erickson, J.L.; Schattat, M.H.; Jan, G.; Frank, H. Using intron position conservation for homology-based gene prediction. Nucleic Acids Res. 2016, 44, e89. [Google Scholar] [CrossRef]
- Keilwagen, J.; Hartung, F.; Paulini, M.; Twardziok, S.O.; Grau, J. Combining RNA-seq data and homology-based gene prediction for plants, animals and fungi. BMC Bioinform. 2018, 19, 189. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Lomsadze, A.; Borodovsky, M. Identification of protein coding regions in RNA transcripts. Nucleic Acids Res. 2015, 43, e78. [Google Scholar] [CrossRef] [PubMed]
- Campbell, M.A.; Haas, B.J.; Hamilton, J.P.; Mount, S.M.; Buell, C.R. Comprehensive analysis of alternative splicing in rice and comparative analyses with Arabidopsis. BMC Genom. 2006, 7, 327. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Lu, S.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; DeWeese-Scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R. CDD: A Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011, 39, D225–D229. [Google Scholar] [CrossRef]
- Koonin, E.V.; Fedorova, N.D.; Jackson, J.D.; Jacobs, A.R.; Krylov, D.M.; Makarova, K.S.; Mazumder, R.; Natale, D.A. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 2004, 5, R7. [Google Scholar] [CrossRef]
- Dimmer, E.C.; Huntley, R.P.; Alam-Faruque, Y.; Sawford, T.; O’Donovan, C.; Martin, M.J.; Bely, B.; Browne, P.; Chan, W.M.; Eberhardt, R. The UniProt-GO annotation database in 2011. Nucleic Acids Res. 2012, 40, D565–D570. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Boeckmann, B.; Bairoch, A.; Apweiler, R.; Blatter, M.C.; Estreicher, A.; Gasteiger, E.; Martin, M.J.; Michoud, K.; O’Donovan, C.; Phan, I. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003, 31, 365–370. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Chu, D.; Zhang, Y.J.; Brown, J.K.; Cong, B.; Xu, B.Y.; Wu, Q.J.; Zhu, G.R. The introduction of the exotic Q biotype of Bemisia tabaci (Gennadius) from the Mediterranean region into China on ornamental crops. Fla. Entomol. 2006, 89, 168–174. [Google Scholar] [CrossRef]
- Chu, D.; Wan, F.H.; Zhang, Y.J.; Brown, J.K. Change in the biotype composition of Bemisia tabaci in Shandong Province of China from 2005 to 2008. Environ. Entomol. 2010, 39, 1028–1036. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (−ΔΔCT) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Xie, W.; Yang, X.; Wang, S.L.; Wu, Q.J.; Zhang, Y.J. Identification of glutathione S-transferases in Bemisia tabaci (Hemiptera: Aleyrodidae) and evidence that GSTd7 helps explain the difference in insecticide susceptibility between B. tabaci Middle East-minor Asia 1 and Mediterranean. Insect Mol. Biol. 2018, 27, 22–35. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).