DNA Methylation Analysis Identifies Novel Epigenetic Loci in Dilated Murine Heart upon Exposure to Volume Overload
Abstract
1. Introduction
2. Results
2.1. LV Dilatation and Contractile Dysfunction in 16-Week-VO-Exposed Mice
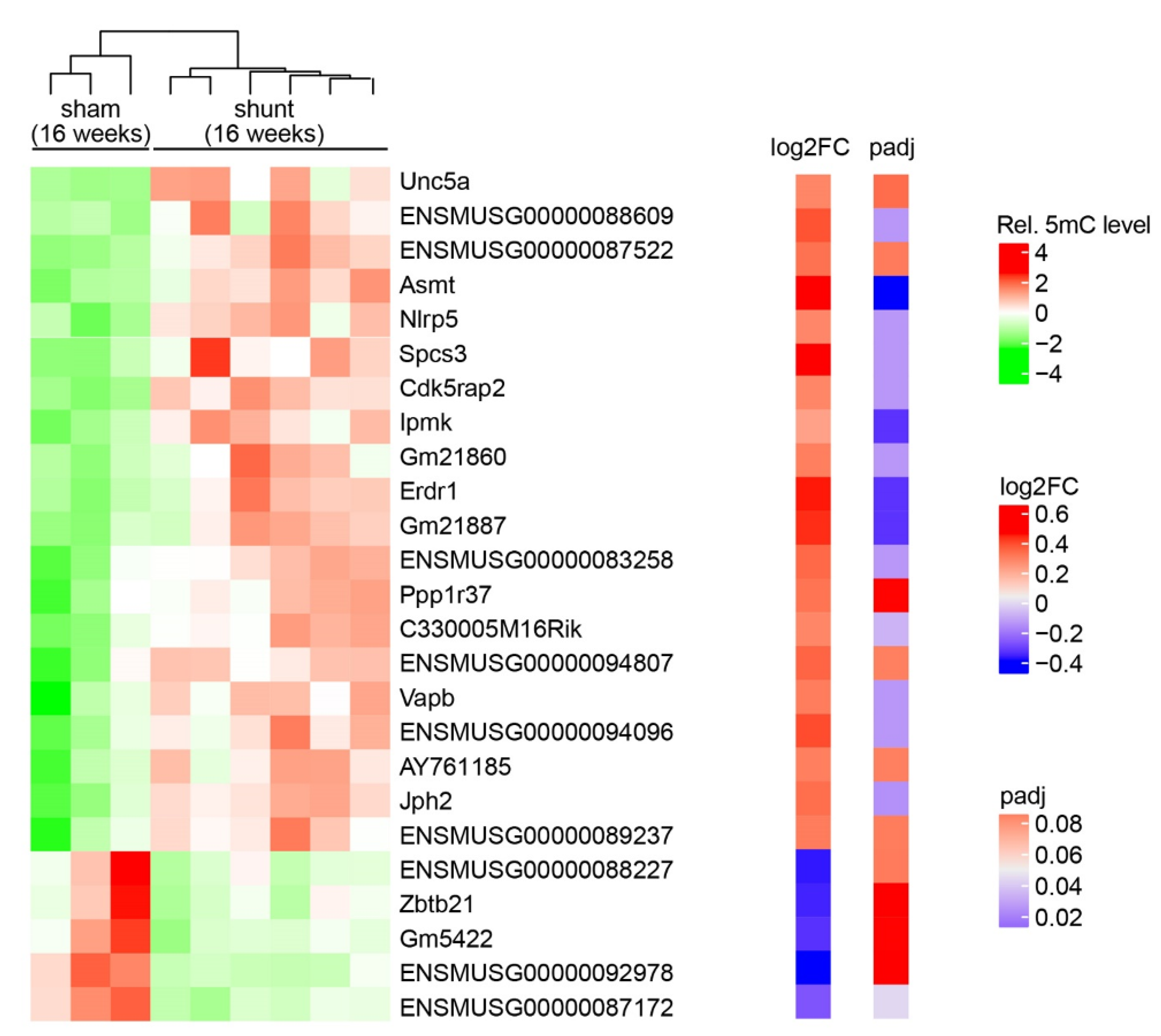
2.2. Landscape of DNA Methylation in Volume-Overloaded Dilated LV
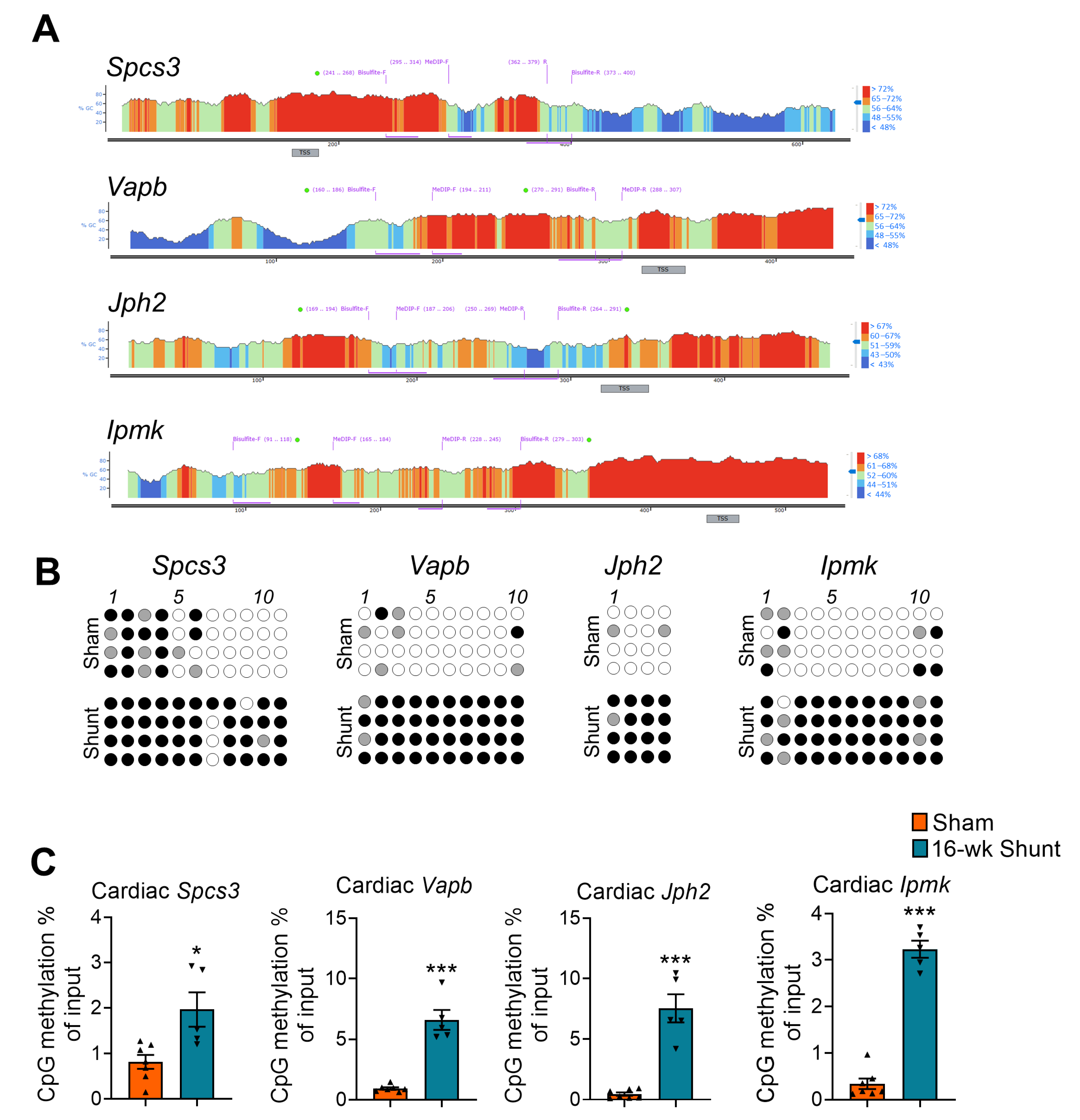
2.3. Validation of Promoter Methylation
2.4. Conserved Methylation Pattern in Circulating DNA
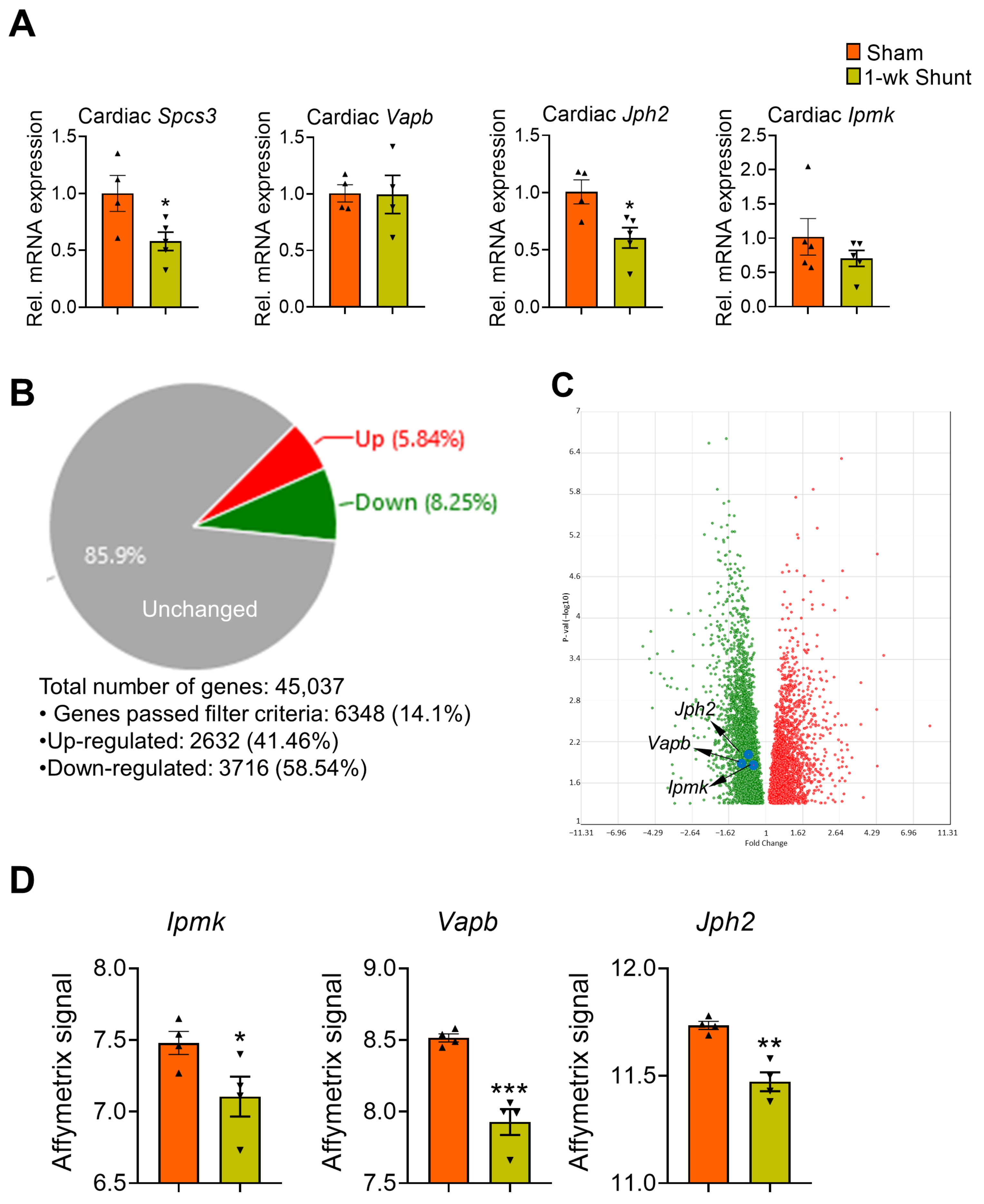
2.5. Association of Loci-Specific Differential Methylation with the Corresponding Gene Expression
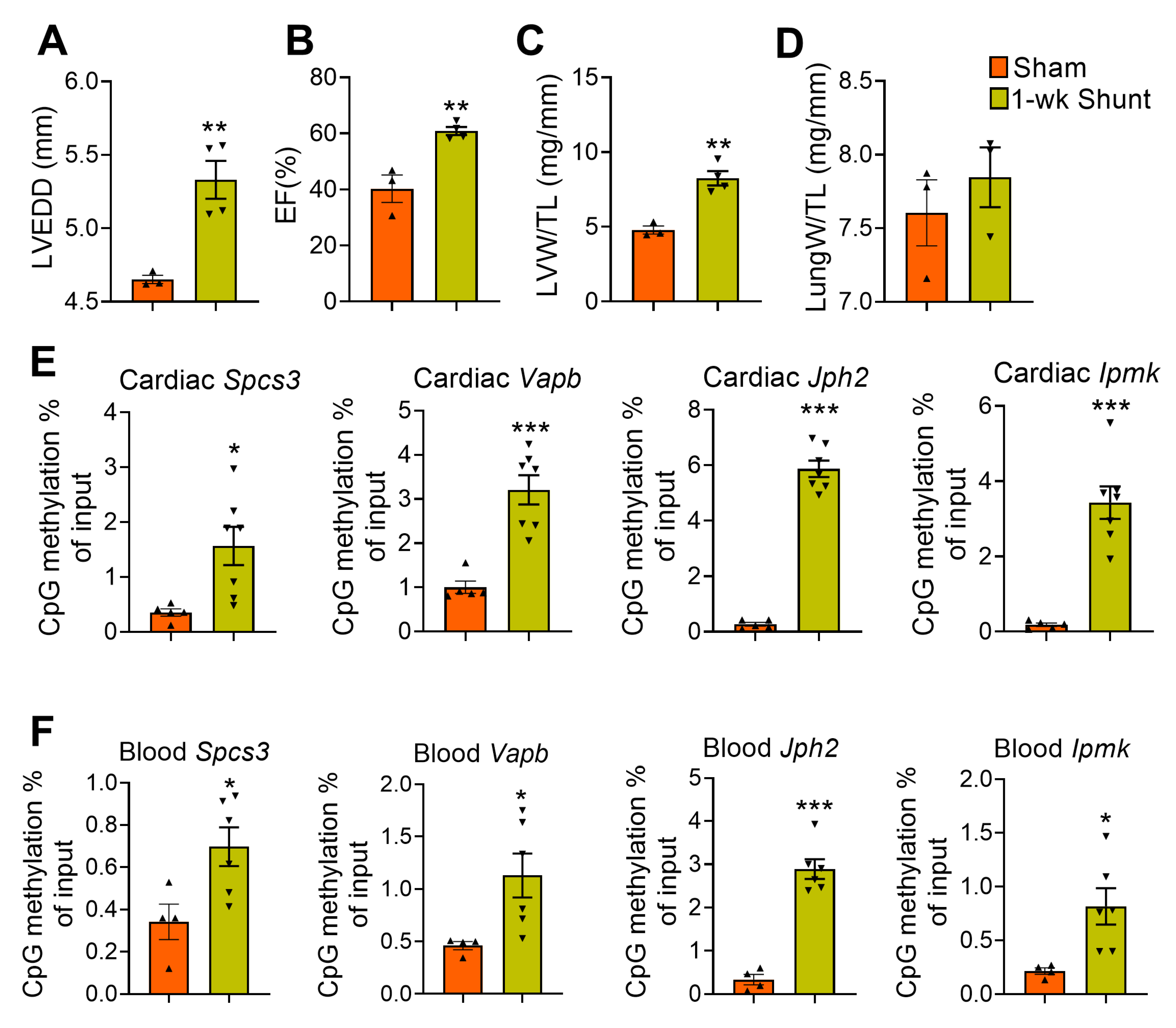
2.6. Similar Methylation Changes Occurred in Dilated LV Early after Shunt before Functional Deterioration
2.7. No Major Changes in the Expression Levels of the DNA Methylation-Modifying Enzymes in Volume-Overloaded Dilated Remodeled LV
3. Discussion
Study Limitations
4. Materials and Methods
4.1. Aortocaval Shunt
4.2. Genomic DNA Isolation
4.3. Reduced Representation Bisulfite Sequencing (RRBS)
4.4. Data Processing and Mapping to the Mouse Genome
4.5. Methylated DNA Immunoprecipitation (MeDIP)-Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
4.6. Bisulfite Sequencing
4.7. Microarray Data Resources
4.8. Quantitative RT-PCR
4.9. Echocardiography
4.10. Histology
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 5mC | 5-methylcytosine |
| Asmt | acetylserotonin O-methyltransferase |
| Cdk5rap2 | CDK5 regulatory subunit-associated protein 2 |
| CpG | Cytosine–guanine dinucleotides |
| DMRs | differentially methylated regions |
| DNMTs | DNA methyltransferases |
| ER | endoplasmic reticulum |
| Erdr1 | erythroid differentiation regulator 1 |
| HF | heart failure |
| Ipmk | inositol polyphosphate multikinase |
| Jph2 | junctophilin-2 |
| LV | left ventricle |
| MeDIP | methylated DNA immunoprecipitation |
| MI | myocardial infarction |
| Nlrp5 | NLR family, pyrin domain containing 5 |
| PO | pressure overload |
| qRT-PCR | quantitative real-time polymerase chain reaction |
| RRBS | reduced representation bisulfite sequencing |
| Spcs3 | signal peptidase complex subunit 3 |
| TET | ten-eleven translocation methylcytosine dioxygenases |
| Vapb | vesicle-associated membrane protein-associated protein B |
| VO | volume overload |
| WGA | wheat germ agglutinin |
| WT | wild type |
References
- Cohn, J.N.; Ferrari, R.; Sharpe, N. Cardiac remodeling—Concepts and clinical implications: A consensus paper from an international forum on cardiac remodeling. Behalf of an International Forum on Cardiac Remodeling. J. Am. Coll. Cardiol. 2000, 35, 569–582. [Google Scholar] [CrossRef] [PubMed]
- Jacob, R.; Gülch, R.W. Functional significance of ventricular dilatation: Reconsideration of Linzbach’s concept of chronic heart failure. Basic Res. Cardiol. 1988, 83, 461–475. [Google Scholar] [CrossRef] [PubMed]
- Vasan, R.S.; Larson, M.G.; Benjamin, E.J.; Evans, J.C.; Levy, D. Left ventricular dilatation and the risk of congestive heart failure in people without myocardial infarction. N. Engl. J. Med. 1997, 336, 1350–1355. [Google Scholar] [CrossRef] [PubMed]
- Haas, G.J.; McCune, S.A.; Brown, D.M.; Cody, R.J. Echocardiographic characterization of left ventricular adaptation in a genetically determined heart failure rat model. Am. Heart J. 1995, 130, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, K.; Reinier, K.; Teodorescu, C.; Uy-Evanado, A.; Aleong, R.; Chugh, H.; Nichols, G.A.; Gunson, K.; London, B.; Jui, J.; et al. Left ventricular diameter and risk stratification for sudden cardiac death. J. Am. Heart Assoc. 2014, 3, e001193. [Google Scholar] [CrossRef] [PubMed]
- Aleong, R.G.; Mulvahill, M.J.; Halder, I.; Carlson, N.E.; Singh, M.; Bloom, H.L.; Dudley, S.C.; Ellinor, P.T.; Shalaby, A.; Weiss, R.; et al. Left Ventricular Dilatation Increases the Risk of Ventricular Arrhythmias in Patients With Reduced Systolic Function. J. Am. Heart Assoc. 2015, 4, e001566. [Google Scholar] [CrossRef]
- Greene, S.J.; Fonarow, G.C.; Butler, J. Risk profiles in heart failure: Baseline, residual, worsening, and advanced heart failure risk. Circ. Heart Fail. 2020, 13, e007132. [Google Scholar] [CrossRef]
- Cohn, J.N. Structural basis of heart failure: Ventricular remodeling and its pharmacologic inhibition. Circulation 1995, 91, 2504–2507. [Google Scholar] [CrossRef]
- Tan, F.L.; Moravec, C.S.; Li, J.; Apperson-Hansen, C.; McCarthy, P.M.; Young, J.B.; Bond, M. The gene expression fingerprint of human heart failure. Proc. Natl. Acad. Sci. USA 2002, 99, 11387–11392. [Google Scholar] [CrossRef]
- Russell-Hallinan, A.; Watson, C.J.; Baugh, J.A. Epigenetics of Aberrant Cardiac Wound Healing. Compr. Physiol. 2018, 8, 451–491. [Google Scholar]
- Feil, R.; Fraga, M.F. Epigenetics and the environment: Emerging patterns and implications. Nat. Rev. Genet. 2012, 13, 97–109. [Google Scholar] [CrossRef]
- Smith, Z.D.; Meissner, A. DNA methylation: Roles in mammalian development. Nat. Rev. Genet. 2013, 14, 204–220. [Google Scholar] [CrossRef] [PubMed]
- Hermann, A.; Goyal, R.; Jeltsch, A. The Dnmt1 DNA-(cytosine-C5)-methyltransferase methylates DNA processively with high preference for hemimethylated target sites. J. Biol. Chem. 2004, 279, 48350–48359. [Google Scholar] [CrossRef] [PubMed]
- Okano, M.; Bell, D.W.; Haber, D.A.; Li, E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 1999, 99, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Ito, S.; D’Alessio, A.C.; Taranova, O.V.; Hong, K.; Sowers, L.C.; Zhang, Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 2010, 466, 1129–1133. [Google Scholar] [CrossRef]
- Tahiliani, M.; Koh, K.P.; Shen, Y.; Pastor, W.A.; Bandukwala, H.; Brudno, Y.; Agarwal, S.; Iyer, L.M.; Liu, D.R.; Aravind, L.; et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 2009, 324, 930–935. [Google Scholar] [CrossRef]
- Domcke, S.; Bardet, A.F.; Adrian Ginno, P.; Hartl, D.; Burger, L.; Schübeler, D. Competition between DNA methylation and transcription factors determines binding of NRF1. Nature 2015, 528, 575–579. [Google Scholar] [CrossRef]
- Schübeler, D. Function and information content of DNA methylation. Nature 2015, 517, 321–326. [Google Scholar] [CrossRef]
- Zhu, H.; Wang, G.; Qian, J. Transcription factors as readers and effectors of DNA methylation. Nat. Rev. Genet. 2016, 17, 551–565. [Google Scholar] [CrossRef]
- Movassagh, M.; Vujic, A.; Foo, R. Genome-wide DNA methylation in human heart failure. Epigenomics 2011, 3, 103–109. [Google Scholar] [CrossRef]
- Haas, J.; Frese, K.S.; Park, Y.J.; Keller, A.; Vogel, B.; Lindroth, A.M.; Weichenhan, D.; Franke, J.; Fischer, S.; Bauer, A.; et al. Alterations in cardiac DNA methylation in human dilated cardiomyopathy. EMBO Mol. Med. 2013, 5, 413–429. [Google Scholar] [CrossRef] [PubMed]
- Meder, B.; Haas, J.; Sedaghat-Hamedani, F.; Kayvanpour, E.; Frese, K.; Lai, A.; Nietsch, R.; Scheiner, C.; Mester, S.; Bordalo, D.M.; et al. Epigenome-Wide Association Study Identifies Cardiac Gene Patterning and a Novel Class of Biomarkers for Heart Failure. Circulation 2017, 136, 1528–1544. [Google Scholar] [CrossRef] [PubMed]
- Nakatochi, M.; Ichihara, S.; Yamamoto, K.; Naruse, K.; Yokota, S.; Asano, H.; Matsubara, T.; Yokota, M. Epigenome-wide association of myocardial infarction with DNA methylation sites at loci related to cardiovascular disease. Clin. Epigenetics 2017, 9, 54. [Google Scholar] [CrossRef]
- Gilsbach, R.; Schwaderer, M.; Preissl, S.; Grüning, B.A.; Kranzhöfer, D.; Schneider, P.; Nührenberg, T.G.; Mulero-Navarro, S.; Weichenhan, D.; Braun, C.; et al. Distinct epigenetic programs regulate cardiac myocyte development and disease in the human heart in vivo. Nat. Commun. 2018, 9, 391. [Google Scholar] [CrossRef] [PubMed]
- Glezeva, N.; Moran, B.; Collier, P.; Moravec, C.S.; Phelan, D.; Donnellan, E.; Russell-Hallinan, A.; O’Connor, D.P.; Gallagher, W.M.; Gallagher, J.; et al. Targeted DNA Methylation Profiling of Human Cardiac Tissue Reveals Novel Epigenetic Traits and Gene Deregulation Across Different Heart Failure Patient Subtypes. Circ. Heart Fail. 2019, 12, e005765. [Google Scholar] [CrossRef] [PubMed]
- Pepin, M.E.; Ha, C.M.; Crossman, D.K.; Litovsky, S.H.; Varambally, S.; Barchue, J.P.; Pamboukian, S.V.; Diakos, N.A.; Drakos, S.G.; Pogwizd, S.M.; et al. Genome-wide DNA methylation encodes cardiac transcriptional reprogramming in human ischemic heart failure. Lab. Investig. 2019, 99, 371–386. [Google Scholar] [CrossRef] [PubMed]
- Gilsbach, R.; Preissl, S.; Grüning, B.A.; Schnick, T.; Burger, L.; Benes, V.; Würch, A.; Bönisch, U.; Günther, S.; Backofen, R.; et al. Dynamic DNA methylation orchestrates cardiomyocyte development, maturation and disease. Nat. Commun. 2014, 5, 5288. [Google Scholar] [CrossRef]
- Watson, C.J.; Horgan, S.; Neary, R.; Glezeva, N.; Tea, I.; Corrigan, N.; McDonald, K.; Ledwidge, M.; Baugh, J. Epigenetic Therapy for the Treatment of Hypertension-Induced Cardiac Hypertrophy and Fibrosis. J. Cardiovasc. Pharmacol. Ther. 2016, 21, 127–137. [Google Scholar] [CrossRef]
- Stenzig, J.; Schneeberger, Y.; Löser, A.; Peters, B.S.; Schaefer, A.; Zhao, R.R.; Ng, S.L.; Höppner, G.; Geertz, B.; Hirt, M.N.; et al. Pharmacological inhibition of DNA methylation attenuates pressure overload-induced cardiac hypertrophy in rats. J. Mol. Cell Cardiol. 2018, 120, 53–63. [Google Scholar] [CrossRef]
- Russell-Hallinan, A.; Neary, R.; Watson, C.J.; Baugh, J.A. Repurposing From Oncology to Cardiology: Low-Dose 5-Azacytidine Attenuates Pathological Cardiac Remodeling in Response to Pressure Overload Injury. J. Cardiovasc. Pharmacol. Ther. 2021, 26, 375–385. [Google Scholar] [CrossRef]
- Luo, X.; Hu, Y.; Shen, J.; Liu, X.; Wang, T.; Li, L.; Li, J. Integrative analysis of DNA methylation and gene expression reveals key molecular signatures in acute myocardial infarction. Clin. Epigenetics 2022, 14, 46. [Google Scholar] [CrossRef] [PubMed]
- Kehat, I.; Molkentin, J.D. Molecular pathways underlying cardiac remodeling during pathophysiological stimulation. Circulation 2010, 122, 2727–2735. [Google Scholar] [CrossRef] [PubMed]
- You, J.; Wu, J.; Zhang, Q.; Ye, Y.; Wang, S.; Huang, J.; Liu, H.; Wang, X.; Zhang, W.; Bu, L.; et al. Differential cardiac hypertrophy and signaling pathways in pressure versus volume overload. Am. J. Physiol. Heart Circ. Physiol. 2018, 314, H552–H562. [Google Scholar] [CrossRef]
- Pitoulis, F.G.; Terracciano, C.M. Heart Plasticity in Response to Pressure- and Volume-Overload: A Review of Findings in Compensated and Decompensated Phenotypes. Front. Physiol. 2020, 11, 92. [Google Scholar] [CrossRef] [PubMed]
- Toischer, K.; Rokita, A.G.; Unsöld, B.; Zhu, W.; Kararigas, G.; Sossalla, S.; Reuter, S.P.; Becker, A.; Teucher, N.; Seidler, T.; et al. Differential cardiac remodeling in preload versus afterload. Circulation 2010, 122, 993–1003. [Google Scholar] [CrossRef]
- Mohamed, B.A.; Schnelle, M.; Khadjeh, S.; Lbik, D.; Herwig, M.; Linke, W.A.; Hasenfuss, G.; Toischer, K. Molecular and structural transition mechanisms in long-term volume overload. Eur. J. Heart Fail. 2016, 18, 362–371. [Google Scholar] [CrossRef]
- Zhang, C.; Chen, B.; Guo, A.; Zhu, Y.; Miller, J.D.; Gao, S.; Yuan, C.; Kutschke, W.; Zimmerman, K.; Weiss, R.M.; et al. Microtubule-mediated defects in junctophilin-2 trafficking contribute to myocyte transverse-tubule remodeling and Ca2+ handling dysfunction in heart failure. Circulation 2014, 129, 1742–1750. [Google Scholar] [CrossRef]
- Poulet, C.; Sanchez-Alonso, J.; Swiatlowska, P.; Mouy, F.; Lucarelli, C.; Alvarez-Laviada, A.; Gross, P.; Terracciano, C.; Houser, S.; Gorelik, J. Junctophilin-2 tethers T-tubules and recruits functional L-type calcium channels to lipid rafts in adult cardiomyocytes. Cardiovasc. Res. 2021, 117, 149–161. [Google Scholar] [CrossRef]
- Guo, A.; Zhang, X.; Iyer, V.R.; Chen, B.; Zhang, C.; Kutschke, W.J.; Weiss, R.M.; Franzini-Armstrong, C.; Song, L.S. Overexpression of junctophilin-2 does not enhance baseline function but attenuates heart failure development after cardiac stress. Proc. Natl. Acad. Sci. USA 2014, 111, 12240–12245. [Google Scholar] [CrossRef]
- Wie, S.; Guo, A.; Chen, B.; Kutschke, W.; Xie, Y.P.; Zimmerman, K.; Weiss, R.M.; Anderson, M.E.; Cheng, H.; Song, L.S. T-tubule remodeling during transition from hypertrophy to heart failure. Circ. Res. 2010, 107, 520–531. [Google Scholar]
- Wagner, E.; Lauterbach, M.A.; Kohl, T.; Westphal, V.; Williams, G.S.; Steinbrecher, J.H.; Streich, J.H.; Korff, B.; Tuan, H.T.; Hagen, B.; et al. Stimulated emission depletion live-cell super-resolution imaging shows proliferative remodeling of T-tubule membrane structures after myocardial infarction. Circ. Res. 2012, 111, 402–414. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.D.; Xu, M.; Li, R.C.; Guo, L.; Lai, Y.S.; Xu, S.M.; Li, S.F.; Lu, Q.L.; Li, L.L.; Zhang, H.B.; et al. Ultrastructural remodelling of Ca2+ signalling apparatus in failing heart cells. Cardiovasc. Res. 2012, 95, 430–438. [Google Scholar] [CrossRef] [PubMed]
- Landstrom, A.P.; Kellen, C.A.; Dixit, S.S.; van Oort, R.J.; Garbino, A.; Weisleder, N.; Ma, J.; Wehrens, X.H.; Ackerman, M.J. Junctophilin-2 expression silencing causes cardiocyte hypertrophy and abnormal intracellular calcium-handling. Circ. Heart Fail. 2011, 4, 214–223. [Google Scholar] [CrossRef] [PubMed]
- Minamisawa, S.; Oshikawa, J.; Takeshima, H.; Hoshijima, M.; Wang, Y.; Chien, K.R.; Ishikawa, Y.; Matsuoka, R. Junctophilin type 2 is associated with caveolin-3 and is down-regulated in the hypertrophic and dilated cardiomyopathies. Biochem. Biophys. Res. Commun. 2004, 325, 852–856. [Google Scholar] [CrossRef] [PubMed]
- Van Oort, R.J.; Garbino, A.; Wang, W.; Dixit, S.S.; Landstrom, A.P.; Gaur, N.; De Almeida, A.C.; Skapura, D.G.; Rudy, Y.; Burns, A.R.; et al. Disrupted junctional membrane complexes and hyperactive ryanodine receptors after acute junctophilin knockdown in mice. Circulation 2011, 123, 979–988. [Google Scholar] [CrossRef]
- Landstrom, A.P.; Weisleder, N.; Batalden, K.B.; Bos, J.M.; Tester, D.J.; Ommen, S.R.; Wehrens, X.H.; Claycomb, W.C.; Ko, J.K.; Hwang, M.; et al. Mutations in JPH2-encoded junctophilin-2 associated with hypertrophic cardiomyopathy in humans. J. Mol. Cell. Cardiol. 2007, 42, 1026–1035. [Google Scholar] [CrossRef]
- Beavers, D.L.; Wang, W.; Ather, S.; Voigt, N.; Garbino, A.; Dixit, S.S.; Landstrom, A.P.; Li, N.; Wang, Q.; Olivotto, I.; et al. Mutation E169K in junctophilin-2 causes atrial fibrillation due to impaired RyR2 stabilization. J. Am. Coll. Cardiol. 2013, 62, 2010–2019. [Google Scholar] [CrossRef]
- Lee, B.; Park, S.J.; Hong, S.; Kim, K.; Kim, S. Inositol Polyphosphate Multikinase Signaling: Multifaceted Functions in Health and Disease. Mol. Cells 2021, 44, 187–194. [Google Scholar] [CrossRef]
- Maag, D.; Maxwell, M.J.; Hardesty, D.A.; Boucher, K.L.; Choudhari, N.; Hanno, A.G.; Ma, J.F.; Snowman, A.S.; Pietropaoli, J.W.; Xu, R.; et al. Inositol polyphosphate multikinase is a physiologic PI3-kinase that activates Akt/PKB. Proc. Natl. Acad. Sci. USA 2011, 108, 1391–1396. [Google Scholar] [CrossRef]
- Resnick, A.C.; Snowman, A.M.; Kang, B.N.; Hurt, K.J.; Snyder, S.H.; Saiardi, A. Inositol polyphosphate multikinase is a nuclear PI3-kinase with transcriptional regulatory activity. Proc. Natl. Acad. Sci. USA 2005, 102, 12783–12788. [Google Scholar] [CrossRef]
- Stoica, R.; De Vos, K.J.; Paillusson, S.; Mueller, S.; Sancho, R.M.; Lau, K.F.; Vizcay-Barrena, G.; Lin, W.L.; Xu, Y.F.; Lewis, J.; et al. ER-mitochondria associations are regulated by the VAPB-PTPIP51 interaction and are disrupted by ALS/FTD-associated TDP-43. Nat. Commun. 2014, 5, 3996. [Google Scholar] [CrossRef] [PubMed]
- Silbernagel, N.; Walecki, M.; Schäfer, M.K.; Kessler, M.; Zobeiri, M.; Rinné, S.; Kiper, A.K.; Komadowski, M.A.; Vowinkel, K.S.; Wemhöner, K.; et al. The VAMP-associated protein VAPB is required for cardiac and neuronal pacemaker channel function. FASEB J. 2018, 32, 6159–6173. [Google Scholar] [CrossRef] [PubMed]
- Zanotti, A.; Coelho, J.P.L.; Kaylani, D.; Singh, G.; Tauber, M.; Hitzenberger, M.; Avci, D.; Zacharias, M.; Russell, R.B.; Lemberg, M.K.; et al. The human signal peptidase complex acts as a quality control enzyme for membrane proteins. Science 2022, 378, 996–1000. [Google Scholar] [CrossRef]
- Wang, Y.; Huang, Y.; Zhang, M.; Zhang, X.; Tang, X.; Kang, Y. Bioinformatic Analysis of the Possible Regulative Network of miR-30a/e in Cardiomyocytes 2 Days Post Myocardial Infarction. Acta Cardiol. Sin. 2018, 34, 175–188. [Google Scholar]
- Sheng, X.; Jin, X.; Liu, Y.; Fan, T.; Zhu, Z.; Jin, J.; Zheng, G.; Chen, Z.; Lu, M.; Wang, Z. The Bioinformatical Identification of Potential Biomarkers in Heart Failure Diagnosis and Treatment. Genet. Res. 2022, 2022, 8727566. [Google Scholar] [CrossRef] [PubMed]
- Dujardin, K.S.; Enriquez-Sarano, M.; Schaff, H.V.; Bailey, K.R.; Seward, J.B.; Tajik, A.J. Mortality and morbidity of aortic regurgitation in clinical practice. A long-term follow-up study. Circulation 1999, 99, 1851–1857. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, R.A.; Otto, C.M.; Bonow, R.O.; Carabello, B.A.; Erwin, J.P., III; Fleisher, L.A.; Jneid, H.; Mack, M.J.; McLeod, C.J.; O’Gara, P.T.; et al. 2017 AHA/ACC Focused Update of the 2014 AHA/ACC Guideline for the Management of Patients With Valvular Heart Disease: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. J. Am. Coll. Cardiol. 2017, 70, 252–289. [Google Scholar] [CrossRef]
- Zemmour, H.; Planer, D.; Magenheim, J.; Moss, J.; Neiman, D.; Gilon, D.; Korach, A.; Glaser, B.; Shemer, R.; Landesberg, G. Non-invasive detection of human cardiomyocyte death using methylation patterns of circulating DNA. Nat. Commun. 2018, 9, 1443. [Google Scholar] [CrossRef]
- Stroun, M.; Maurice, P.; Vasioukhin, V.; Lyautey, J.; Lederrey, C.; Lefort, F.; Rossier, A.; Chen, X.Q.; Anker, P. The origin and mechanism of circulating DNA. Ann. N. Y. Acad. Sci. 2000, 906, 161–168. [Google Scholar] [CrossRef]
- Ren, J.; Jiang, L.; Liu, X.; Liao, Y.; Zhao, X.; Tang, F.; Yu, H.; Shao, Y.; Wang, J.; Wen, L.; et al. Heart-specific DNA methylation analysis in plasma for the investigation of myocardial damage. J. Transl. Med. 2022, 20, 36. [Google Scholar] [CrossRef]
- Nassar, F.J.; Msheik, Z.S.; Nasr, R.R.; Temraz, S.N. Methylated circulating tumor DNA as a biomarker for colorectal cancer diagnosis, prognosis, and prediction. Clin. Epigenetics 2021, 13, 111. [Google Scholar] [CrossRef] [PubMed]
- Amatu, A.; Schirripa, M.; Tosi, F.; Lonardi, S.; Bencardino, K.; Bonazzina, E.; Palmeri, L.; Patanè, D.A.; Pizzutilo, E.G.; Mussolin, B.; et al. High Circulating Methylated DNA Is a Negative Predictive and Prognostic Marker in Metastatic Colorectal Cancer Patients Treated With Regorafenib. Front. Oncol 2019, 9, 622. [Google Scholar] [CrossRef]
- Yang, X.; Wen, X.; Guo, Q.; Zhang, Y.; Liang, Z.; Wu, Q.; Li, Z.; Ruan, W.; Ye, Z.; Wang, H.; et al. Predicting disease-free survival in colorectal cancer by circulating tumor DNA methylation markers. Clin. Epigenetics 2022, 14, 160. [Google Scholar] [CrossRef]
- Fernández-Sanlés, A.; Sayols-Baixeras, S.; Subirana, I.; Sentí, M.; Pérez-Fernández, S.; de Castro Moura, M.; Esteller, M.; Marrugat, J.; Elosua, R. DNA methylation biomarkers of myocardial infarction and cardiovascular disease. Clin. Epigenetics 2021, 13, 86. [Google Scholar] [CrossRef] [PubMed]
- Konki, M.; Lindgren, N.; Kyläniemi, M.; Venho, R.; Laajala, E.; Ghimire, B.; Lahesmaa, R.; Kaprio, J.; Rinne, J.O.; Lund, R.J. Plasma cell-free DNA methylation marks for episodic memory impairment: A pilot twin study. Sci. Rep. 2020, 10, 14192. [Google Scholar] [CrossRef] [PubMed]
- Dawlaty, M.M.; Breiling, A.; Le, T.; Raddatz, G.; Barrasa, M.I.; Cheng, A.W.; Gao, Q.; Powell, B.E.; Li, Z.; Xu, M.; et al. Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Dev. Cell 2013, 24, 310–323. [Google Scholar] [CrossRef]
- Cardoso-Júnior, C.A.M.; Yagound, B.; Ronai, I.; Remnant, E.J.; Hartfelder, K.; Oldroyd, B.P. DNA methylation is not a driver of gene expression reprogramming in young honey bee workers. Mol. Ecol. 2021, 30, 4804–4818. [Google Scholar] [CrossRef]
- Joshi, K.; Liu, S.; Breslin, S.J.P.; Zhang, J. Mechanisms that regulate the activities of TET proteins. Cell. Mol. Life Sci. 2022, 79, 363. [Google Scholar] [CrossRef]
- Trichon, B.H.; Felker, G.M.; Shaw, L.K.; Cabell, C.H.; O’Connor, C.M. Relation of frequency and severity of mitral regurgitation to survival among patients with left ventricular systolic dysfunction and heart failure. Am. J. Cardiol. 2003, 91, 538–543. [Google Scholar] [CrossRef]
- Houser, S.R.; Margulies, K.B.; Murphy, A.M.; Spinale, F.G.; Francis, G.S.; Prabhu, S.D.; Rockman, H.A.; Kass, D.A.; Molkentin, J.D.; Sussman, M.A.; et al. Animal models of heart failure: A scientific statement from the American Heart Association. Circ. Res. 2012, 111, 131–150. [Google Scholar] [CrossRef]
- Meissner, A.; Gnirke, A.; Bell, G.W.; Ramsahoye, B.; Lander, E.S.; Jaenisch, R. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005, 33, 5868–5877. [Google Scholar] [CrossRef]
- Seiler Vellame, D.; Castanho, I.; Dahir, A.; Mill, J.; Hannon, E. Characterizing the properties of bisulfite sequencing data: Maximizing power and sensitivity to identify between-group differences in DNA methylation. BMC Genom. 2021, 22, 446. [Google Scholar] [CrossRef] [PubMed]
- Krueger, F.; Andrews, S.R. Bismark: A flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 2011, 27, 1571–1572. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Redondo, I.; Planells, B.; Cánovas, S.; Ivanova, E.; Kelsey, G.; Gutiérrez-Adán, A. Genome-wide DNA methylation dynamics during epigenetic reprogramming in the porcine germline. Clin. Epigenetics 2021, 13, 27. [Google Scholar] [CrossRef] [PubMed]
- Zacchigna, S.; Paldino, A.; Falcão-Pires, I.; Daskalopoulos, E.P.; Dal Ferro, M.; Vodret, S.; Lesizza, P.; Cannatà, A.; Miranda-Silva, D.; Lourenço, A.P.; et al. function in adult rodents: A position paper of the ESC Working Group on Myocardial Function. Cardiovasc. Res. 2021, 117, 43–59. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, X.; Elkenani, M.; Tan, X.; Hain, J.k.; Cui, B.; Schnelle, M.; Hasenfuss, G.; Toischer, K.; Mohamed, B.A. DNA Methylation Analysis Identifies Novel Epigenetic Loci in Dilated Murine Heart upon Exposure to Volume Overload. Int. J. Mol. Sci. 2023, 24, 5885. https://doi.org/10.3390/ijms24065885
Xu X, Elkenani M, Tan X, Hain Jk, Cui B, Schnelle M, Hasenfuss G, Toischer K, Mohamed BA. DNA Methylation Analysis Identifies Novel Epigenetic Loci in Dilated Murine Heart upon Exposure to Volume Overload. International Journal of Molecular Sciences. 2023; 24(6):5885. https://doi.org/10.3390/ijms24065885
Chicago/Turabian StyleXu, Xingbo, Manar Elkenani, Xiaoying Tan, Jara katharina Hain, Baolong Cui, Moritz Schnelle, Gerd Hasenfuss, Karl Toischer, and Belal A. Mohamed. 2023. "DNA Methylation Analysis Identifies Novel Epigenetic Loci in Dilated Murine Heart upon Exposure to Volume Overload" International Journal of Molecular Sciences 24, no. 6: 5885. https://doi.org/10.3390/ijms24065885
APA StyleXu, X., Elkenani, M., Tan, X., Hain, J. k., Cui, B., Schnelle, M., Hasenfuss, G., Toischer, K., & Mohamed, B. A. (2023). DNA Methylation Analysis Identifies Novel Epigenetic Loci in Dilated Murine Heart upon Exposure to Volume Overload. International Journal of Molecular Sciences, 24(6), 5885. https://doi.org/10.3390/ijms24065885





