Lacticaseibacillus rhamnosus ATCC 53103 and Limosilactobacillus reuteri ATCC 53608 Synergistically Boost Butyrate Levels upon Tributyrin Administration Ex Vivo
Abstract
1. Introduction
2. Results
2.1. Microbiota of Study Subjects Covered Interpersonal Differences in Gut Microbiota Composition
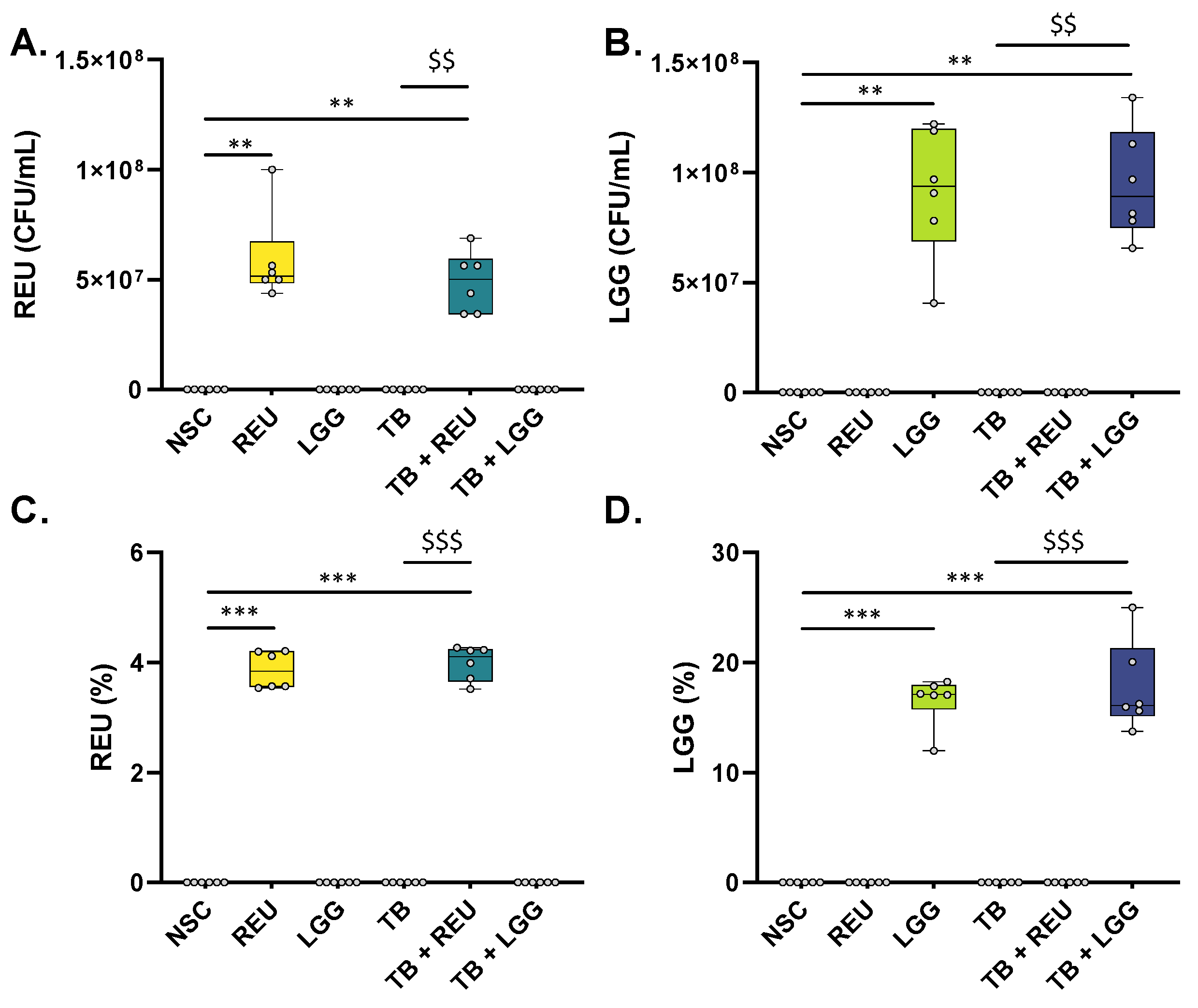
2.2. REU and LGG Remained Viable throughout the Entire Duration of the 48 h Ex Vivo Experiment
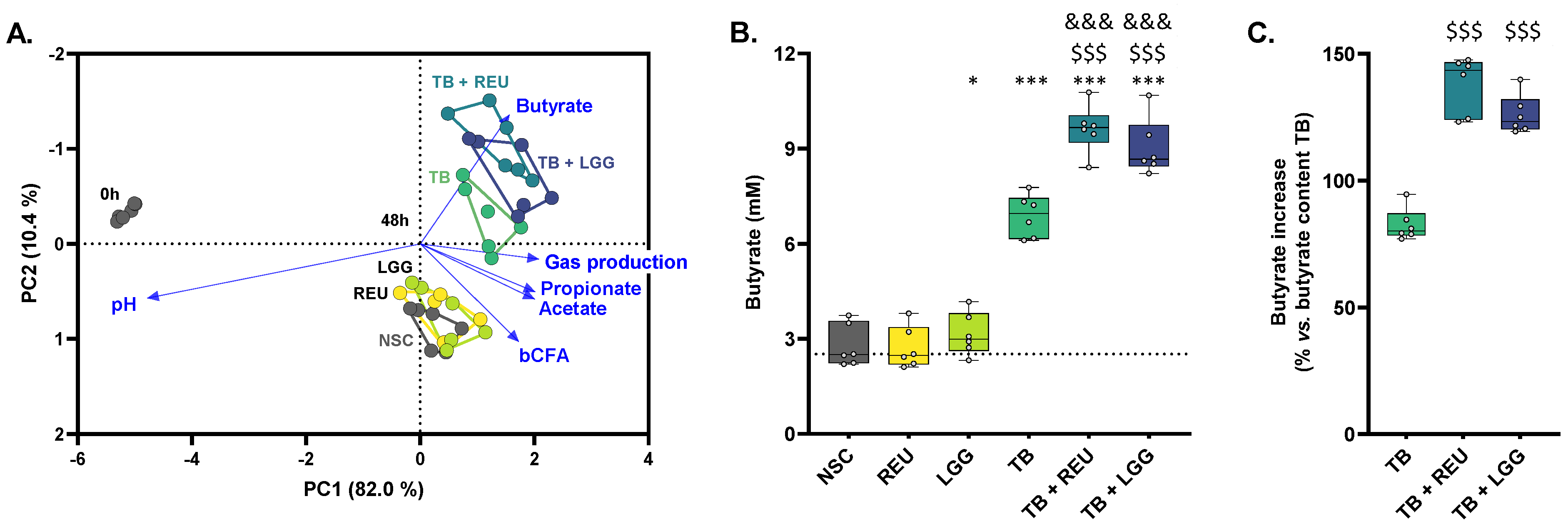
2.3. TB Increased Butyrate Levels, Which Were Further Enhanced upon Probiotic Co-Administration
2.4. The Combination of TB with REU Increased Microbial Diversity
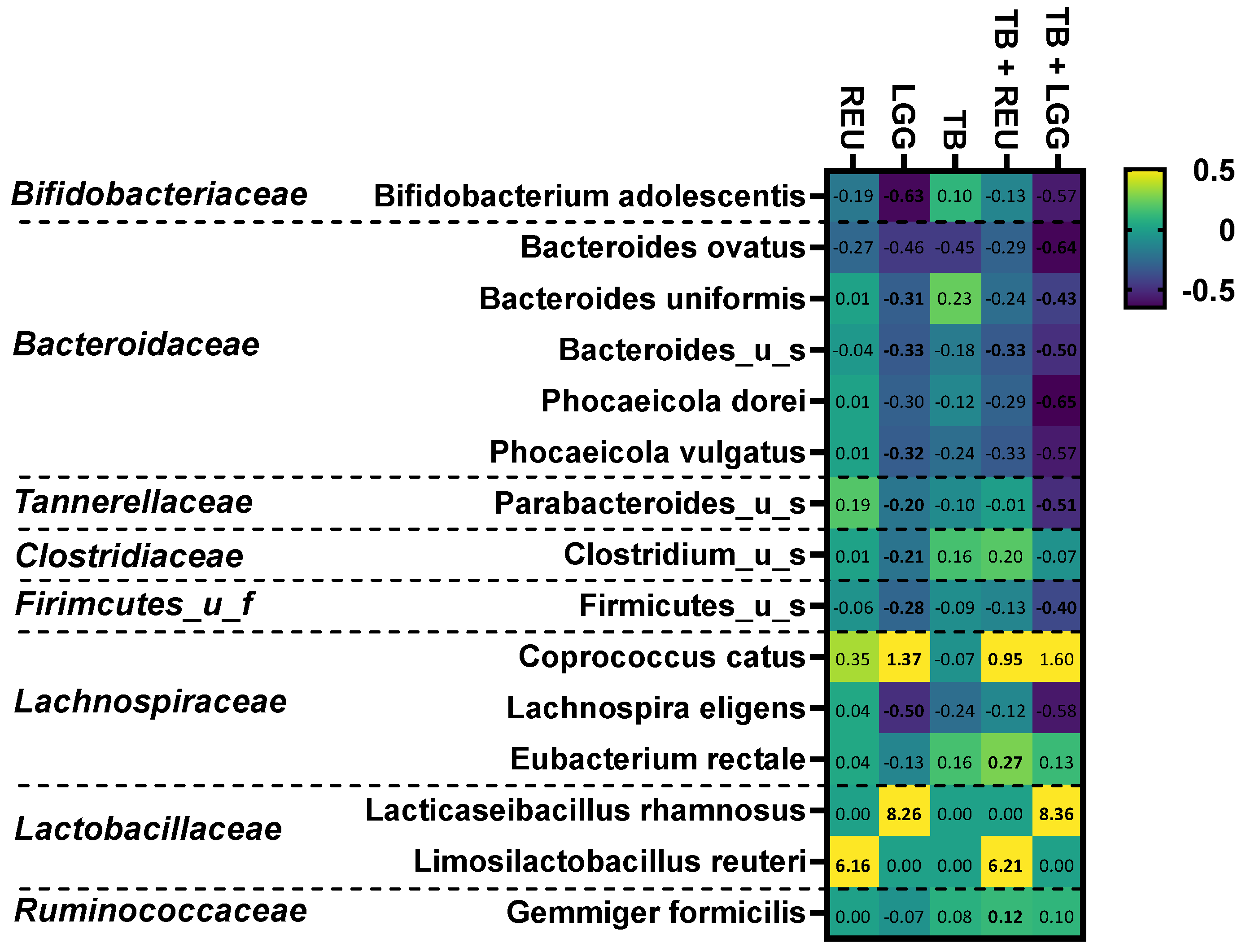
2.5. The Combination of TB with LGG and Especially REU Increased the Abundance of Specific Butyrate-Producing Species
3. Discussion
4. Materials and Methods
4.1. Test Products: TB, LGG, and REU
4.2. SIFR® Technology
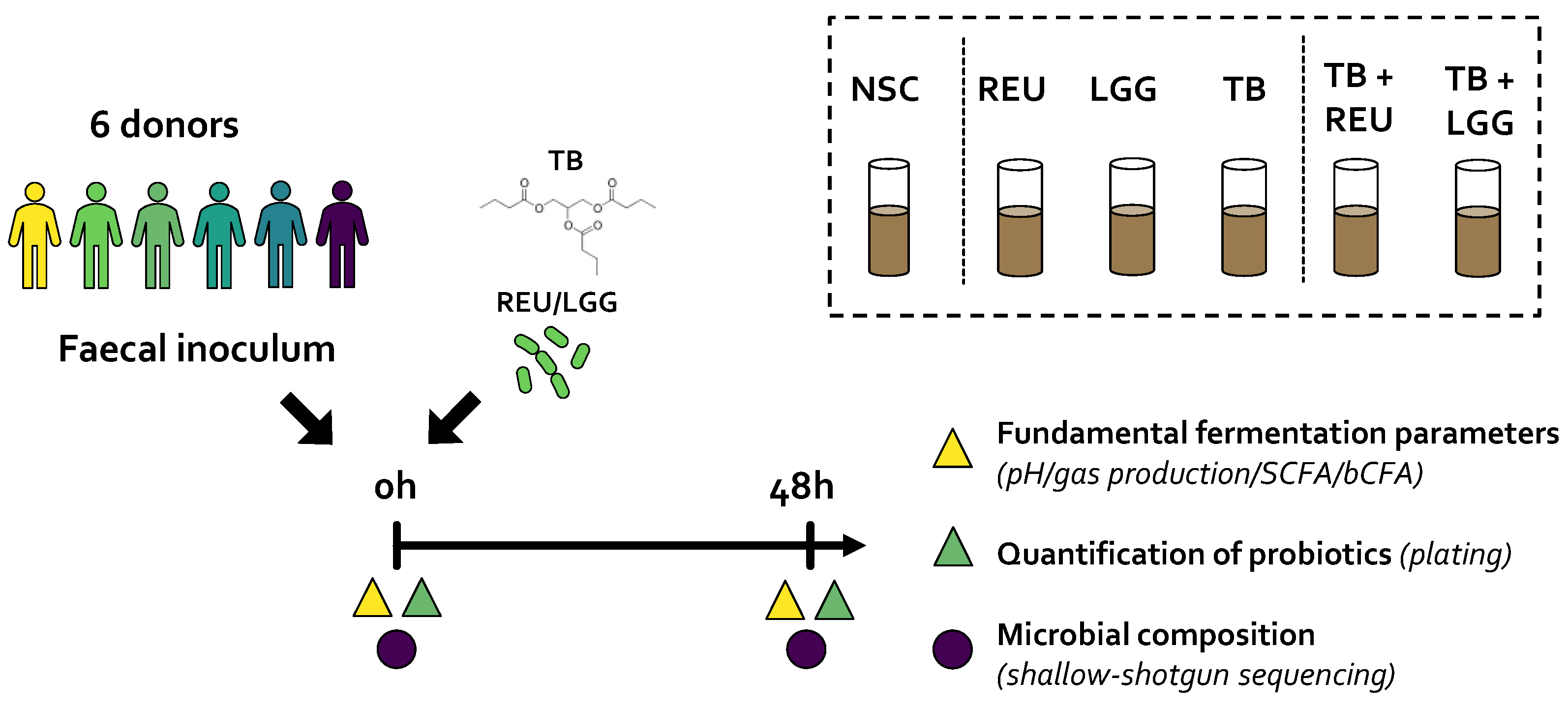
4.3. Study Design
4.4. Fundamental Fermentation Parameters
4.5. Selective Enumeration of Lactobacillaceae species (LGG and REU)
4.6. Microbiota Phylogenetic Analysis via Shallow Shotgun Sequencing
4.7. Data Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Alshehri, D.; Saadah, O.; Mosli, M.; Edris, S.; Alhindi, R.; Bahieldin, A. Dysbiosis of Gut Microbiota in Inflammatory Bowel Disease: Current Therapies and Potential for Microbiota-Modulating Therapeutic Approaches. Bosn. J. Basic Med. Sci. 2021, 21, 270. [Google Scholar] [CrossRef] [PubMed]
- Konturek, P.C.; Harsch, I.A.; Konturek, K.; Schink, M.; Konturek, T.; Neurath, M.F.; Zopf, Y. Gut–Liver Axis: How Do Gut Bacteria Influence the Liver? Med. Sci. 2018, 6, 79. [Google Scholar] [CrossRef] [PubMed]
- Nishiwaki, H.; Ito, M.; Ishida, T.; Hamaguchi, T.; Maeda, T.; Kashihara, K.; Tsuboi, Y.; Ueyama, J.; Shimamura, T.; Mori, H. Meta-analysis of Gut Dysbiosis in Parkinson’s Disease. Mov. Disord. 2020, 35, 1626–1635. [Google Scholar] [CrossRef] [PubMed]
- Serino, M.; Blasco-Baque, V.; Nicolas, S.; Burcelin, R. Far from the Eyes, Close to the Heart: Dysbiosis of Gut Microbiota and Cardiovascular Consequences. Curr. Cardiol. Rep. 2014, 16, 540. [Google Scholar] [CrossRef]
- Zhu, Q.; Gao, R.; Zhang, Y.; Pan, D.; Zhu, Y.; Zhang, X.; Yang, R.; Jiang, R.; Xu, Y.; Qin, H. Dysbiosis Signatures of Gut Microbiota in Coronary Artery Disease. Physiol. Genom. 2018, 50, 893–903. [Google Scholar] [CrossRef]
- Zou, S.; Fang, L.; Lee, M.-H. Dysbiosis of Gut Microbiota in Promoting the Development of Colorectal Cancer. Gastroenterol. Rep. 2018, 6, 1–12. [Google Scholar] [CrossRef]
- Koh, A.; De Vadder, F.; Kovatcheva-Datchary, P.; Bäckhed, F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 2016, 165, 1332–1345. [Google Scholar] [CrossRef]
- van der Hee, B.; Wells, J.M. Microbial Regulation of Host Physiology by Short-Chain Fatty Acids. Trends Microbiol. 2021, 29, 700–712. [Google Scholar] [CrossRef]
- Saleri, R.; Borghetti, P.; Ravanetti, F.; Cavalli, V.; Ferrari, L.; De Angelis, E.; Andrani, M.; Martelli, P. Effects of Different Short-Chain Fatty Acids (SCFA) on Gene Expression of Proteins Involved in Barrier Function in IPEC-J2. Porc. Health Manag. 2022, 8, 21. [Google Scholar] [CrossRef]
- Giromini, C.; Baldi, A.; Rebucci, R.; Lanzoni, D.; Policardi, M.; Sundaram, T.S.; Purup, S. Role of Short Chain Fatty Acids to Counteract Inflammatory Stress and Mucus Production in Human Intestinal HT29-MTX-E12 Cells. Foods 2022, 11, 1983. [Google Scholar] [CrossRef]
- Liu, H.; Wang, J.; He, T.; Becker, S.; Zhang, G.; Li, D.; Ma, X. Butyrate: A Double-Edged Sword for Health? Adv. Nutr. 2018, 9, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Guilloteau, P.; Martin, L.; Eeckhaut, V.; Ducatelle, R.; Zabielski, R.; Van Immerseel, F. From the Gut to the Peripheral Tissues: The Multiple Effects of Butyrate. Nutr. Res. Rev. 2010, 23, 366–384. [Google Scholar] [CrossRef] [PubMed]
- Canani, R.B.; Di Costanzo, M.; Leone, L.; Pedata, M.; Meli, R.; Calignano, A. Potential Beneficial Effects of Butyrate in Intestinal and Extraintestinal Diseases. World J. Gastroenterol. 2011, 17, 1519. [Google Scholar] [CrossRef]
- Peng, L.; Li, Z.-R.; Green, R.S.; Holzman, I.R.; Lin, J. Butyrate Enhances the Intestinal Barrier by Facilitating Tight Junction Assembly via Activation of AMP-Activated Protein Kinase in Caco-2 Cell Monolayers. J. Nutr. 2009, 139, 1619–1625. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-B.; Wang, P.-Y.; Wang, X.; Wan, Y.-L.; Liu, Y.-C. Butyrate Enhances Intestinal Epithelial Barrier Function via Up-Regulation of Tight Junction Protein Claudin-1 Transcription. Dig. Dis. Sci. 2012, 57, 3126–3135. [Google Scholar] [CrossRef]
- Mátis, G.; Mackei, M.; Boomsma, B.; Fébel, H.; Nadolna, K.; Szymański, Ł.; Edwards, J.E.; Neogrády, Z.; Kozłowski, K. Dietary Protected Butyrate Supplementation of Broilers Modulates Intestinal Tight Junction Proteins and Stimulates Endogenous Production of Short Chain Fatty Acids in the Caecum. Animals 2022, 12, 1940. [Google Scholar] [CrossRef]
- Liu, L.; Ling, H.; Zhang, W.; Zhou, Y.; Li, Y.; Peng, N.; Zhao, S. Functional Comparison of Clostridium Butyricum and Sodium Butyrate Supplementation on Growth, Intestinal Health, and the Anti-Inflammatory Response of Broilers. Front. Microbiol. 2022, 13, 914212. [Google Scholar] [CrossRef]
- Kelly, C.J.; Colgan, S.P. Breathless in the Gut: Implications of Luminal O2 for Microbial Pathogenicity. Cell Host Microbe 2016, 19, 427–428. [Google Scholar] [CrossRef]
- Louis, P.; Flint, H.J. Formation of Propionate and Butyrate by the Human Colonic Microbiota. Environ. Microbiol. 2017, 19, 29–41. [Google Scholar] [CrossRef]
- Duncan, S.H.; Louis, P.; Flint, H.J. Lactate-Utilizing Bacteria, Isolated from Human Feces, That Produce Butyrate as a Major Fermentation Product. Appl. Environ. Microbiol. 2004, 70, 5810–5817. [Google Scholar] [CrossRef]
- Boesmans, L.; Valles-Colomer, M.; Wang, J.; Eeckhaut, V.; Falony, G.; Ducatelle, R.; Van Immerseel, F.; Raes, J.; Verbeke, K. Butyrate Producers as Potential Next-Generation Probiotics: Safety Assessment of the Administration of Butyricicoccus Pullicaecorum to Healthy Volunteers. Msystems 2018, 3, e00094-18. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Li, Y.; Wen, Z.; Liu, W.; Meng, L.; Huang, H. Oscillospira-a Candidate for the next-Generation Probiotics. Gut Microbes 2021, 13, 1987783. [Google Scholar] [CrossRef] [PubMed]
- McOrist, A.L.; Miller, R.B.; Bird, A.R.; Keogh, J.B.; Noakes, M.; Topping, D.L.; Conlon, M.A. Fecal Butyrate Levels Vary Widely among Individuals but Are Usually Increased by a Diet High in Resistant Starch. J. Nutr. 2011, 141, 883–889. [Google Scholar] [CrossRef] [PubMed]
- Cresci, G.; Nagy, L.E.; Ganapathy, V. Lactobacillus GG and Tributyrin Supplementation Reduce Antibiotic-induced Intestinal Injury. J. Parenter. Enter. Nutr. 2013, 37, 763–774. [Google Scholar] [CrossRef]
- Seyhan, A.A. Lost in Translation: The Valley of Death across Preclinical and Clinical Divide—Identification of Problems and Overcoming Obstacles. Transl. Med. Commun. 2019, 4, 18. [Google Scholar] [CrossRef]
- O’Donnell, M.M.; Rea, M.C.; Shanahan, F.; Ross, R.P. The Use of a Mini-Bioreactor Fermentation System as a Reproducible, High-Throughput Ex Vivo Batch Model of the Distal Colon. Front. Microbiol. 2018, 9, 1844. [Google Scholar] [CrossRef]
- Van den Abbeele, P.; Verstrepen, L.; Ghyselinck, J.; Albers, R.; Marzorati, M.; Mercenier, A. A Novel Non-Digestible, Carrot-Derived Polysaccharide (CRG-I) Selectively Modulates the Human Gut Microbiota While Promoting Gut Barrier Integrity: An Integrated In Vitro Approach. Nutrients 2020, 12, 1917. [Google Scholar] [CrossRef]
- Biagini, F.; Calvigioni, M.; Lapomarda, A.; Vecchione, A.; Magliaro, C.; De Maria, C.; Montemurro, F.; Celandroni, F.; Mazzantini, D.; Mattioli-Belmonte, M.; et al. A Novel 3D in Vitro Model of the Human Gut Microbiota. Sci. Rep. 2020, 10, 21499. [Google Scholar] [CrossRef]
- Gaisawat, M.B.; MacPherson, C.W.; Tremblay, J.; Piano, A.; Iskandar, M.M.; Tompkins, T.A.; Kubow, S. Probiotic Supplementation in a Clostridium Difficile-Infected Gastrointestinal Model Is Associated with Restoring Metabolic Function of Microbiota. Microorganisms 2019, 8, 60. [Google Scholar] [CrossRef]
- Rajilić-Stojanović, M.; Maathuis, A.; Heilig, H.G.H.J.; Venema, K.; de Vos, W.M.; Smidt, H. Evaluating the Microbial Diversity of an in Vitro Model of the Human Large Intestine by Phylogenetic Microarray Analysis. Microbiology 2010, 156, 3270–3281. [Google Scholar] [CrossRef]
- Van den Abbeele, P.; Grootaert, C.; Marzorati, M.; Possemiers, S.; Verstraete, W.; Gérard, P.; Rabot, S.; Bruneau, A.; El Aidy, S.; Derrien, M.; et al. Microbial Community Development in a Dynamic Gut Model Is Reproducible, Colon Region Specific, and Selective for Bacteroidetes and Clostridium Cluster IX. Appl. Environ. Microbiol. 2010, 76, 5237–5246. [Google Scholar] [CrossRef] [PubMed]
- Van den Abbeele, P.; Belzer, C.; Goossens, M.; Kleerebezem, M.; De Vos, W.M.; Thas, O.; De Weirdt, R.; Kerckhof, F.-M.; Van de Wiele, T. Butyrate-Producing Clostridium Cluster XIVa Species Specifically Colonize Mucins in an In Vitro Gut Model. ISME J. 2013, 7, 949–961. [Google Scholar] [CrossRef]
- Lavelle, A.; Lennon, G.; O’Sullivan, O.; Docherty, N.; Balfe, A.; Maguire, A.; Mulcahy, H.E.; Doherty, G.; O’Donoghue, D.; Hyland, J.; et al. Spatial Variation of the Colonic Microbiota in Patients with Ulcerative Colitis and Control Volunteers. Gut 2015, 64, 1553–1561. [Google Scholar] [CrossRef] [PubMed]
- Healey, G.R.; Murphy, R.; Brough, L.; Butts, C.A.; Coad, J. Interindividual Variability in Gut Microbiota and Host Response to Dietary Interventions. Nutr. Rev. 2017, 75, 1059–1080. [Google Scholar] [CrossRef] [PubMed]
- Capurso, L. Thirty Years of Lactobacillus rhamnosus GG: A Review. J. Clin. Gastroenterol. 2019, 53 (Suppl. S1), S1–S41. [Google Scholar] [CrossRef]
- Van den Abbeele, P.; Marzorati, M.; Derde, M.; De Weirdt, R.; Joan, V.; Possemiers, S.; Van de Wiele, T. Arabinoxylans, Inulin and Lactobacillus reuteri 1063 Repress the Adherent-Invasive Escherichia coli from Mucus in a Mucosa-Comprising Gut Model. NPJ Biofilms Microbiomes 2016, 2, 16016. [Google Scholar] [CrossRef]
- Van den Abbeele, P.; Deyaert, S.; Thabuis, C.; Perreau, C.; Bajic, D.; Wintergerst, E.; Joossens, M.; Firrman, J.; Walsh, D.; Baudot, A. Bridging Preclinical and Clinical Gut Microbiota Research Using the Ex Vivo SIFR Technology. Front. Microbiol. 2023. submitted. [Google Scholar]
- Costea, P.I.; Hildebrand, F.; Arumugam, M.; Bäckhed, F.; Blaser, M.J.; Bushman, F.D.; de Vos, W.M.; Ehrlich, S.D.; Fraser, C.M.; Hattori, M.; et al. Enterotypes in the Landscape of Gut Microbial Community Composition. Nat. Microbiol. 2018, 3, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Yang, H.; Xu, Z.; Li, X.; Leng, X. Dietary Supplementation of Tributyrin Improved the Growth, Feed Utilization and Intestinal Histology of Grass Carp (Ctenopharyngodon idella). Aquac. Nutr. 2021, 27, 2007–2018. [Google Scholar] [CrossRef]
- Xie, D.; Dai, Q.; Xu, C.; Li, Y. Dietary Tributyrin Modifies Intestinal Function by Altering Morphology, Gene Expression and Microbiota Profile in Common Carp (Cyprinus carpio) Fed All-Plant Diets. Aquac. Nutr. 2021, 27, 439–453. [Google Scholar] [CrossRef]
- Rizzatti, G.; Lopetuso, L.R.; Gibiino, G.; Binda, C.; Gasbarrini, A. Proteobacteria: A Common Factor in Human Diseases. BioMed Res. Int. 2017, 2017, 9351507. [Google Scholar] [CrossRef]
- Shin, N.-R.; Whon, T.W.; Bae, J.-W. Proteobacteria: Microbial Signature of Dysbiosis in Gut Microbiota. Trends Biotechnol. 2015, 33, 496–503. [Google Scholar] [CrossRef]
- Louis, P.; Flint, H.J. Diversity, Metabolism and Microbial Ecology of Butyrate-Producing Bacteria from the Human Large Intestine. FEMS Microbiol. Lett. 2009, 294, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Reichardt, N.; Duncan, S.H.; Young, P.; Belenguer, A.; McWilliam Leitch, C.; Scott, K.P.; Flint, H.J.; Louis, P. Phylogenetic Distribution of Three Pathways for Propionate Production within the Human Gut Microbiota. ISME J. 2014, 8, 1323–1335. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wu, J.; Lv, M.; Shao, Z.; Hungwe, M.; Wang, J.; Bai, X.; Xie, J.; Wang, Y.; Geng, W. Metabolism Characteristics of Lactic Acid Bacteria and the Expanding Applications in Food Industry. Front. Bioeng. Biotechnol. 2021, 9, 612285. [Google Scholar] [CrossRef] [PubMed]
- Sheridan, P.O.; Louis, P.; Tsompanidou, E.; Shaw, S.; Harmsen, H.J.; Duncan, S.H.; Flint, H.J.; Walker, A.W.Y. Distribution, Organization and Expression of Genes Concerned with Anaerobic Lactate Utilization in Human Intestinal Bacteria. Microb. Genom. 2022, 8, 000739. [Google Scholar] [CrossRef]
- Lozupone, C.A.; Stombaugh, J.I.; Gordon, J.I.; Jansson, J.K.; Knight, R. Diversity, Stability and Resilience of the Human Gut Microbiota. Nature 2012, 489, 220–230. [Google Scholar] [CrossRef]
- Kriss, M.; Hazleton, K.Z.; Nusbacher, N.M.; Martin, C.G.; Lozupone, C.A. Low Diversity Gut Microbiota Dysbiosis: Drivers, Functional Implications and Recovery. Curr. Opin. Microbiol. 2018, 44, 34–40. [Google Scholar] [CrossRef]
- Cleusix, V.; Lacroix, C.; Vollenweider, S.; Duboux, M.; Le Blay, G. Inhibitory Activity Spectrum of Reuterin Produced by Lactobacillus Reuteri against Intestinal Bacteria. BMC Microbiol. 2007, 7, 101. [Google Scholar] [CrossRef]
- Cleusix, V.; Lacroix, C.; Vollenweider, S.; Le Blay, G. Glycerol Induces Reuterin Production and Decreases Escherichia coli Population in an In Vitro Model of Colonic Fermentation with Immobilized Human Feces. FEMS Microbiol. Ecol. 2008, 63, 56–64. [Google Scholar] [CrossRef]
- Chen, L.; Bromberger, P.D.; Nieuwenhuiys, G.; Hatti-Kaul, R. Redox Balance in Lactobacillus Reuteri DSM20016: Roles of Iron-Dependent Alcohol Dehydrogenases in Glucose/Glycerol Metabolism. PLoS ONE 2016, 11, e0168107. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wang, K.; Oh, J.-H.; Zhang, S.; van Pijkeren, J.-P.; Cheng, C.C.; Ren, D.; Wei, H.; Gänzle, M.G.; Walter, J. A Phylogenetic View on the Role of Glycerol for Growth Enhancement and Reuterin Formation in Limosilactobacillus Reuteri. Front. Microbiol. 2020, 11, 601422. [Google Scholar] [CrossRef] [PubMed]
- Axelsson, L.T.; Chung, T.C.; Dobrogosz, W.J.; Lindgren, S.E. Production of a Broad Spectrum Antimicrobial Substance by Lactobacillus Reuteri. Microb. Ecol. Health Dis. 1989, 2, 131–136. [Google Scholar]
- Schaefer, L.; Auchtung, T.A.; Hermans, K.E.; Whitehead, D.; Borhan, B.; Britton, R.A. The Antimicrobial Compound Reuterin (3-Hydroxypropionaldehyde) Induces Oxidative Stress via Interaction with Thiol Groups. Microbiology 2010, 156, 1589. [Google Scholar] [CrossRef]
- Kastl, A.J.; Terry, N.A.; Wu, G.D.; Albenberg, L.G. The Structure and Function of the Human Small Intestinal Microbiota: Current Understanding and Future Directions. Cell. Mol. Gastroenterol. Hepatol. 2019, 9, 33–45. [Google Scholar] [CrossRef]
- Stephen, A.M.; Champ, M.M.-J.; Cloran, S.J.; Fleith, M.; van Lieshout, L.; Mejborn, H.; Burley, V.J. Dietary Fibre in Europe: Current State of Knowledge on Definitions, Sources, Recommendations, Intakes and Relationships to Health. Nutr. Res. Rev. 2017, 30, 149–190. [Google Scholar] [CrossRef] [PubMed]
- Quagliani, D.; Felt-Gunderson, P. Closing America’s Fiber Intake Gap: Communication Strategies from a Food and Fiber Summit. Am. J. Lifestyle Med. 2017, 11, 80–85. [Google Scholar] [CrossRef]
- Elmadfa, I.; Meyer, A. Overview of Dietary Fibre Intake across European Countries. 2021. Available online: https://knowledge4policy.ec.europa.eu/health-promotion-knowledge-gateway/dietary-fibre-overview-3_en (accessed on 8 March 2023).
- Rivera-Chávez, F.; Zhang, L.F.; Faber, F.; Lopez, C.A.; Byndloss, M.X.; Olsan, E.E.; Xu, G.; Velazquez, E.M.; Lebrilla, C.B.; Winter, S.E. Depletion of Butyrate-Producing Clostridia from the Gut Microbiota Drives an Aerobic Luminal Expansion of Salmonella. Cell Host Microbe 2016, 19, 443–454. [Google Scholar] [CrossRef] [PubMed]
- Sitkin, S.; Pokrotnieks, J. Clinical Potential of Anti-Inflammatory Effects of Faecalibacterium prausnitzii and Butyrate in Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2019, 25, e40–e41. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Xie, S.; Lv, D.; Zhang, Y.; Deng, J.; Zeng, L.; Chen, Y. A Reduction in the Butyrate Producing Species Roseburia Spp. and Faecalibacterium prausnitzii Is Associated with Chronic Kidney Disease Progression. Antonie Van Leeuwenhoek 2016, 109, 1389–1396. [Google Scholar] [CrossRef]
- Ceccarani, C.; Bassanini, G.; Montanari, C.; Casiraghi, M.C.; Ottaviano, E.; Morace, G.; Biasucci, G.; Paci, S.; Borghi, E.; Verduci, E. Proteobacteria Overgrowth and Butyrate-Producing Taxa Depletion in the Gut Microbiota of Glycogen Storage Disease Type 1 Patients. Metabolites 2020, 10, 133. [Google Scholar] [CrossRef] [PubMed]
- Umirah, F.; Neoh, C.F.; Ramasamy, K.; Lim, S.M. Differential Gut Microbiota Composition between Type 2 Diabetes Mellitus Patients and Healthy Controls: A Systematic Review. Diabetes Res. Clin. Pract. 2021, 173, 108689. [Google Scholar] [CrossRef] [PubMed]
- Hartemink, R.; Domenech, V.R.; Rombouts, F.M. LAMVAB—A New Selective Medium for the Isolation of Lactobacilli from Faeces. J. Microbiol. Methods 1997, 29, 77–84. [Google Scholar] [CrossRef]
- De Weirdt, R.; Possemiers, S.; Vermeulen, G.; Moerdijk-Poortvliet, T.C.W.; Boschker, H.T.S.; Verstraete, W.; Van de Wiele, T. Human Faecal Microbiota Display Variable Patterns of Glycerol Metabolism. FEMS Microbiol. Ecol. 2010, 74, 601–611. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, K.; Maki, K.A.; Vizioli, C.; Carnell, S.; Goodman, E.; Hurley, M.; Harris, C.; Colwell, R.; Steele, K.; Joseph, P.V. The Neuro-Endo-Microbio-Ome Study: A Pilot Study of Neurobiological Alterations Pre- Versus Post-Bariatric Surgery. Biol. Res. Nurs. 2022, 24, 362–378. [Google Scholar] [CrossRef]
- Hasan, N.A.; Young, B.A.; Minard-Smith, A.T.; Saeed, K.; Li, H.; Heizer, E.M.; McMillan, N.J.; Isom, R.; Abdullah, A.S.; Bornman, D.M.; et al. Microbial Community Profiling of Human Saliva Using Shotgun Metagenomic Sequencing. PLoS ONE 2014, 9, e97699. [Google Scholar] [CrossRef]
- Rohart, F.; Gautier, B.; Singh, A.; Cao, K.-A.L. MixOmics: An R Package for ’omics Feature Selection and Multiple Data Integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Van den Abbeele, P.; Goggans, M.; Deyaert, S.; Baudot, A.; Van de Vliet, M.; Calatayud Arroyo, M.; Lelah, M. Lacticaseibacillus rhamnosus ATCC 53103 and Limosilactobacillus reuteri ATCC 53608 Synergistically Boost Butyrate Levels upon Tributyrin Administration Ex Vivo. Int. J. Mol. Sci. 2023, 24, 5859. https://doi.org/10.3390/ijms24065859
Van den Abbeele P, Goggans M, Deyaert S, Baudot A, Van de Vliet M, Calatayud Arroyo M, Lelah M. Lacticaseibacillus rhamnosus ATCC 53103 and Limosilactobacillus reuteri ATCC 53608 Synergistically Boost Butyrate Levels upon Tributyrin Administration Ex Vivo. International Journal of Molecular Sciences. 2023; 24(6):5859. https://doi.org/10.3390/ijms24065859
Chicago/Turabian StyleVan den Abbeele, Pieter, Mallory Goggans, Stef Deyaert, Aurélien Baudot, Michiel Van de Vliet, Marta Calatayud Arroyo, and Michael Lelah. 2023. "Lacticaseibacillus rhamnosus ATCC 53103 and Limosilactobacillus reuteri ATCC 53608 Synergistically Boost Butyrate Levels upon Tributyrin Administration Ex Vivo" International Journal of Molecular Sciences 24, no. 6: 5859. https://doi.org/10.3390/ijms24065859
APA StyleVan den Abbeele, P., Goggans, M., Deyaert, S., Baudot, A., Van de Vliet, M., Calatayud Arroyo, M., & Lelah, M. (2023). Lacticaseibacillus rhamnosus ATCC 53103 and Limosilactobacillus reuteri ATCC 53608 Synergistically Boost Butyrate Levels upon Tributyrin Administration Ex Vivo. International Journal of Molecular Sciences, 24(6), 5859. https://doi.org/10.3390/ijms24065859






