Gene Structure, Expression and Function Analysis of MEF2 in the Pacific White Shrimp Litopenaeus vannamei
Abstract
1. Introduction
2. Results
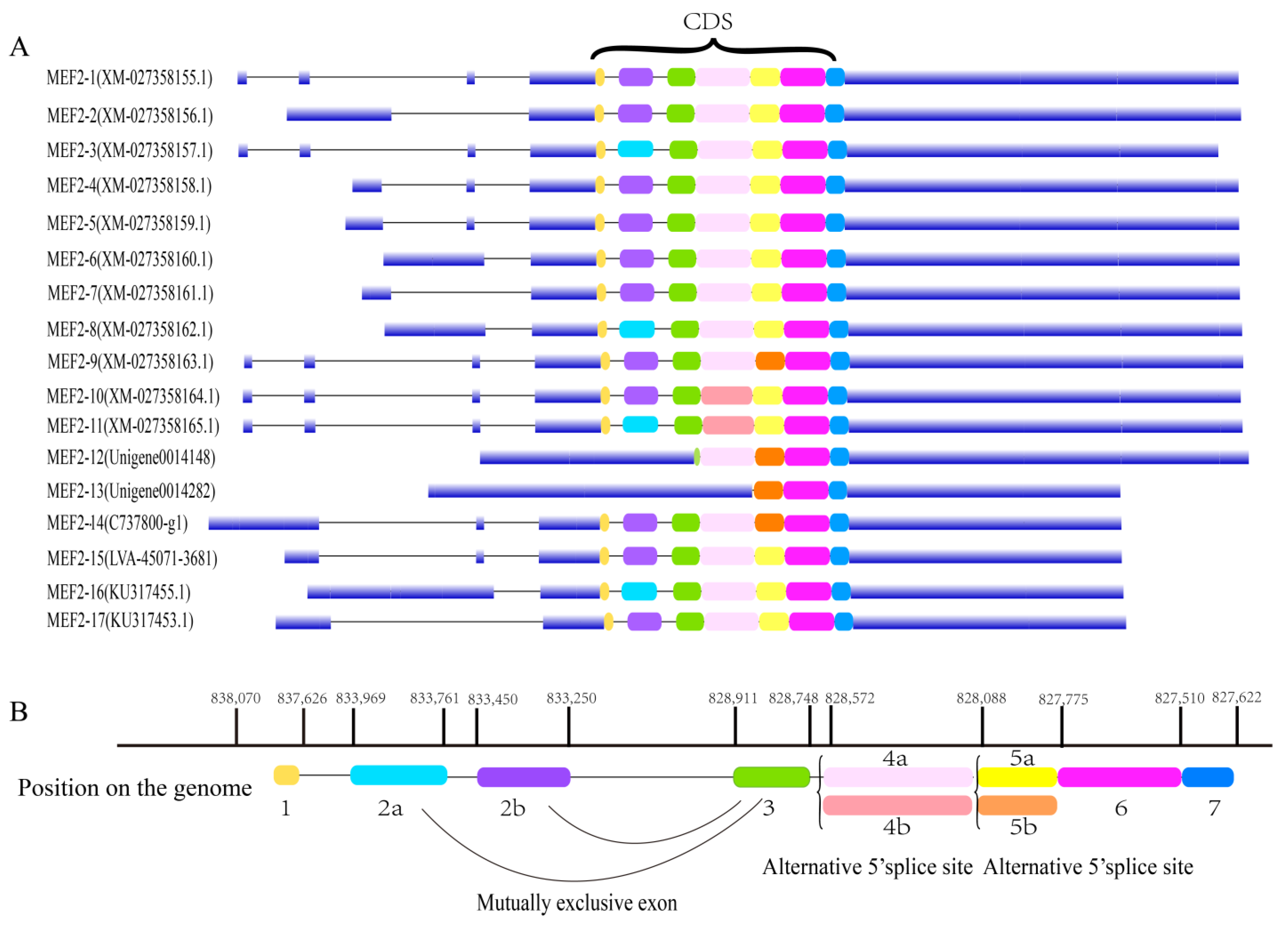
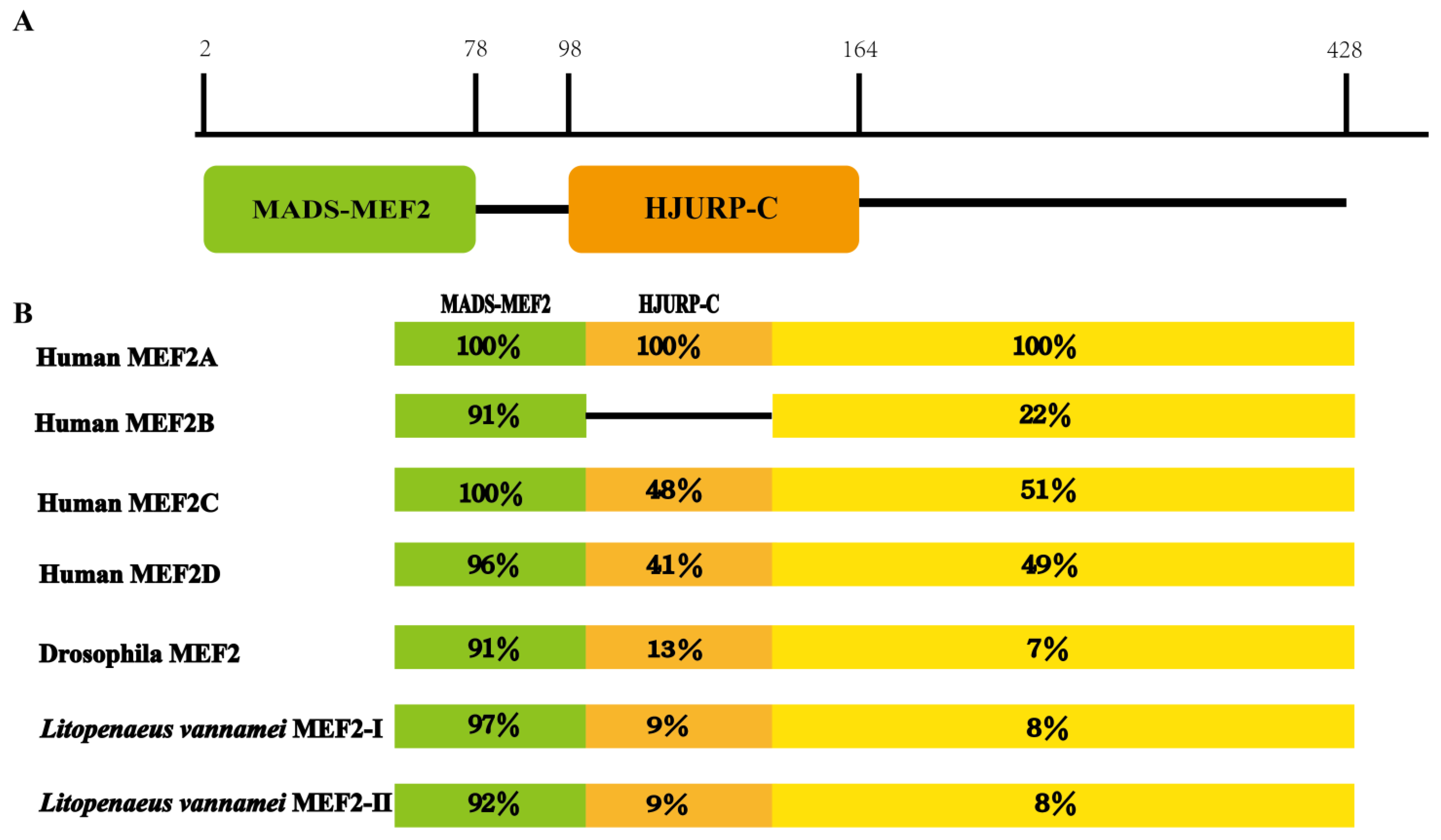
2.1. Structural Analysis of the LvMEF2 Gene
2.2. Phylogenetic Analysis of LvMEF2 Gene
2.3. LvMEF2 Gene Expression Profiles
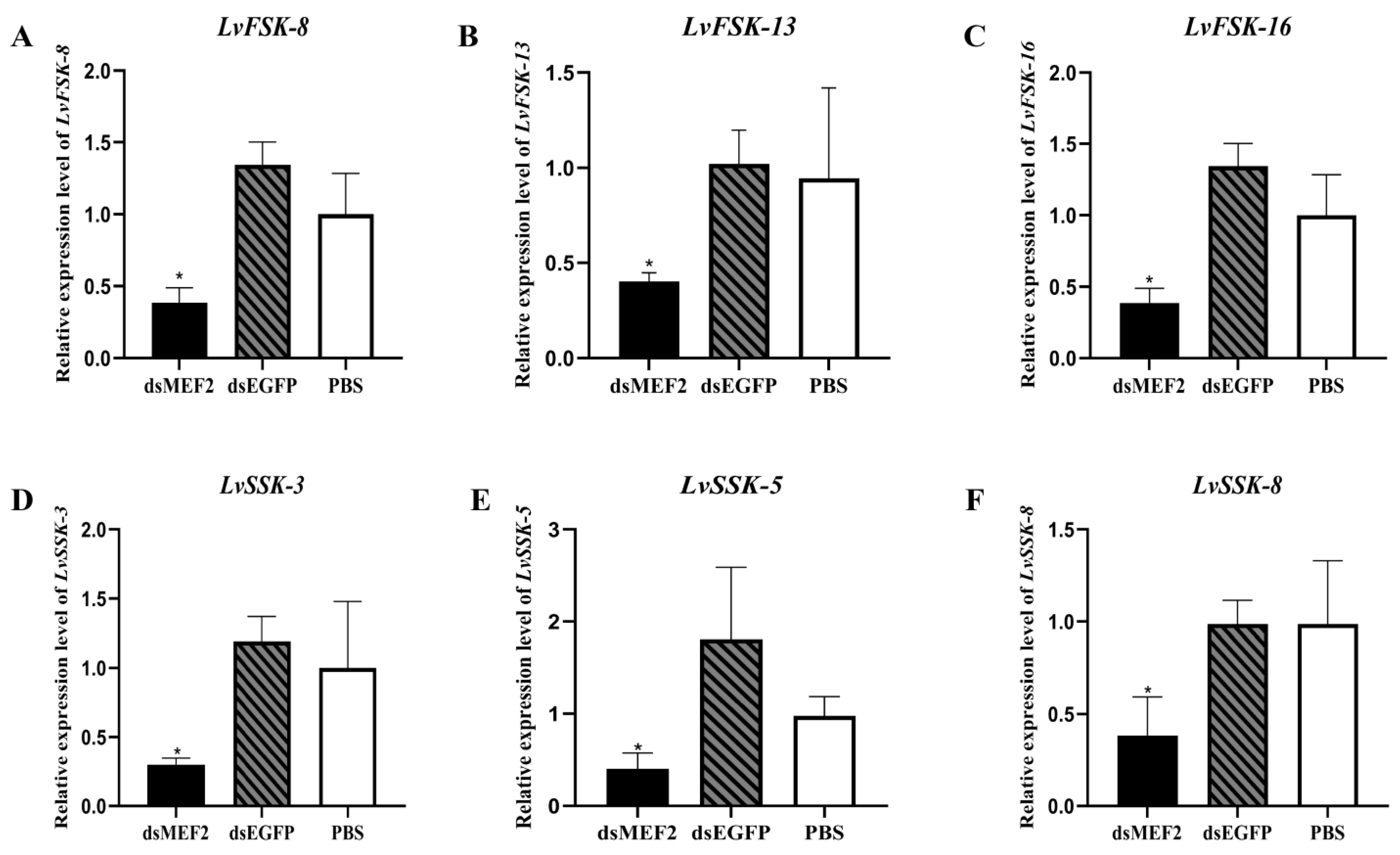
2.4. LvMEF2 RNA Interference
2.5. Histology and Total Viable Bacteria Count after LvMEF2 Knockdown
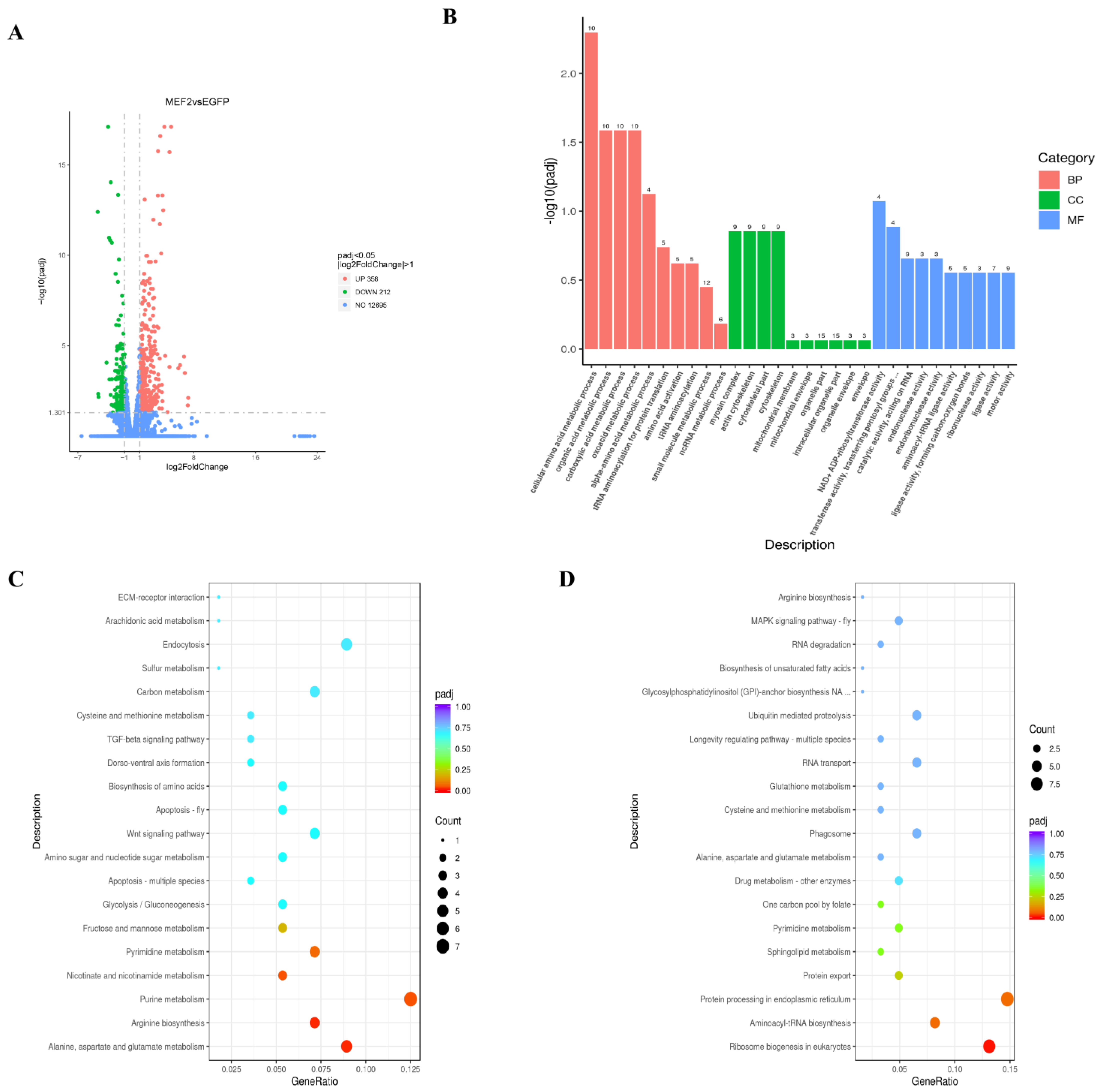
2.6. Differentially Expressed Genes (DEGs) in Muscle after LvMEF2 Knockdown
3. Discussion
3.1. LvMEF2 Has Multiple Splice Variants
3.2. LvMEF2 Has Complex Expression Profiles
3.3. LvMEF2 Affects the Growth and Immunity of the Shrimp
4. Materials and Methods
4.1. Experimental Animals
4.2. Identification and Analysis of MEF2 Gene in L. vannamei
4.3. Construction of Phylogenetic Tree
4.4. RNA Extraction and cDNA Synthesis
4.5. Gene Expression Pattern Analysis
4.6. Double-Stranded RNA Synthesis and Interference Experiments of LvMEF2
4.7. Tissue Section and Total Viable Bacteria Count after LvMEF2 Gene Knowdown
4.8. Transcriptome Sequencing Analysis of Muscle Samples after LvMEF2 Interference
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tian, Z.; Jiao, C. Muscle atrophy and growth induced by molting in Crustacean: A review. Fish. Sci. 2016, 35, 603–606. [Google Scholar] [CrossRef]
- Mykles, D.L.; Medler, S. Skeletal Muscle Differentiation, Growth, and Plasticity. In The Natural History of Crustacea, Volume 4: Physiology, 2nd ed.; Ernest, S., Martin, T., Eds.; Oxford University: Oxford, UK, 2015; pp. 134–167. [Google Scholar]
- Nurrish, S.J.; Treisman, R. DNA binding specificity determinants in MADS-box transcription factors. Mol. Cell. Biol. 1995, 15, 4076–4085. [Google Scholar] [CrossRef]
- Gossett, L.A.; Kelvin, D.J.; Sternberg, E.A.; Olson, E.N. A new myocyte-specific enhancer-binding factor that recognizes a conserved element associated with multiple muscle-specific gene. Mol. Cell. Biol. 1989, 9, 5022–5033. [Google Scholar] [CrossRef]
- Paulin, R.; Sutendra, G.; Gurtu, V.; Dromparis, P.; Haromy, A.; Provencher, S.; Bonnet, S.; Michelakis, E.D. A miR-208–Mef2 axis drives the decompensation of right ventricular function in pulmonary hypertension. Circ. Res. 2015, 116, 56–69. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Zhou, D.; Ding, N.-Z.; Teng, C.-B.; Yan, X.-C.; Liang, Y. Common carp mef2 genes: Evolution and expression. Genes 2019, 10, 588. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; de Folter, S.; Shen, X.; Zhang, W.; Tao, S. Vertebrate paralogous MEF2 genes: Origin, conservation, and evolution. PLoS ONE 2011, 6, e17334. [Google Scholar] [CrossRef]
- Genikhovich, G.; Technau, U. Complex functions of MEF2 splice variants in the differentiation of endoderm and of a neuronal cell type in a Sea anemone. Development 2011, 138, 4911–4919. [Google Scholar] [CrossRef]
- Shore, P.; Sharrocks, A.D. The MADS-Box family of transcription factors. Chem. Eur. J. 1995, 229, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.V.; Hughes, S.M. Mef2 and the skeletal muscle differentiation program. Semin. Cell Dev. Biol. 2017, 72, 33–44. [Google Scholar] [CrossRef]
- Hennebry, A.; Berry, C.; Siriett, V.; O′Callaghan, P.; Chau, L.; Watson, T.; Sharma, M.; Kambadur, R. Myostatin regulates fiber-type composition of skeletal muscle by regulating MEF2 and MyoD gene expression. Am. J. Physiol. Physiol. 2009, 296, C525–C534. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, G.; Wang, J.; Ji, Z.; Dong, F.; Chao, T. Analysis of differential gene expression and novel transcript units of ovine muscle transcriptomes. PLoS ONE 2014, 9, e89817. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, G.; Wang, J.; Ji, Z.; Liu, Z.; Pi, X.; Chen, C. Characterization and comparative analyses of muscle transcriptomes in dorper and small-tailed han sheep using RNA-Seq technique. PLoS ONE 2013, 8, e72686. [Google Scholar] [CrossRef]
- Jung, S.-Y.; Ko, Y.-G. TRIM72, a novel negative feedback regulator of myogenesis, is transcriptionally activated by the synergism of MyoD (or myogenin) and MEF2. Biochem. Biophys. Res. Commun. 2010, 396, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Schiaffino, S.; Dyar, K.A.; Calabria, E. Skeletal muscle mass is controlled by the MRF4-MEF2 axis. Curr. Opin. Clin. Nutr. Metab. Care 2018, 21, 164–167. [Google Scholar] [CrossRef] [PubMed]
- Crittenden, J.R.; Skoulakis, E.M.C.; Goldstein, E.S.; Davis, R.L. Drosophila mef2 is essential for normal mushroom body and wing development. Biol. Open 2018, 7, bio035618. [Google Scholar] [CrossRef]
- Sun, P.S.; Soderlund, M.; Venzon, N.C.; Ye, D.; Lu, Y. Isolation and characterization of two actins of the Pacific white shrimp, Litopenaeus vannamei. Mar. Biol. 2007, 151, 2145–2151. [Google Scholar] [CrossRef]
- Zhang, X.; Yuan, J.; Zhang, X.; Liu, C.; Li, F.; Xiang, J. Genome-wide identification and expression profiles of myosin genes in the pacific white shrimp, Litopenaeus vannamei. Front. Physiol. 2019, 10, 610. [Google Scholar] [CrossRef]
- Lee, J.-H.; Momani, J.; Kim, Y.M.; Kang, C.-K.; Choi, J.-H.; Baek, H.-J.; Kim, H.-W. Effective RNA-silencing strategy of Lv-MSTN/GDF11 gene and its effects on the growth in shrimp, Litopenaeus vannamei. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2015, 179, 9–16. [Google Scholar] [CrossRef]
- Yan, B.; Xue, B.; Zhao, L.; Gao, H. Progress on the research of the growth-related genes in Crustaceans. J. Oceanol. Limnol. 2016, 151, 97–104. [Google Scholar] [CrossRef]
- Sellars, M.J.; Trewin, C.; McWilliam, S.M.; Glaves, R.S.; Hertzler, P.L. Transcriptome profiles of Penaeus (Marsupenaeus) japonicus animal and vegetal half-embryos: Identification of sex determination, germ line, mesoderm, and other developmental genes. Mar. Biotechnol. 2015, 17, 252–265. [Google Scholar] [CrossRef]
- Wei, J.K.; Zhang, X.J.; Yu, Y.; Huang, H.; Li, F.H.; Xiang, J.H. Comparative transcriptomic characterization of the early development in Pacific White Shrimp Litopenaeus vannamei. PLoS ONE 2014, 9, e106201. [Google Scholar] [CrossRef]
- Price, A.L.; Patel, N.H. Investigating divergent mechanisms of mesoderm development in arthropods: The expression ofPh-twist andPh-mef2 inParhyale hawaiensis. J. Exp. Zool. Part B Mol. Dev. Evol. 2008, 310B, 24–40. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, R.Q.; Zhou, T.T.; Yang, S.P.; Chan, S.F. Characterization of a molt-related myostatin gene (FmMstn) from the banana shrimp Fenneropenaeus Merguiensis. Gen. Comp. Endocrinol. 2017, 248, 55–68. [Google Scholar] [CrossRef]
- Martin, J.F.; Miano, J.M.; Hustad, C.M.; Copeland, N.G.; Jenkins, N.A.; Olson, E.N. A MEF2 gene that generates a muscle-specific isoform via alternative messenger-RNA splicing Mol. Cell. Biol. 1994, 14, 1647–1656. [Google Scholar] [CrossRef]
- Molkentin, J.D.; Firulli, A.B.; Black, B.L.; Martin, J.F.; Hustad, C.M.; Copeland, N.; Jenkins, N.; Lyons, G.; Olson, E.N. MEF2B is a potent transactivator expressed in early myogenic lineages. Mol. Cell. Biol. 1996, 16, 3814–3824. [Google Scholar] [CrossRef]
- Wu, W.; Huang, X.; Cheng, J.; Li, Z.; de Folter, S.; Huang, Z.; Jiang, X.; Pang, H.; Tao, S. Conservation and evolution in and among SRF- and MEF2-type MADS domains and their binding sites. Mol. Biol. Evol. 2011, 28, 501–511. [Google Scholar] [CrossRef]
- Qi, Y.; Zhang, X.; Pang, Y.; Wang, Y.; Liao, h. Expression patterns of MyoD and MEF2 genes during the development of Nanyang cattle longissimus dorsi muscle. J. Henan Agric. Univ. 2012, 46, 558–561. [Google Scholar] [CrossRef]
- Xuefei, L.I.U.; Yuexiang, W.; Qiu, J.; Linxi, Q.; Guangwen, G.A.O.; Yongxin, D.; Tao, Z.; Houyan, S. MEF2C gene expression profile during zebrafish embryogenesis. Fudan Univ. J. Med. Sci. 2006, 33, 89–91. [Google Scholar]
- Zhu, K.; Liu, B.; Guo, H.; Zhang, N.; Zhang, D. Molecular cloning and expression characterization analysis of MEF2Cs in Chlamys nobilis. Freshw. Biol. 2018, 48, 32–38. [Google Scholar] [CrossRef]
- Moretti, I.; Ciciliot, S.; Dyar, K.A.; Abraham, R.; Murgia, M.; Agatea, L.; Akimoto, T.; Bicciato, S.; Forcato, M.; Pierre, P.; et al. MRF4 negatively regulates adult skeletal muscle growth by repressing MEF2 activity. Nat. Commun. 2016, 7, 12397. [Google Scholar] [CrossRef]
- Bird, L. Immunometabolism: MEF2 in sickness and in health. Nat. Rev. Immunol. 2013, 13, 845. [Google Scholar] [CrossRef] [PubMed]
- Pon, J.R.; Marra, M.A. MEF2 transcription factors: Developmental regulators and emerging cancer genes. OncoTargets Ther. 2016, 7, 2297–2312. [Google Scholar] [CrossRef]
- Wang, W.Y.; Montel-Hagen, A.; Sasidharan, R.; Mikkola, H.K.A. MEF2C maintains B cell homeostasis through the regulation of DNA repair machinery. Blood 2012, 120, 278. [Google Scholar] [CrossRef]
- Potthoff, M.J.; Arnold, M.A.; McAnally, J.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. Regulation of skeletal muscle sarcomere integrity and postnatal muscle function by Mef2c. Mol. Cell. Biol. 2007, 27, 8143–8151. [Google Scholar] [CrossRef]
- Schlesinger, J.; Schueler, M.; Grunert, M.; Fischer, J.J.; Zhang, Q.; Krueger, T.; Lange, M.; Tönjes, M.; Dunkel, I.; Sperling, S.R. The cardiac transcription network modulated by Gata4, Mef2a, Nkx2.5, Srf, histone modifications, and MicroRNAs. PLoS Genet. 2011, 7, e1001313. [Google Scholar] [CrossRef]
- Callegari, S.; McKinnon, R.A.; Andrews, S.; Lopes, M.A.D.B. The MEF2 gene is essential for yeast longevity, with a dual role in cell respiration and maintenance of mitochondrial membrane potential. FEBS Lett. 2011, 585, 1140–1146. [Google Scholar] [CrossRef]
- Nath, S.R.; Lieberman, M.L.; Yu, Z.; Marchioretti, C.; Jones, S.T.; Danby, E.C.E.; Van Pelt, K.M.; Sorarù, G.; Robins, D.M.; Bates, G.P.; et al. MEF2 impairment underlies skeletal muscle atrophy in polyglutamine disease. Acta Neuropathol. 2020, 140, 63–80. [Google Scholar] [CrossRef]
- Lilly, B.; Zhao, B.; Ranganayakulu, G.; Paterson, B.M.; Schulz, R.A.; Olson, E.N. Requirement of MADS domain transcription factor D-MEF2 for muscle formation in Drosophila. Science 1995, 267, 688–693. [Google Scholar] [CrossRef]
- Bryantsev, A.L.; Baker, P.W.; Lovato, T.L.; Jaramillo, M.S.; Cripps, R.M. Differential requirements for myocyte enhancer factor-2 during adult myogenesis in Drosophila. Dev. Biol. 2012, 361, 191–207. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, M.; Kim, S.W.; Imanaka-Yoshida, K.; Yoshida, T.; Abel, E.D.; Eliceiri, B.; Yang, Y.; Ulevitch, R.J.; Lee, J.D. Targeted deletion of BMK1/ERK5 in adult mice perturbs vascular integrity and leads to endothelial failure. J. Clin. Investig. 2004, 113, 1138–1148. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Linseman, D.A.; Allen, M.P.; Meintzer, M.K.; Wang, X.; Laessig, T.; Wierman, M.E.; Heidenreich, K.A. Myocyte enhancer factor 2A and 2D undergo phosphorylation and caspase-mediated degradation during apoptosis of rat cerebellar granule neurons. J. Neurosci. 2001, 21, 6544–6552. [Google Scholar] [CrossRef]
- Clark, R.I.; Tan, S.W.; Pean, C.B.; Roostalu, U.; Vivancos, V.; Bronda, K.; Pilatova, M.; Fu, J.; Walker, D.W.; Berdeaux, R.; et al. MEF2 is an in vivo immune-metabolic switch. Cell 2013, 155, 435–447. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Zhang, X.; Wei, J.; Sun, X.; Yuan, J.; Li, F.; Xiang, J. Whole transcriptome analysis provides insights into molecular mechanisms for molting in Litopenaeus vannamei. PLoS ONE 2015, 10, e0144350. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yuan, J.; Sun, Y.; Li, S.; Gao, Y.; Yu, Y.; Liu, C.; Wang, Q.; Lv, X.; Zhang, X.; et al. Penaeid shrimp genome provides insights into benthic adaptation and frequent molting. Nat. Commun. 2019, 10, 356. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.; Park, C.; Bennett, C.; Salzberg, S. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 1. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, stringtie and ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
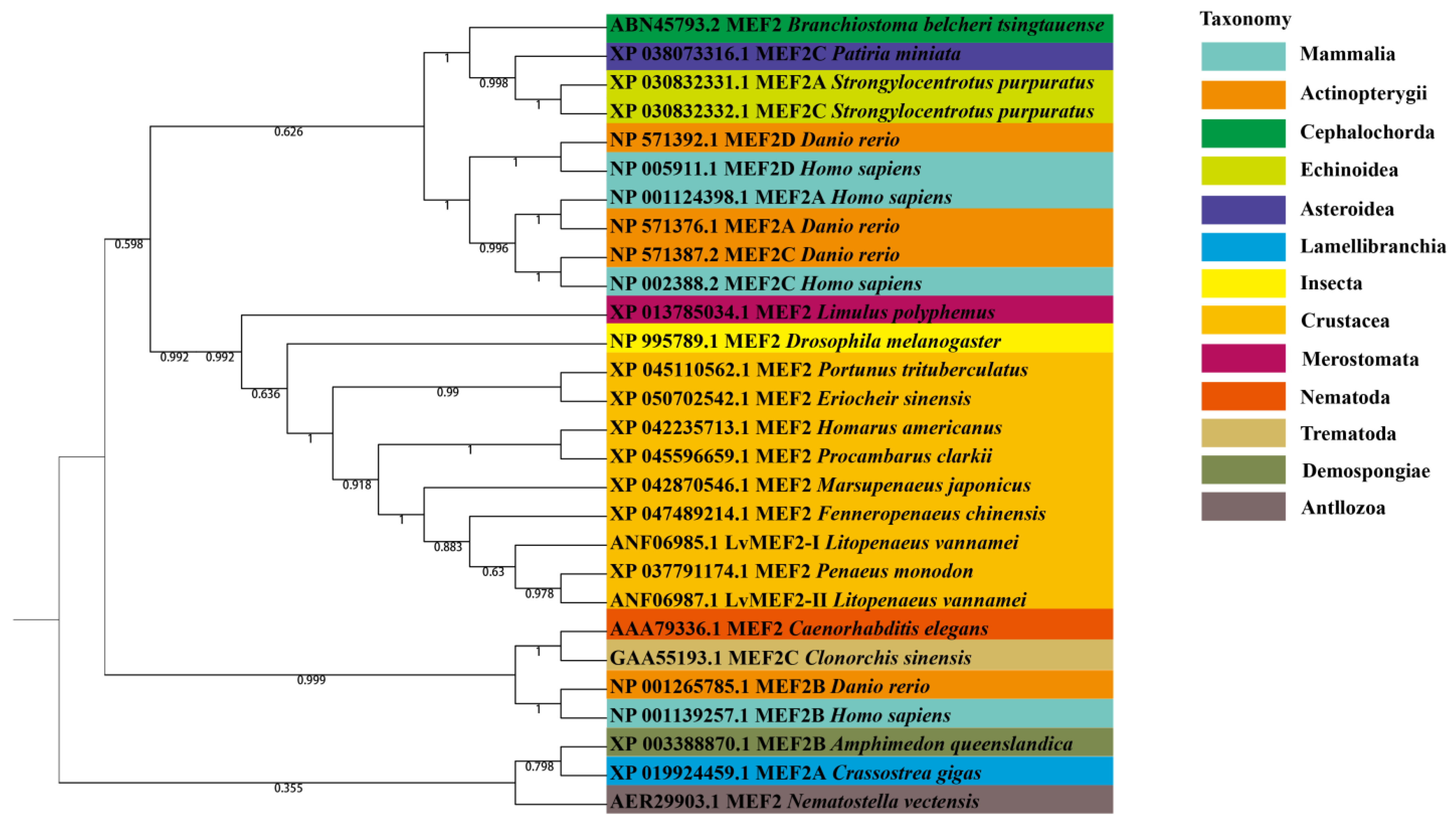



| Phylum | Class | Species | Accession Number | Name |
|---|---|---|---|---|
| Chordata | Mammalia | Homo sapiens | NP_001124398.1 | MEF2A |
| NP_001139257.1 | MEF2B | |||
| NP_002388.2 | MEF2C | |||
| NP_005911.1 | MEF2D | |||
| Actinopterygii | Danio rerio | NP_571376.1 | MEF2A | |
| NP_001265785.1 | MEF2B | |||
| NP_571387.2 | MEF2C | |||
| NP_571392.1 | MEF2D | |||
| Cephalochorda | Branchiostoma | ABN45793.2 | MEF2 | |
| Echinodermata | Echinoidea | Strongylocentrotus purpuratus | XP_030832331.1 | MEF2A |
| XP_030832332.1 | MEF2C | |||
| Asteroidea | Patiria miniata | XP_038073316.1 | MEF2 | |
| Mollusca | Lamellibranchia | Crassostrea gigas | XP_019924459.1 | MEF2A |
| Arthropod | Insecta | Drosophila melanogaster | NP_995789.1 | MEF2 |
| Crustacea | Litopenaeus vannamei | ANF06985.1 | LvMEF2-Ι | |
| ANF06987.1 | LvMEF2-ΙI | |||
| Fenneropenaeus chinensis | XP_047489214.1 | MEF2 | ||
| Marsupenaeus japonicus | XP_042870546.1 | MEF2 | ||
| Penaeus monodon | XP_037791174.1 | MEF2 | ||
| Procambarus clarkii | XP_045596659.1 | MEF2 | ||
| Homarus americanus | XP_042235713.1 | MEF2 | ||
| Eriocheir sinensis | XP_050702542.1 | MEF2 | ||
| Portunus trituberculatus | XP_045110562.1 | MEF2 | ||
| Merostomata | Limulus polyphemus | XP_013785034.1 | MEF2 | |
| Nematoda | Nematoda | Caenorhabditis elegans | AAA79336.1 | MEF2 |
| Platyhelminthes | Trematoda | Clonorchis sinensis | GAA55193.1 | MEF2 |
| Coelenterata | Antllozoa | Nematostella vectensis | AER29903.1 | MEF2 |
| Porifera | Demospongiae | Amphimedon queenslandica | XP_003388870.1 | MEF2 |
| Gene ID | Gene Description | log2Fold Change | |
|---|---|---|---|
| Muscle-related | novel.2155 | Myosin N-terminal SH3-like domain | −4.509773837 |
| LVAN04176 | Myosin-4 | −2.978206791 | |
| novel.4121 | Myosin heavy chain 13, skeletal muscle | −1.772697548 | |
| LVAN16923 | ACT2_BOMMO Actin, muscle-type | −4.386672998 | |
| LVAN16916 | ACT3_DROME Actin-57 | −2.098122282 | |
| LVAN16917 | ACT_MANSE Actin, muscle | −2.036265474 | |
| Immunity and cellular-stress-related | LVAN23842 | Heat shock protein 22 | −2.237662541 |
| LVAN23843 | Heat shock protein 20 | −2.036169236 | |
| LVAN17308 | Heat shock protein 70 | −1.753724055 | |
| LVAN17629 | Heat shock protein 60 | −1.44127178 | |
| LVAN23851 | Heat shock protein 20 | −1.939231582 | |
| novel.751 | Heat shock protein 10, mitochondrial | −1.216804722 | |
| LVAN09991 | Heat shock protein 90 | −1.75361939 | |
| LVAN17366 | Heat shock protein 70 | −1.2135562 | |
| novel.2459 | Heat shock protein 70 | −1.068649747 | |
| novel.386 | Heat shock protein 70 | −1.019314088 | |
| LVAN24387 | Heat shock protein 27 | −1.40954243 | |
| LVAN23841 | Heat shock protein 20/alpha-crystallin family | −1.853351031 | |
| LVAN14669 | Heat shock protein 20/alpha-crystallin family | −1.839008177 | |
| LVAN16387 | Immunoglobulin I-set domain | 1.62053716 | |
| LVAN04048 | Immunoglobulin I-set domain | 1.716095306 | |
| LVAN04045 | Immunoglobulin I-set domain | 3.467470081 | |
| LVAN10981 | Immunoglobulin I-set domain | 1.13315602 | |
| LVAN19532 | Immunoglobulin I-set domain | 1.175834599 | |
| LVAN14378 | Immunoglobulin I-set domain | 1.8356356 | |
| LVAN12953 | Ras-like GTP-binding protein Rho1 | 1.946771427 | |
| LVAN25515 | Ras-related protein Rab-2A | 1.317082979 | |
| novel.5097 | NFX1-type zinc finger-containing protein 1 | 2.242125717 | |
| LVAN22842 | NFX1-type zinc finger-containing protein 1 | 1.523548395 | |
| LVAN00304 | NFX1-type zinc finger-containing protein 1 | 3.012657901 | |
| novel.1678 | Interferon-related developmental regulator 1 | −4.437000125 | |
| novel.1677 | Interferon-related developmental regulator 2 | −1.275543979 | |
| LVAN11504 | MFS-1 | −2.807489076 | |
| Protein synthesis-related | LVAN04287 | Glutamate–cysteine ligase catalytic subunit | −1.984075309 |
| LVAN14777 | Methionine synthase | −1.947132225 | |
| novel.4193 | Aspartate–tRNA ligase | −1.771238279 | |
| LVAN22403 | Eukaryotic translation initiation factor 2 subunit 1 | −1.674467809 | |
| novel.1790 | Tubulin beta-1 chain | −1.648057083 | |
| LVAN06976 | Glutaminyl–tRNA synthetase | −1.506118061 | |
| novel.2347 | E3 ubiquitin–protein ligase | −1.489605619 | |
| novel.1785 | Glutamic–pyruvic transaminase | −1.444998457 | |
| LVAN22801 | Serine/threonine–protein kinase | −1.278572371 | |
| LVAN07472 | Tyrosine–tRNA ligase | −1.25618501 | |
| novel.3677 | Serine/threonine–protein kinase | −1.193831772 | |
| LVAN13244 | Phenylalanine–tRNA ligase beta subunit | −1.114841361 | |
| novel.2206 | Valine–tRNA ligase | −1.072706294 | |
| LVAN15077 | Casein kinase II subunit | −1.026426438 | |
| Mitochondrial-related | LVAN00040 | Mitochondrial import inner membrane translocase subunit | −2.067899986 |
| LVAN06526 | Mitochondrial glycine transporter | −1.79981545 | |
| novel.716 | Mitochondrial pyruvate carrier 1 | −1.480260336 | |
| novel.2831 | Mitochondrial import receptor subunit | −1.018535966 | |
| novel.2485 | Mitochondrial-processing peptidase subunit alpha | −1.017153841 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xia, Y.; Zhong, X.; Zhang, X.; Zhang, X.; Yuan, J.; Liu, C.; Sha, Z.; Li, F. Gene Structure, Expression and Function Analysis of MEF2 in the Pacific White Shrimp Litopenaeus vannamei. Int. J. Mol. Sci. 2023, 24, 5832. https://doi.org/10.3390/ijms24065832
Xia Y, Zhong X, Zhang X, Zhang X, Yuan J, Liu C, Sha Z, Li F. Gene Structure, Expression and Function Analysis of MEF2 in the Pacific White Shrimp Litopenaeus vannamei. International Journal of Molecular Sciences. 2023; 24(6):5832. https://doi.org/10.3390/ijms24065832
Chicago/Turabian StyleXia, Yanting, Xiaoyun Zhong, Xiaoxi Zhang, Xiaojun Zhang, Jianbo Yuan, Chengzhang Liu, Zhenxia Sha, and Fuhua Li. 2023. "Gene Structure, Expression and Function Analysis of MEF2 in the Pacific White Shrimp Litopenaeus vannamei" International Journal of Molecular Sciences 24, no. 6: 5832. https://doi.org/10.3390/ijms24065832
APA StyleXia, Y., Zhong, X., Zhang, X., Zhang, X., Yuan, J., Liu, C., Sha, Z., & Li, F. (2023). Gene Structure, Expression and Function Analysis of MEF2 in the Pacific White Shrimp Litopenaeus vannamei. International Journal of Molecular Sciences, 24(6), 5832. https://doi.org/10.3390/ijms24065832





