The Drp1-Mediated Mitochondrial Fission Protein Interactome as an Emerging Core Player in Mitochondrial Dynamics and Cardiovascular Disease Therapy
Abstract
1. Introduction
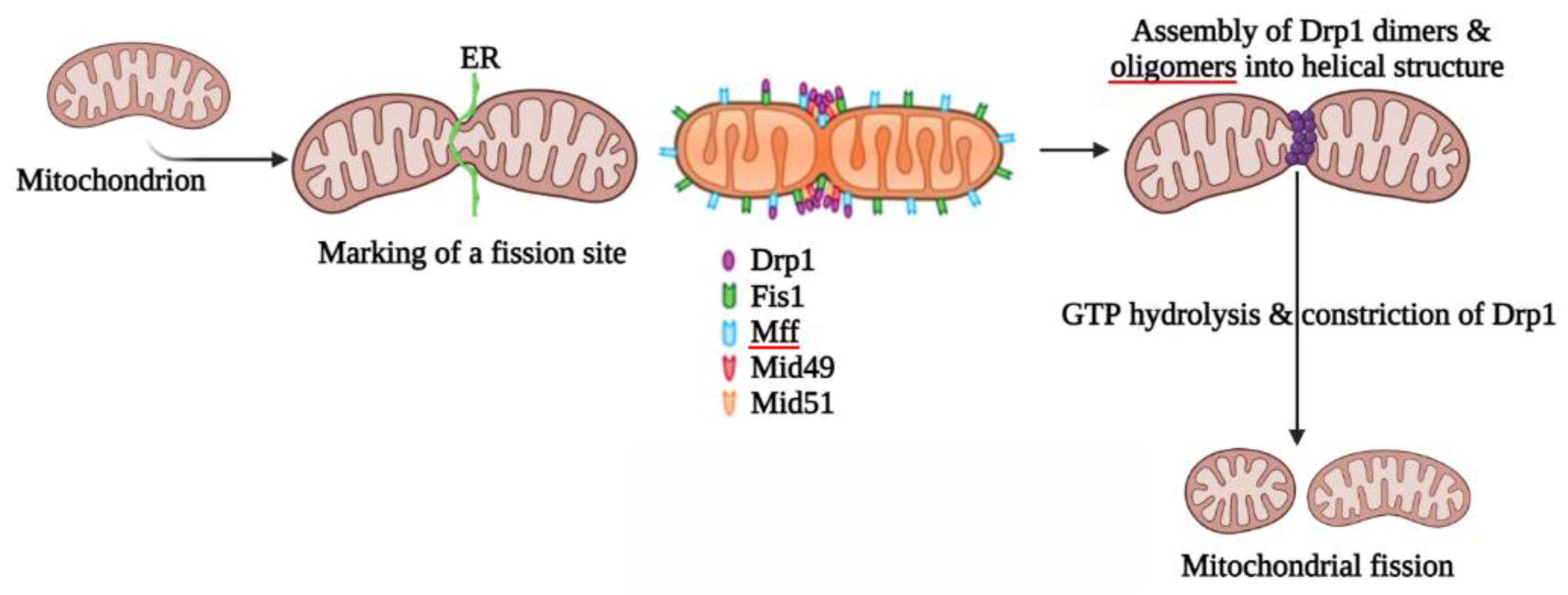
2. Mitochondrial Fission
2.1. Proteins Involved in Mitochondrial Fission
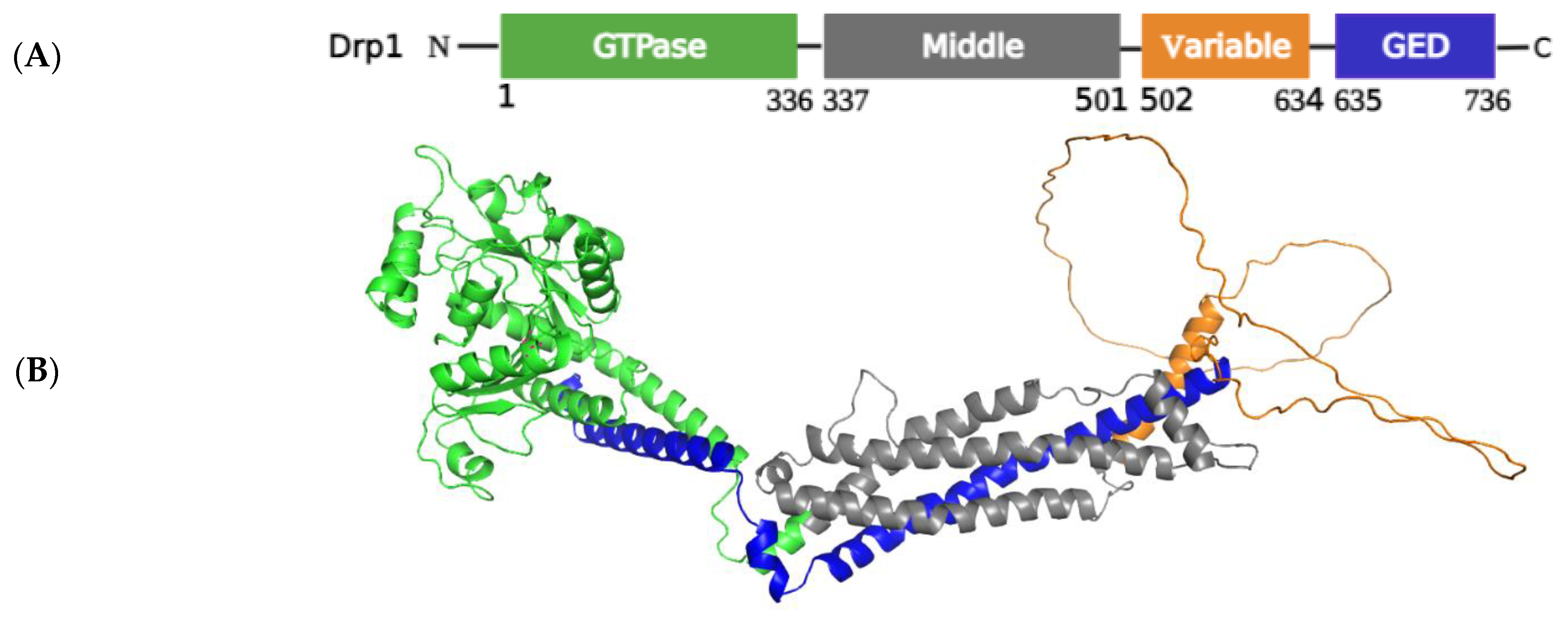
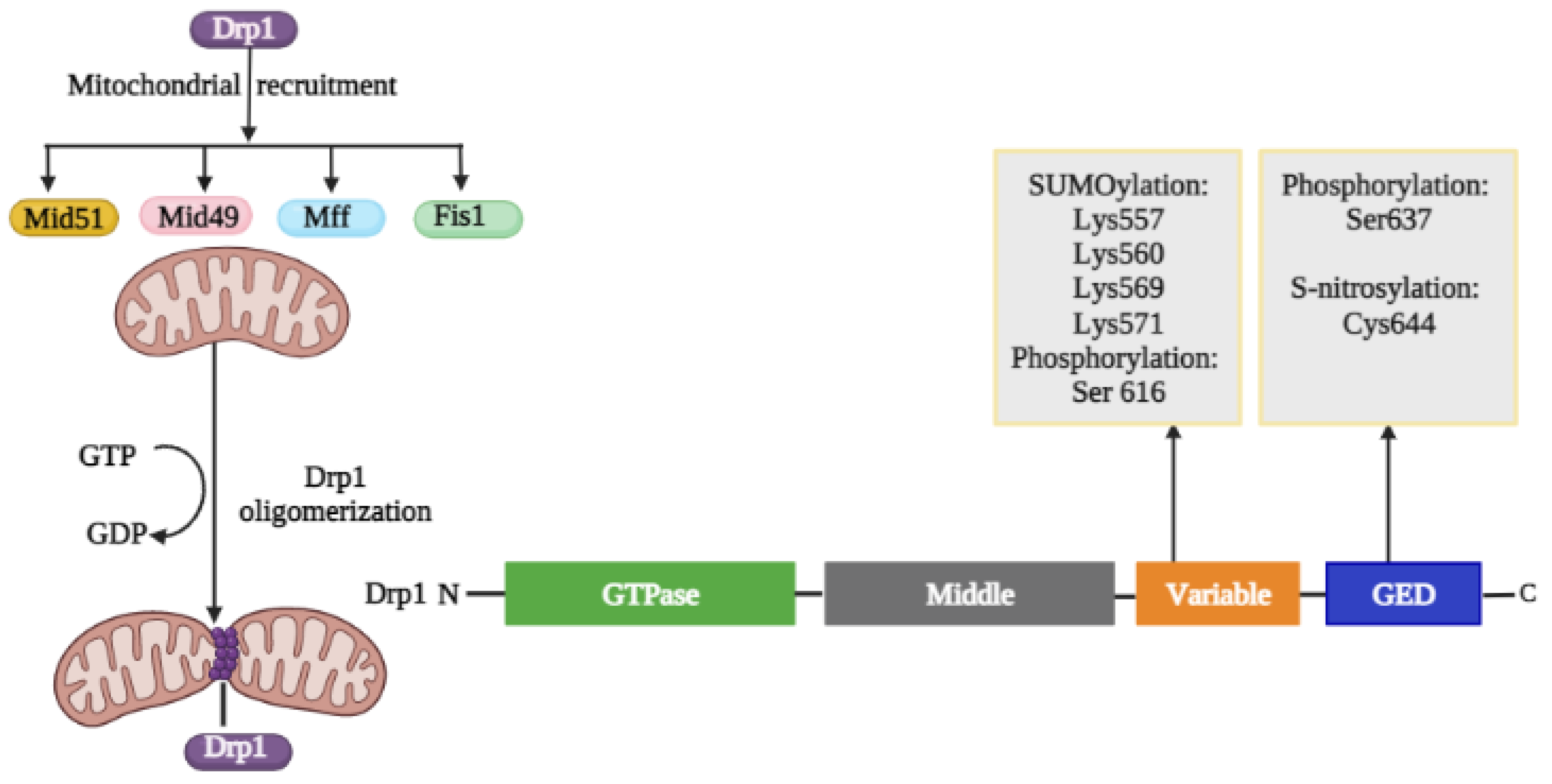
2.1.1. Structural Elements and Roles of Drp1 in Mitochondrial Fission
Regulation of Drp1 via Post-Translational Modifications
2.1.2. Structural Elements and Roles of Fis1 in Mitochondrial Fission
2.1.3. Structural Elements and Roles of Mff in Mitochondrial Fission
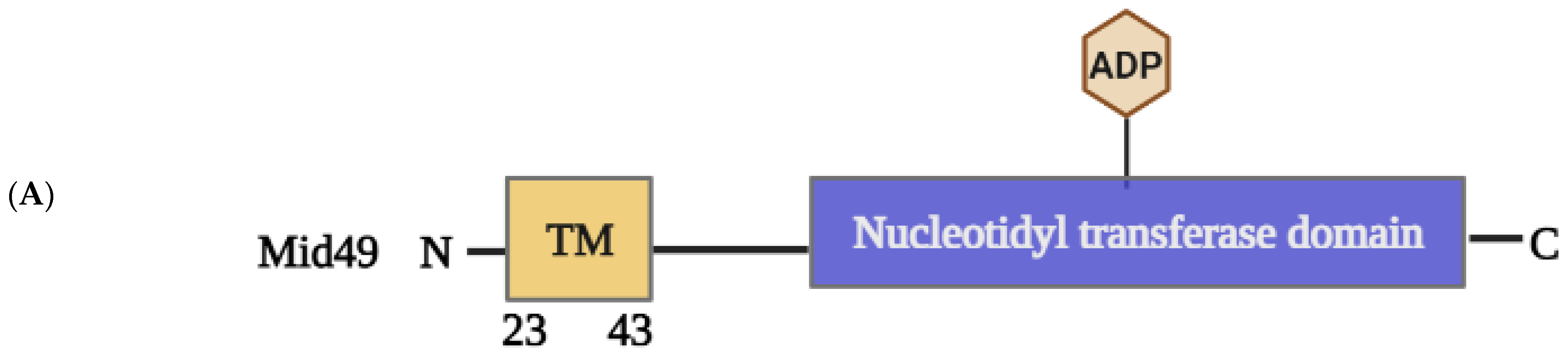
2.1.4. Structural Elements and Roles of Mid49 and Mid51 in Mitochondrial Fission
3. New Insights into Mitochondrial Fission Protein Interaction Networks
3.1. Role of Distribution and Oligomerization of Drp1 in PPI Networking
3.2. Lack of Membrane-Anchored Domains in Drp1 Facilitates its PPI Networking
3.3. Role of Mff in GTPase Activity of Drp1 and Oligomerization
3.4. Role of Actin Filaments in Drp1 Recruitment
3.5. Role of Drp1 Phosphorylation Status in Mitochondrial Fission-Mediated Interactome
3.6. Role of Fis1 in Drp1-Mediated Fission Networking
3.7. Mid51-Mediated Recruitment of Drp1
3.8. Role of Mid51 Interaction with Fis1 in Drp1-Induced Fission
3.9. Mid49/Mid51-Orchestrated Drp1-Mediated Fission
3.10. Mff-, Fis1-, Mid49-, and Mid51-Mediated Drp1 PPI Networking in Fission Regulation
4. Inhibition of Mitochondrial Fission Protein Interactions
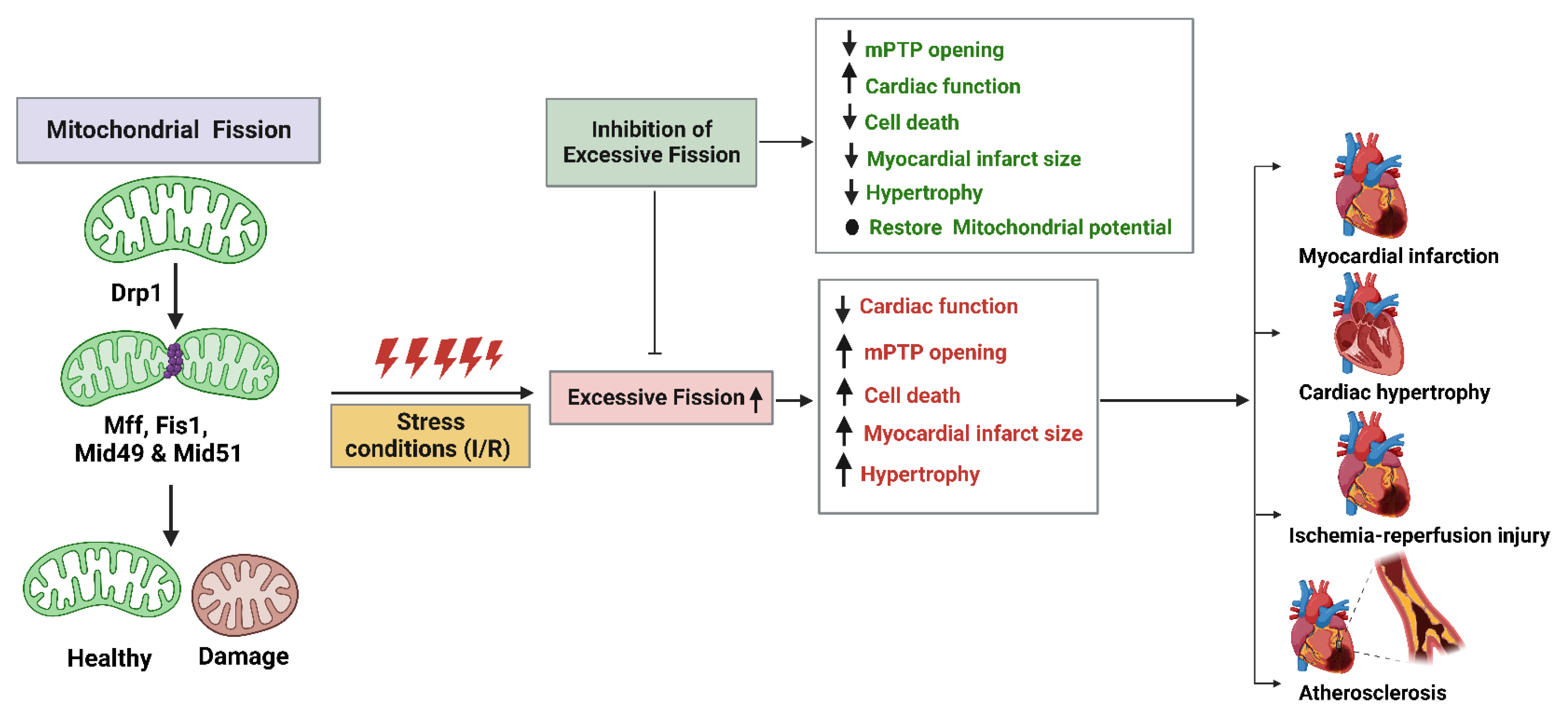
5. Mitochondrial Fission Proteins and Cardiovascular Diseases
5.1. Atherosclerosis
5.2. Cardiac Hypertrophy
5.3. Myocardial Infarction (MI)
5.4. Myocardial Ischemic/Reperfusion (I/R) Injury
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cogliati, S.; Enriquez, J.A.; Scorrano, L. Mitochondrial Cristae: Where Beauty Meets Functionality. Trends Biochem. Sci. 2016, 41, 261–273. [Google Scholar] [CrossRef]
- Formosa, L.E.; Ryan, M.T. Mitochondrial OXPHOS complex assembly lines. Nat. Cell Biol. 2018, 20, 511–513. [Google Scholar] [CrossRef]
- Tamura, Y.; Kawano, S.; Endo, T. Organelle contact zones as sites for lipid transfer. J. Biochem. 2019, 165, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Chial, H.; Craig, J. mtDNA and mitochondrial diseases. Nat. Educ. 2008, 1, 217. [Google Scholar]
- Detmer, S.A.; Chan, D.C. Functions and dysfunctions of mitochondrial dynamics. Nat. Rev. Mol. Cell Biol. 2007, 8, 870–879. [Google Scholar] [CrossRef]
- Mendl, N.; Occhipinti, A.; Müller, M.; Wild, P.; Dikic, I.; Reichert, A.S. Mitophagy in yeast is independent of mitochondrial fission and requires the stress response gene WHI2. J. Cell Sci. 2011, 124, 1339–1350. [Google Scholar] [CrossRef] [PubMed]
- Adaniya, S.M.; O-Uchi, J.; Cypress, M.W.; Kusakari, Y.; Jhun, B.S. Mitochondrial function and actin regulate dynamin-related protein 1-dependent mitochondrial fission. Curr. Biol. 2005, 15, 678–683. [Google Scholar]
- Adaniya, S.M.; O-Uchi, J.; Cypress, M.W.; Kusakari, Y.; Jhun, B.S. Posttranslational modifications of mitochondrial fission and fusion proteins in cardiac physiology and pathophysiology. Am. J. Physiol.-Cell Physiol. 2019, 316, C583–C604. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Chen, J.-X. Sirtuin 3, Endothelial Metabolic Reprogramming, and Heart Failure With Preserved Ejection Fraction. J. Cardiovasc. Pharmacol. 2019, 74, 315. [Google Scholar] [CrossRef]
- Serasinghe, M.N.; Chipuk, J.E. Mitochondrial Fission in Human Diseases. Pharmacol. Mitochondria 2016, 240, 159–188. [Google Scholar] [CrossRef]
- Cho, B.; Choi, S.Y.; Cho, H.M.; Kim, H.J.; Sun, H.J.K.A.W. Physiological and Pathological Significance of Dynamin-Related Protein 1 (Drp1)-Dependent Mitochondrial Fission in the Nervous System. Exp. Neurobiol. 2013, 22, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Seo, B.J.; Yoon, S.H.; Do, J.T. Mitochondrial Dynamics in Stem Cells and Differentiation. Int. J. Mol. Sci. 2018, 19, 3893. [Google Scholar] [CrossRef] [PubMed]
- Brown, G.C.; Murphy, M.P.; Scott, I.; Youle, R.J. Mitochondrial fission and fusion. Essays Biochem. 2010, 47, 85–98. [Google Scholar] [CrossRef]
- Labrousse, A.M.; Zappaterra, M.D.; Rube, D.A.; van der Bliek, A.M. C. elegans Dynamin-Related Protein DRP-1 Controls Severing of the Mitochondrial Outer Membrane. Mol. Cell 1999, 4, 815–826. [Google Scholar] [CrossRef]
- Bleazard, W.; McCaffery, J.M.; King, E.J.; Bale, S.; Mozdy, A.; Tieu, Q.; Nunnari, J.; Shaw, J.M. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat. Cell Biol. 1999, 1, 298–304. [Google Scholar] [CrossRef]
- Smirnova, E.; Griparic, L.; Shurland, D.-L.; van der Bliek, A.M.; Schiavon, C.R.; Turn, R.E.; Newman, L.E.; Kahn, R.A.; Spang, M.E.A.; Ganesan, V.; et al. Dynamin-related Protein Drp1 Is Required for Mitochondrial Division in Mammalian Cells. Mol. Biol. Cell 2001, 12, 2245–2256. [Google Scholar] [CrossRef] [PubMed]
- Kraus, F.; Ryan, M.T. The constriction and scission machineries involved in mitochondrial fission. J. Cell Sci. 2017, 130, 2953–2960. [Google Scholar] [CrossRef] [PubMed]
- Koch, A.; Thiemann, M.; Grabenbauer, M.; Yoon, Y.; McNiven, M.A.; Schrader, M. Dynamin-like Protein 1 Is Involved in Peroxisomal Fission. J. Biol. Chem. 2003, 278, 8597–8605. [Google Scholar] [CrossRef]
- Ishihara, N.; Nomura, M.; Jofuku, A.; Kato, H.; Suzuki, S.O.; Masuda, K.; Otera, H.; Nakanishi, Y.; Nonaka, I.; Goto, Y.-I.; et al. Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nat. Cell Biol. 2009, 11, 958–966. [Google Scholar] [CrossRef]
- Ji, W.K.; Hatch, A.L.; Merrill, R.A.; Strack, S.; Higgs, H.N. Actin filaments target the oligomeric maturation of the dynamin GTPase Drp1 to mitochondrial fission sites. eLife 2015, 4, e11553. [Google Scholar] [CrossRef]
- Friedman, J.R.; Lackner, L.L.; West, M.; DiBenedetto, J.R.; Nunnari, J.; Voeltz, G.K. ER Tubules Mark Sites of Mitochondrial Division. Science 2011, 334, 358–362. [Google Scholar] [CrossRef]
- Hu, C.; Huang, Y.; Li, L. Drp1-dependent mitochondrial fission plays critical roles in physiological and pathological progresses in mammals. Int. J. Mol. Sci. 2017, 18, 144. [Google Scholar] [CrossRef] [PubMed]
- Otera, H.; Mihara, K. Discovery of the membrane receptor for mitochondrial fission GTPase Drp1. Small GTPases 2011, 2, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Giacomello, M.; Pyakurel, A.; Glytsou, C.; Scorrano, L. The cell biology of mitochondrial membrane dynamics. Nat. Rev. Mol. Cell Biol. 2020, 21, 204–224. [Google Scholar] [CrossRef]
- Atkins, K.; Dasgupta, A.; Chen, K.-H.; Mewburn, J.; Archer, S.L. The role of Drp1 adaptor proteins MiD49 and MiD51 in mitochondrial fission: Implications for human disease. Clin. Sci. 2016, 130, 1861–1874. [Google Scholar] [CrossRef]
- Korobova, F.; Ramabhadran, V.; Higgs, H.N. An Actin-Dependent Step in Mitochondrial Fission Mediated by the ER-Associated Formin INF2. Science 2013, 339, 464–467. [Google Scholar] [CrossRef] [PubMed]
- Bera, A.; Lavanya, G.; Reshmi, R.; Dev, K.; Kumar, R. Mechanistic and therapeutic role of Drp1 in the pathogenesis of Alzheimer’s disease. Eur. J. Neurosci. 2022, 56, 5516–5531. [Google Scholar] [CrossRef] [PubMed]
- Bamshad, C.; Najafi-Ghalehlou, N.; Pourmohammadi-Bejarpasi, Z.; Tomita, K.; Kuwahara, Y.; Sato, T.; Feizkhah, A.; Roushnadeh, A.M.; Roudkenar, M.H. Mitochondria: How eminent in ageing and neurodegenerative disorders? Hum. Cell 2023, 36, 41–61. [Google Scholar] [CrossRef] [PubMed]
- Baltz, A.G.; Munschauer, M.; Schwanhäusser, B.; Vasile, A.; Murakawa, Y.; Schueler, M.; Youngs, N.; Penfold-Brown, D.; Drew, K.; Milek, M.; et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell 2012, 46, 674–690. [Google Scholar] [CrossRef]
- Gaudet, P.; Livstone, M.S.; Lewis, S.E.; Thomas, P.D. Phylogenetic-based propagation of functional annotations within the Gene Ontology consortium. Brief. Bioinform. 2011, 12, 449–462. [Google Scholar] [CrossRef]
- A Kenniston, J.; A Lemmon, M. Dynamin GTPase regulation is altered by PH domain mutations found in centronuclear myopathy patients. EMBO J. 2010, 29, 3054–3067. [Google Scholar] [CrossRef] [PubMed]
- Yonashiro, R.; Ishido, S.; Kyo, S.; Fukuda, T.; Goto, E.; Matsuki, Y.; Ohmura-Hoshino, M.; Sada, K.; Hotta, H.; Yamamura, H.; et al. A novel mitochondrial ubiquitin ligase plays a critical role in mitochondrial dynamics. EMBO J. 2006, 25, 3618–3626. [Google Scholar] [CrossRef] [PubMed]
- Imoto, M.; Tachibana, I.; Urrutia, R. Identification and functional characterization of a novel human protein highly related to the yeast dynamin-like GTPase Vps1p. J. Cell Sci. 1998, 111, 1341–1349. [Google Scholar] [CrossRef] [PubMed]
- Reubold, T.F.; Faelber, K.; Plattner, N.; Posor, Y.; Ketel, K.; Curth, U.; Schlegel, J.; Anand, R.; Manstein, D.J.; Noé, F.; et al. Crystal structure of the dynamin tetramer. Nature 2015, 525, 404–408. [Google Scholar] [CrossRef]
- Gandre-Babbe, S.; van der Bliek, A.M. The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol. Biol. Cell 2008, 19, 2402–2412. [Google Scholar] [CrossRef]
- Serasinghe, M.N.; Yoon, Y. The mitochondrial outer membrane protein hFis1 regulates mitochondrial morphology and fission through self-interaction. Exp. Cell Res. 2008, 314, 3494–3507. [Google Scholar] [CrossRef]
- Niemann, A.; Ruegg, M.; La Padula, V.; Schenone, A.; Suter, U. Ganglioside-induced differentiation associated protein 1 is a regulator of the mitochondrial network: New implications for Charcot-Marie-Tooth disease. J. Cell Biol. 2005, 170, 1067–1078. [Google Scholar] [CrossRef]
- Pedrola, L.; Espert, A.; Wu, X.; Claramunt, R.; Shy, M.; Palau, F. GDAP1, the protein causing Charcot–Marie–Tooth disease type 4A, is expressed in neurons and is associated with mitochondria. Hum. Mol. Genet. 2005, 14, 1087–1094. [Google Scholar] [CrossRef]
- Shaw, J.M.; Nunnari, J. Mitochondrial dynamics and division in budding yeast. Trends Cell Biol. 2002, 12, 178–184. [Google Scholar] [CrossRef]
- Zeng, L.; Xiao, Q.; Chen, M.; Margariti, A.; Martin, D.; Ivetic, A.; Xu, H.; Mason, J.; Wang, W.; Cockerill, G.; et al. Vascular Endothelial Cell Growth–Activated XBP1 Splicing in Endothelial Cells Is Crucial for Angiogenesis. Circulation 2013, 127, 1712–1722. [Google Scholar] [CrossRef]
- Yamana, S.; Tokiyama, A.; Mizutani, K.; Hirata, K.I.; Takai, Y.; Rikitake, Y. The cell adhesion molecule Necl-4/CADM4 serves as a novel regulator for contact inhibition of cell movement and proliferation. PLoS ONE 2015, 10, e0124259. [Google Scholar] [CrossRef] [PubMed]
- Kendall, R.L.; Rutledge, R.Z.; Mao, X.; Tebben, A.J.; Hungate, R.W.; Thomas, K.A. Vascular endothelial growth factor receptor KDR tyrosine kinase activity is increased by autophosphorylation of two activation loop tyrosine residues. J. Biol. Chem. 1999, 274, 6453–6460. [Google Scholar] [CrossRef] [PubMed]
- Dougher, M.; I Terman, B. Autophosphorylation of KDR in the kinase domain is required for maximal VEGF-stimulated kinase activity and receptor internalization. Oncogene 1999, 18, 1619–1627. [Google Scholar] [CrossRef] [PubMed]
- Khayati, F.; Pérez-Cano, L.; Maouche, K.; Sadoux, A.; Boutalbi, Z.; Podgorniak, M.-P.; Maskos, U.; Setterblad, N.; Janin, A.; Calvo, F.; et al. EMMPRIN/CD147 is a novel coreceptor of VEGFR-2 mediating its activation by VEGF. Oncotarget 2015, 6, 9766–9780. [Google Scholar] [CrossRef] [PubMed]
- De Stefani, D.; Raffaello, A.; Teardo, E.; Szabò, I.; Rizzuto, R. A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature 2011, 476, 336–340. [Google Scholar] [CrossRef]
- Baughman, J.M.; Perocchi, F.; Girgis, H.S.; Plovanich, M.; Belcher-Timme, C.A.; Sancak, Y.; Bao, X.R.; Strittmatter, L.; Goldberger, O.; Bogorad, R.L.; et al. Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature 2011, 476, 341–345. [Google Scholar] [CrossRef]
- Luck, K.; Kim, D.-K.; Lambourne, L.; Spirohn, K.; Begg, B.E.; Bian, W.; Brignall, R.; Cafarelli, T.; Campos-Laborie, F.J.; Charloteaux, B.; et al. A reference map of the human binary protein interactome. Nature 2020, 580, 402–408. [Google Scholar] [CrossRef]
- Koirala, S.; Guo, Q.; Kalia, R.; Bui, H.T.; Eckert, D.M.; Frost, A.; Shaw, J.M. Interchangeable adaptors regulate mitochondrial dynamin assembly for membrane scission. Proc. Natl. Acad. Sci. USA 2013, 110, E1342–E1351. [Google Scholar] [CrossRef]
- Richter, V.; Palmer, C.; Osellame, L.D.; Singh, A.P.; Elgass, K.; Stroud, D.; Sesaki, H.; Kvansakul, M.; Ryan, M.T. Structural and functional analysis of MiD51, a dynamin receptor required for mitochondrial fission. J. Cell Biol. 2014, 204, 477–486. [Google Scholar] [CrossRef]
- Zhao, J.; Liu, T.; Jin, S.; Wang, X.; Qu, M.; Uhlén, P.; Tomilin, N.; Shupliakov, O.; Lendahl, U.; Nistér, M. Human MIEF1 recruits Drp1 to mitochondrial outer membranes and promotes mitochondrial fusion rather than fission. EMBO J. 2011, 30, 2762–2778. [Google Scholar] [CrossRef]
- Palmer, C.S.; Osellame, L.D.; Laine, D.; Koutsopoulos, O.S.; Frazier, A.E.; Ryan, M.T. MiD49 and MiD51, new components of the mitochondrial fission machinery. EMBO Rep. 2011, 12, 565–573. [Google Scholar] [CrossRef] [PubMed]
- Losón, O.C.; Song, Z.; Chen, H.; Chan, D.C. Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol. Biol. Cell 2013, 24, 659–667. [Google Scholar] [CrossRef] [PubMed]
- Bartsakoulia, M.; Pyle, A.; Troncoso-Chandía, D.; Vial-Brizzi, J.; Paz-Fiblas, M.V.; Duff, J.; Griffin, H.; Boczonadi, V.; Lochmüller, H.; Kleinle, S.; et al. A novel mechanism causing imbalance of mitochondrial fusion and fission in human myopathies. Hum. Mol. Genet. 2018, 27, 1186–1195. [Google Scholar] [CrossRef]
- Tondera, D.; Czauderna, F.; Paulick, K.; Schwarzer, R.; Kaufmann, J.; Santel, A. The mitochondrial protein MTP18 contributes to mitochondrial fission in mammalian cells. J. Cell Sci. 2005, 118, 3049–3059. [Google Scholar] [CrossRef]
- Kroemer, G.; Galluzzi, L.; Vandenabeele, P.; Abrams, J.; Alnemri, E.S.; Baehrecke, E.H.; Blagosklonny, M.V.; El-Deiry, W.S.; Golstein, P.; Green, D.R.; et al. Classification of cell death: Recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ. 2009, 16, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Pashkova, N.; Gakhar, L.; Winistorfer, S.C.; Yu, L.; Ramaswamy, S.; Piper, R.C. WD40 Repeat Propellers Define a Ubiquitin-Binding Domain that Regulates Turnover of F Box Proteins. Mol. Cell 2010, 40, 433–443. [Google Scholar] [CrossRef] [PubMed]
- Strack, S.; Cribbs, J.T. Allosteric Modulation of Drp1 Mechanoenzyme Assembly and Mitochondrial Fission by the Variable Domain. J. Biol. Chem. 2012, 287, 10990–11001. [Google Scholar] [CrossRef]
- Fröhlich, C.; Grabiger, S.; Schwefel, D.; Faelber, K.; Rosenbaum, E.; Mears, J.; Rocks, O.; Daumke, O. Structural insights into oligomerization and mitochondrial remodelling of dynamin 1-like protein. EMBO J. 2013, 32, 1280–1292. [Google Scholar] [CrossRef]
- Kishida, H.; Sugio, S. Crystal structure of GTPase domain fused with minimal stalks from human dynamin-1-like protein (Dlp1) in complex with several nucleotide analogues. Curr. Top. Pept. Protein Res. 2013, 14, 67–77. [Google Scholar]
- Ramachandran, R.; Schmid, S.L. The dynamin superfamily. Curr. Biol. 2018, 28, R411–R416. [Google Scholar] [CrossRef]
- Joshi, A.U.; Saw, N.L.; Shamloo, M.; Mochly-Rosen, D. Drp1/Fis1 interaction mediates mitochondrial dysfunction, bioenergetic failure and cognitive decline in Alzheimer’s disease. Oncotarget 2018, 9, 6128. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.-Y.; Wei, X.-X.; Zhi, X.-L.; Wang, X.-H.; Meng, D. Drp1-dependent mitochondrial fission in cardiovascular disease. Acta Pharmacol. Sin. 2021, 42, 655–664. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.; Liu, T.; Jin, S.B.; Ning, C.; Lendahl, U.; Nistér, M.; Zhao, J. MIEF1/2 function as adaptors to recruit Drp1 to mitochondria and regulate the association of Drp1 with Mff. Sci. Rep. 2017, 7, 880. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger. PyMOL. 2020. Available online: http://www.pymol.org/pymol (accessed on 5 November 2022).
- Peng, Y.; Liu, H.; Liu, J.; Long, J. Post-translational modifications on mitochondrial metabolic enzymes in cancer. Free. Radic. Biol. Med. 2022, 179, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Qi, Z.; Huang, Z.; Xie, F.; Chen, L. Dynaminrelated protein 1: A critical protein in the pathogenesis of neural system dysfunctions and neurodegen-erative diseases. J. Cell. Physiol. 2019, 234, 10032–10046. [Google Scholar] [CrossRef]
- Soundararajan, R.; Hernández-Cuervo, H.; Stearns, T.M.; Griswold, A.J.; Patil, S.S.; Fukumoto, J.; Narala, V.R.; Galam, L.; Lockey, R.; Kolliputi, N. A-Kinase Anchor Protein 1 deficiency causes mitochondrial dysfunction in mouse model of hyperoxia induced acute lung injury. Front. Pharmacol. 2022, 13, 980723. [Google Scholar] [CrossRef]
- Cribbs, J.T.; Strack, S. Reversible phosphorylation of Drp1 by cyclic AMP-dependent protein kinase and calcineurin regulates mitochondrial fission and cell death. EMBO Rep. 2007, 8, 939–944. [Google Scholar] [CrossRef]
- Chang, C.-R.; Blackstone, C. Dynamic regulation of mitochondrial fission through modification of the dynamin-related protein Drp1. Ann. N. Y. Acad. Sci. 2010, 1201, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Kandimalla, R.; Reddy, P.H. Multiple faces of dynamin-related protein 1 and its role in Alzheimer’s disease pathogenesis. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2016, 1862, 814–828. [Google Scholar] [CrossRef]
- Bo, T.; Yamamori, T.; Suzuki, M.; Sakai, Y.; Yamamoto, K.; Inanami, O. Calmodulin-dependent protein kinase II (CaMKII) mediates radiation-induced mitochondrial fission by regulating the phosphorylation of dynamin-related protein 1 (Drp1) at serine 616. Biochem. Biophys. Res. Commun.-Tions 2018, 495, 1601–1607. [Google Scholar] [CrossRef] [PubMed]
- Brand, C.S.; Tan, V.P.; Brown, J.H.; Miyamoto, S. RhoA regulates Drp1 mediated mitochondrial fission through ROCK to protect cardiomyocytes. Cell. Signal. 2018, 50, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Guo, S.; Tang, Y.; Mou, C.; Hu, X.; Shao, F.; Yan, W.; Wu, Q. Mitochondrial Fusion and Fission in Neuronal Death Induced by Cerebral Ischemia-Reperfusion and Its Clinical Application: A Mini-Review. Med. Sci. Monit. 2020, 26, e928651. [Google Scholar] [CrossRef] [PubMed]
- Li, H.B.; Wang, R.X.; Jiang, H.B.; Zhang, E.D.; Tan, J.Q.; Xu, H.Z.; Zhou, R.R.; Xia, X.B. Mitochondrial ribosomal protein L10 associates with cyclin B1/Cdk1 activity and mitochondrial function. DNA Cell Biol. 2016, 35, 680–690. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Nakamura, T.; Lipton, S.A. Redox reactions induced by nitrosative stress mediate protein misfolding and mitochondrial dysfunction in neurodegenerative diseases. Mol. Neurobiol. 2010, 41, 55–72. [Google Scholar] [CrossRef]
- Haun, F.; Nakamura, T.; Shiu, A.D.; Cho, D.H.; Tsunemi, T.; Holland, E.A.; La Spada, A.R.; Lipton, S.A. S-nitrosylation of dynamin-related protein 1 mediates mutant huntingtin-induced mitochondrial fragmentation and neuronal injury in Huntington’s disease. Antioxid. Redox Signal. 2013, 19, 1173–1184. [Google Scholar] [CrossRef]
- Bossy, B.; Petrilli, A.; Klinglmayr, E.; Chen, J.; Lütz-Meindl, U.; Knott, A.B.; Masliah, E.; Schwarzenbacher, R.; Bossy-Wetzel, E. S-Nitrosylation of DRP1 Does Not Affect Enzymatic Activity and is Not Specific to Alzheimer’s Disease. J. Alzheimer’s Dis. 2010, 20, S513–S526. [Google Scholar] [CrossRef]
- Pagliuso, A.; Cossart, P.; Stavru, F. The ever-growing complexity of the mitochondrial fission machinery. Cell. Mol. Life Sci. 2018, 75, 355–374. [Google Scholar] [CrossRef]
- Wang, H.; Song, P.; Du, L.; Tian, W.; Yue, W.; Liu, M.; Li, D.; Wang, B.; Zhu, Y.; Cao, C.; et al. Parkin ubiquitinates Drp1 for proteasome-dependent degradation: Implication of dysregulated mitochondrial dynamics in Parkinson disease. J. Biol. Chem. 2011, 286, 11649–11658. [Google Scholar] [CrossRef]
- Williams, J.A. The Role of Parkin and Mitophagy in Acetaminophen and Alcohol-Induced Liver Injuries. Ph.D. Thesis, University of Kansas, Lawrence, KS, USA, 2015. [Google Scholar]
- Recasens Ibabe, A. Initiation, Progression and Extension of Parkinson’s Disease: Role of α-synuclein. Ph.D. Thesis, Universitat Autònoma de Barcelona, Barcelona, Spain, 2014. [Google Scholar]
- Cherok, E.; Xu, S.; Li, S.; Das, S.; Meltzer, W.A.; Zalzman, M.; Wang, C.; Karbowski, M. Novel regulatory roles of Mff and Drp1 in E3 ubiquitin ligase MARCH5–dependent degradation of MiD49 and Mcl1 and control of mitochondrial dynamics. Mol. Biol. Cell 2017, 28, 396–410. [Google Scholar] [CrossRef]
- Karbowski, M.; Neutzner, A.; Youle, R.J. The mitochondrial E3 ubiquitin ligase MARCH5 is required for Drp1 dependent mitochondrial division. J. Cell Biol. 2007, 178, 71–84. [Google Scholar] [CrossRef]
- Wolf, M.A. Analysis of p97-mediated Ku80 Extraction in DNA Double-Strand Break Repair in Cells. Ph.D. Thesis, University of Duisburg-Essen, Duisburg, Germany, 2021. [Google Scholar]
- Mendler, L.; Braun, T.; Müller, S. The ubiquitin-like SUMO system and heart function: From development to disease. Circ. Res. 2016, 118, 132–144. [Google Scholar] [CrossRef] [PubMed]
- Camoes, F.; Bonekamp, N.A.; Delille, H.K.; Schrader, M. Organelle dynamics and dysfunction: A closer link between peroxisomes and mitochondria. J. Inherit. Metab. Dis. 2009, 32, 163–180. [Google Scholar] [CrossRef]
- Yoon, Y.; Krueger, E.W.; Oswald, B.J.; McNiven, M.A. The Mitochondrial Protein hFis1 Regulates Mitochondrial Fission in Mammalian Cells through an Interaction with the Dynamin-Like Protein DLP1. Mol. Cell. Biol. 2003, 23, 5409–5420. [Google Scholar] [CrossRef] [PubMed]
- Tieu, Q.; Nunnari, J. Mdv1p Is a Wd Repeat Protein That Interacts with the Dynamin-Related Gtpase, Dnm1p, to Trigger Mitochondrial Division. J. Cell Biol. 2000, 151, 353–366. [Google Scholar] [CrossRef]
- Yang, F.; Moss, L.G.; Phillips, G.N., Jr. The molecular structure of green fluorescent protein. Nat. Biotechnol. 1996, 14, 1246–1251. [Google Scholar] [CrossRef]
- Zhao, J.; Lendahl, U.; Nistér, M. Regulation of mitochondrial dynamics: Convergences and divergences between yeast and vertebrates. Cell. Mol. Life Sci. 2013, 70, 951–976. [Google Scholar] [CrossRef] [PubMed]
- Yoon, Y.; Pitts, K.R.; McNiven, M.A. Mammalian Dynamin-like Protein DLP1 Tubulates Membranes. Mol. Biol. Cell 2001, 12, 2894–2905. [Google Scholar] [CrossRef] [PubMed]
- Dohm, J.A.; Lee, S.J.; Hardwick, J.M.; Hill, R.B.; Gittis, A.G. Cytosolic domain of the human mitochondrial fission protein fis1 adopts a TPR fold. Proteins Struct. Funct. Bioinform. 2004, 54, 153–156. [Google Scholar] [CrossRef] [PubMed]
- Scheel, H.; Hofmann, K. Prediction of a common structural scaffold for proteasome lid, COP9-signalosome and eIF3 complexes. BMC Bioinform. 2005, 6, 71. [Google Scholar] [CrossRef]
- Griffin, E.E.; Graumann, J.; Chan, D.C. The WD40 protein Caf4p is a component of the mitochondrial fission machinery and recruits Dnm1p to mitochondria. J. Cell Biol. 2005, 170, 237–248. [Google Scholar] [CrossRef]
- Yu, R.; Lendahl, U.; Nistér, M.; Zhao, J. Regulation of Mammalian Mitochondrial Dynamics: Opportunities and Challenges. Front. Endocrinol. 2020, 11, 374. [Google Scholar] [CrossRef] [PubMed]
- Xian, H.; Liou, Y.-C. Functions of outer mitochondrial membrane proteins: Mediating the crosstalk between mitochondrial dynamics and mitophagy. Cell Death Differ. 2021, 28, 827–842. [Google Scholar] [CrossRef] [PubMed]
- Alirol, E. Role of hFis1 in Mitochondrial Fission and Apoptosis. Ph.D. Thesis, University of Geneva, Geneva, Switzerland, 2006. [Google Scholar] [CrossRef]
- Huybrechts, S. Peroxisome Dynamics in Mammalian Cells: A Fluorescence-Based Approach; Leuven University Press: Leuven, Belgium, 2010. [Google Scholar]
- Dar, B.A.; Lone, A.M.; Shah, W.A.; Qurishi, M.A. Synthesis and screening of ursolic acid-benzylidine derivatives as potential anti-cancer agents. Eur. J. Med. Chem. 2016, 111, 26–32. [Google Scholar] [CrossRef]
- Yu, R.; Liu, T.; Ning, C.; Tan, F.; Jin, S.-B.; Lendahl, U.; Zhao, J.; Nistér, M. The phosphorylation status of Ser-637 in dynamin-related protein 1 (Drp1) does not determine Drp1 recruitment to mitochondria. J. Biol. Chem. 2019, 294, 17262–17277. [Google Scholar] [CrossRef] [PubMed]
- Palmer, C.S.; Elgass, K.D.; Parton, R.G.; Osellame, L.D.; Stojanovski, D.; Ryan, M.T. Adaptor Proteins MiD49 and MiD51 Can Act Independently of Mff and Fis1 in Drp1 Recruitment and Are Specific for Mitochondrial Fission. J. Biol. Chem. 2013, 288, 27584–27593. [Google Scholar] [CrossRef]
- Wang, L.; Tu, Z.; Sun, F. A network-based integrative approach to prioritize reliable hits from multiple genome-wide RNAi screens in Drosophila. BMC Genom. 2009, 10, 220. [Google Scholar] [CrossRef]
- Otera, H.; Ishihara, N.; Mihara, K. New insights into the function and regulation of mitochondrial fission. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2013, 1833, 1256–1268. [Google Scholar] [CrossRef]
- Liu, R.; Chan, D.C. The mitochondrial fission receptor Mff selectively recruits oligomerized Drp1. Mol. Biol. Cell 2015, 26, 4466–4477. [Google Scholar] [CrossRef]
- Liu, T.; Yu, R.; Jin, S.B.; Han, L.; Lendahl, U.; Zhao, J.; Nistér, M. The mitochondrial elongation factors MIEF1 and MIEF2 exert partially distinct functions in mitochondrial dynamics. Exp. Cell Res. 2013, 319, 2893–2904. [Google Scholar] [CrossRef]
- Osellame, L.D.; Singh, A.P.; Stroud, D.A.; Palmer, C.S.; Stojanovski, D.; Ramachandran, R.; Ryan, M.T. Cooperative and independent roles of the Drp1 adaptors Mff, MiD49 and MiD51 in mitochondrial fission. J. Cell Sci. 2016, 129, 2170–2181. [Google Scholar]
- Kalia, R.; Wang, R.Y.-R.; Yusuf, A.; Thomas, P.V.; Agard, D.A.; Shaw, J.M.; Frost, A. Structural basis of mitochondrial receptor binding and constriction by DRP1. Nature 2018, 558, 401–405. [Google Scholar] [CrossRef] [PubMed]
- Losón, O.C.; Meng, S.; Ngo, H.; Liu, R.; Kaiser, J.T.; Chan, D.C. Crystal structure and functional analysis of MiD49, a receptor for the mitochondrial fission protein Drp1. Protein Sci. 2015, 24, 386–394. [Google Scholar] [CrossRef] [PubMed]
- Losón, O.C.; Liu, R.; Rome, M.E.; Meng, S.; Kaiser, J.T.; Shan, S.-O.; Chan, D.C. The Mitochondrial Fission Receptor MiD51 Requires ADP as a Cofactor. Structure 2014, 22, 367–377. [Google Scholar] [CrossRef]
- Basu, K.; Lajoie, D.; Aumentado-Armstrong, T.; Chen, J.; Koning, R.I.; Bossy, B.; Bostina, M.; Sik, A.; Bossy-Wetzel, E.; Rouiller, I. Molecular mechanism of DRP1 assembly studied in vitro by cryo-electron microscopy. PLoS ONE 2017, 12, e0179397. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
- Michalska, B.M.; Kwapiszewska, K.; Szczepanowska, J.; Kalwarczyk, T.; Patalas-Krawczyk, P.; Szczepański, K.; Hołyst, R.; Duszyński, J.; Szymański, J. Insight into the fission mechanism by quantitative characterization of Drp1 protein distribution in the living cell. Sci. Rep. 2018, 8, 8122. [Google Scholar] [CrossRef]
- Schmidt, O.; Pfanner, N.; Meisinger, C. Mitochondrial protein import: From proteomics to functional mechanisms. Nat. Rev. Mol. Cell Biol. 2010, 11, 655–667. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Gao, Y.; Zhang, C.; Li, H.; Pan, S.; Wang, X.; Du, S.; Deng, Z.; Wang, L.; Song, Z.; et al. SAMM50 Affects Mitochondrial Morphology through the Association of Drp1 in Mammalian Cells. FEBS Lett. 2016, 590, 1313–1323. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, L.; Wu, S.; Xing, D. Drp1, Mff, Fis1, and MiD51 are coordinated to mediate mitochondrial fission during UV irradiation-induced apoptosis. FASEB J. 2016, 30, 466–476. [Google Scholar] [CrossRef]
- Koch, A.; Yoon, Y.; Bonekamp, N.A.; McNiven, M.A.; Schrader, M. A Role for Fis1 in Both Mitochondrial and Peroxisomal Fission in Mammalian Cells. Mol. Biol. Cell 2005, 16, 5077–5086. [Google Scholar] [CrossRef]
- Clinton, R.W.; Francy, C.A.; Ramachandran, R.; Qi, X.; Mears, J.A. Dynamin-related Protein 1 Oligomerization in Solution Impairs Functional Interactions with Membrane-anchored Mitochondrial Fission Factor. J. Biol. Chem. 2016, 291, 478–492. [Google Scholar] [CrossRef] [PubMed]
- Lu, B.; Kennedy, B.; Clinton, R.W.; Wang, E.J.; McHugh, D.; Stepanyants, N.; Macdonald, P.J.; Mears, J.A.; Qi, X.; Ramachandran, R. Steric interference from intrinsically disordered regions controls dynamin-related protein 1 self-assembly during mitochondrial fission. Sci. Rep. 2018, 8, 10879. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Kage, F.; Higgs, H.N. Mff oligomerization is required for Drp1 activation and synergy with actin filaments during mitochondrial division. Mol. Biol. Cell 2021, 32, ar5. [Google Scholar] [CrossRef] [PubMed]
- Otera, H.; Wang, C.; Cleland, M.M.; Setoguchi, K.; Yokota, S.; Youle, R.J.; Mihara, K. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J. Cell Biol. 2010, 191, 1141–1158. [Google Scholar] [CrossRef]
- Hatch, A.L.; Ji, W.-K.; Merrill, R.A.; Strack, S.; Higgs, H.N. Actin filaments as dynamic reservoirs for Drp1 recruitment. Mol. Biol. Cell 2016, 27, 3109–3121. [Google Scholar] [CrossRef]
- Valera-Alberni, M.; Joffraud, M.; Miro-Blanch, J.; Capellades, J.; Junza, A.; Dayon, L.; Galindo, A.N.; Sanchez-Garcia, J.L.; Valsesia, A.; Cercillieux, A.; et al. Crosstalk between Drp1 phosphorylation sites during mitochondrial remodeling and their impact on metabolic adaptation. Cell Rep. 2021, 36, 109565. [Google Scholar] [CrossRef]
- Joshi, A.U.; Saw, N.L.; Vogel, H.; Cunnigham, A.D.; Shamloo, M.; Mochly-Rosen, D. Inhibition of Drp1/Fis1 interaction slows progression of amyotrophic lateral sclerosis. EMBO Mol. Med. 2018, 10, e8166. [Google Scholar] [CrossRef]
- Suzuki, M.; Jeong, S.-Y.; Karbowski, M.; Youle, R.J.; Tjandra, N. The Solution Structure of Human Mitochondria Fission Protein Fis1 Reveals a Novel TPR-like Helix Bundle. J. Mol. Biol. 2003, 334, 445–458. [Google Scholar] [CrossRef]
- Yu, T.; Fox, R.J.; Burwell, L.; Yoon, Y. Regulation of mitochondrial fission and apoptosis by the mitochondrial outer membrane protein hFis1. J. Cell Sci. 2005, 118, 4141–4151. [Google Scholar] [CrossRef]
- Hall, A. Mitochondrial fusion and fission proteins: Novel therapeutic targets for combating cardiovascular disease. Br. J. Pharmacol. 2014, 171, 1890–1906. [Google Scholar] [CrossRef]
- Yu, R.; Liu, T.; Jin, S.B.; Ankarcrona, M.; Lendahl, U.; Nistér, M.; Zhao, J. MIEF1/2 orchestrate mitochondrial dynamics through direct engagement with both the fission and fusion machineries. BMC Biol. 2021, 19, 229. [Google Scholar] [CrossRef]
- Elgass, K.D.; Smith, E.A.; LeGros, M.A.; Larabell, C.A.; Ryan, M.T. Analysis of ER–mitochondria contacts using correlative fluorescence microscopy and soft X-ray tomography of mammalian cells. J. Cell Sci. 2015, 128, 2795–2804. [Google Scholar] [PubMed]
- Jokinen, R. Genetic Studies of Tissue-Specific Mitochondrial DNA Segregation in Mammals. Ph.D. Thesis, University of Helsinki, Helsinki, Finland, 2016. [Google Scholar]
- Zimmermann, A.K. A Novel Mechanism Underlying BCL-2 Antioxidant Function: Its Role in Mitochondrial Apoptotic Pathways and Virus-Induced Neuronal Cell Death. Ph.D. Thesis, The University of Colorado, Aurora, CO, USA, 2007. [Google Scholar]
- Koch, J.; Feichtinger, R.G.; Freisinger, P.; Pies, M.; Schrödl, F.; Iuso, A.; Sperl, W.; Mayr, J.A.; Prokisch, H.; Haack, T.B. Disturbed mitochondrial and peroxisomal dynamics due to loss of MFF causes Leigh-like encephalopathy, optic atrophy and peripheral neuropathy. J. Med. Genet. 2016, 53, 270–278. [Google Scholar] [CrossRef]
- Gong, J.; Chen, Y.; Pu, F.; Sun, P.; He, F.; Zhang, L.; Li, Y.; Ma, Z.; Wang, H. Understanding Membrane Protein Drug Targets in Computational Perspective. Curr. Drug Targets 2019, 20, 551–564. [Google Scholar] [CrossRef]
- Li, Y.H.; Yu, C.Y.; Li, X.X.; Zhang, P.; Tang, J.; Yang, Q.; Fu, T.; Zhang, X.; Cui, X.; Tu, G.; et al. Therapeutic target database update 2018: Enriched resource for facilitating bench-to-clinic research of targeted therapeutics. Nucleic Acids Res. 2018, 46, D1121–D1127. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhang, Y.; Lian, X.; Li, F.; Wang, C.; Zhu, F.; Qiu, Y.; Chen, Y. Therapeutic target database update 2022: Facilitating drug discovery with enriched comparative data of targeted agents. Nucleic Acids Res. 2022, 50, D1398–D1407. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zhao, F.; Ma, X.; Perry, G.; Zhu, X. Mitochondria dysfunction in the pathogenesis of Alzheimer’s disease: Recent advances. Mol. Neurodegener. 2020, 15, 30. [Google Scholar] [CrossRef]
- Qi, X.; Qvit, N.; Su, Y.-C.; Mochly-Rosen, D. A Novel Drp1 inhibitor diminishes aberrant mitochondrial fission and neurotoxicity. J. Cell Sci. 2013, 126, 789–802. [Google Scholar] [CrossRef]
- Banerjee, R.; Mukherjee, A.; Nagotu, S. Mitochondrial dynamics and its impact on human health and diseases: Inside the DRP1 blackbox. J. Mol. Med. 2021, 100, 1–21. [Google Scholar] [CrossRef]
- Kornfeld, O.S.; Qvit, N.; Haileselassie, B.; Shamloo, M.; Bernardi, P.; Mochly-Rosen, D. Interaction of mitochondrial fission factor with dynamin related protein 1 governs physiological mitochondrial function in vivo. Sci. Rep. 2018, 8, 14034. [Google Scholar] [CrossRef]
- Wu, Q.; Xia, S.X.; Li, Q.Q.; Gao, Y.; Shen, X.; Ma, L.; Zhang, M.Y.; Wang, T.; Li, Y.S.; Wang, Z.F.; et al. Mitochondrial division inhibitor 1 (Mdivi-1) offers neuroprotection through diminishing cell death and improving functional outcome in a mouse model of traumatic brain injury. Brain Res. 2016, 1630, 134–143. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Wang, H.-Y.; Wu, B.; Cheng, C.-Y.; Xiao, W.; Wang, Z.-Z.; Yang, Y.-Y.; Li, P.; Yang, H. Ginkgolide K attenuates neuronal injury after ischemic stroke by inhibiting mitochondrial fission and GSK-3β-dependent increases in mitochondrial membrane permeability. Oncotarget 2017, 8, 44682–44693. [Google Scholar] [CrossRef] [PubMed]
- Grohm, J.; Kim, S.-W.; Mamrak, U.; Tobaben, S.; Cassidy-Stone, A.; Nunnari, J.; Plesnila, N.; Culmsee, C. Inhibition of Drp1 provides neuroprotection in vitro and in vivo. Cell Death Differ. 2012, 19, 1446–1458. [Google Scholar] [CrossRef] [PubMed]
- Chuang, Y.-C.; Lin, T.-K.; Yang, D.-I.; Yang, J.-L.; Liou, C.-W.; Chen, S.-D. Peroxisome proliferator-activated receptor-gamma dependent pathway reduces the phosphorylation of dynamin-related protein 1 and ameliorates hippocampal injury induced by global ischemia in rats. J. Biomed. Sci. 2016, 23, 44. [Google Scholar] [CrossRef] [PubMed]
- Pisani, D.F.; Barquissau, V.; Chambard, J.-C.; Beuzelin, D.; Ghandour, R.A.; Giroud, M.; Mairal, A.; Pagnotta, S.; Cinti, S.; Langin, D.; et al. Mitochondrial fission is associated with UCP1 activity in human brite/beige adipocytes. Mol. Metab. 2018, 7, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Flippo, K.H.; Gnanasekaran, A.; Perkins, G.A.; Ajmal, A.; Merrill, R.A.; Dickey, A.S.; Taylor, S.S.; McKnight, G.S.; Chauhan, A.K.; Usachev, Y.M.; et al. AKAP1 Protects from Cerebral Ischemic Stroke by Inhibiting Drp1-Dependent Mitochondrial Fission. J. Neurosci. 2018, 38, 8233–8242. [Google Scholar] [CrossRef]
- Aishwarya, R.; Alam, S.; Abdullah, C.S.; Morshed, M.; Nitu, S.S.; Panchatcharam, M.; Miriyala, S.; Kevil, C.G.; Bhuiyan, S. Pleiotropic effects of mdivi-1 in altering mitochondrial dynamics, respiration, and autophagy in cardiomyocytes. Redox Biol. 2020, 36, 101660. [Google Scholar] [CrossRef]
- Ding, M.; Feng, N.A.; Tang, D.; Feng, J.; Li, Z.; Jia, M.; Liu, Z.; Gu, X.; Wang, Y.; Fu, F.; et al. Melatonin prevents D rp1-mediated mitochondrial fission in diabetic hearts through SIRT 1-PGC 1α pathway. J. Pineal Res. 2018, 65, e12491. [Google Scholar] [CrossRef] [PubMed]
- Yin, C.-F.; Chang, Y.-W.; Huang, H.-C.; Juan, H.-F. Targeting protein interaction networks in mitochondrial dynamics for cancer therapy. Drug Discov. Today 2021, 27, 1077–1087. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, Y.; Wang, G.; Ye, X.; Zhang, J.; Cao, G.; Zhao, Y.; Gao, Z.; Zhang, Y.; Yu, B.; et al. YiQiFuMai Powder Injection Protects against Ischemic Stroke via Inhibiting Neuronal Apoptosis and PKCδ/Drp1-Mediated Excessive Mitochondrial Fission. Oxidative Med. Cell. Longev. 2017, 2017, 1832093. [Google Scholar] [CrossRef]
- Ong, S.B.; Kalkhoran, S.B.; Hernández-Reséndiz, S.; Samangouei, P.; Ong, S.G.; Hausenloy, D.J. Mitochondrial-shaping proteins in cardiac health and disease–The long and the short of it! Cardiovasc. Drugs Ther. 2017, 31, 87–107. [Google Scholar] [CrossRef]
- Quiles, J.M.; Gustafsson, B. The role of mitochondrial fission in cardiovascular health and disease. Nat. Rev. Cardiol. 2022, 19, 723–736. [Google Scholar] [CrossRef] [PubMed]
- Uchikado, Y.; Ikeda, Y.; Ohishi, M. Current Understanding of the Pivotal Role of Mitochondrial Dynamics in Cardiovascular Diseases and Senescence. Front. Cardiovasc. Med. 2022, 9, 905072. [Google Scholar] [CrossRef] [PubMed]
- Bisaccia, G. Mitochondrial dysfunction and heart disease: Critical appraisal of an overlooked association. Int. J. Mol. Sci. 2021, 22, 614. [Google Scholar] [CrossRef] [PubMed]
- Veloso, C.D.; Belew, G.D.; Ferreira, L.L.; Grilo, L.F.; Jones, J.G.; Portincasa, P.; Sardão, V.A.; Oliveira, P.J. A Mitochondrial Approach to Cardiovascular Risk and Disease. Curr. Pharm. Des. 2019, 25, 3175–3194. [Google Scholar] [CrossRef] [PubMed]
- Alston, C.L.; Rocha, M.C.; Lax, N.Z.; Turnbull, D.M.; Taylor, R.W. The genetics and pathology of mitochondrial disease. J. Pathol. 2017, 241, 236–250. [Google Scholar] [CrossRef]
- Daiber, A.; Andreadou, I.; Oelze, M.; Davidson, S.M.; Hausenloy, D.J. Discovery of new therapeutic redox targets for cardioprotection against ischemia/reperfusion injury and heart failure. Free. Radic. Biol. Med. 2021, 163, 325–343. [Google Scholar] [CrossRef]
- Nan, J.; Zhu, W.; Rahman, M.; Liu, M.; Li, D.; Su, S.; Zhang, N.; Hu, X.; Yu, H.; Gupta, M.P.; et al. Molecular regulation of mitochondrial dynamics in cardiac disease. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2017, 1864, 1260–1273. [Google Scholar] [CrossRef]
- Liu, X.; Guo, C.; Zhang, Q. Novel insights into the involvement of mitochondrial fission/fusion in heart failure: From molecular mechanisms to targeted therapies. Cell Stress Chaperones 2023, 1–12. [Google Scholar] [CrossRef]
- Ding, M.; Dong, Q.; Liu, Z.; Liu, Z.; Qu, Y.; Li, X.; Huo, C.; Jia, X.; Fu, F.; Wang, X. Inhibition of dynamin-related protein 1 protects against myocardial ischemia–reperfusion injury in diabetic mice. Cardiovasc. Diabetol. 2017, 16, 19. [Google Scholar] [CrossRef]
- Wu, N.N.; Zhang, Y.; Ren, J. Mitophagy, mitochondrial dynamics, and homeostasis in cardiovascular aging. Oxidative Med. Cell. Longev. 2019, 2019, 9825061. [Google Scholar] [CrossRef]
- Chistiakov, D.A.; Shkurat, T.P.; Melnichenko, A.A.; Grechko, A.V.; Orekhov, A.N. The role of mitochondrial dysfunction in cardiovascular disease: A brief review. Ann. Med. 2018, 50, 121–127. [Google Scholar] [CrossRef]
- Yu, E.P.; Reinhold, J.; Yu, H.; Starks, L.; Uryga, A.K.; Foote, K.; Finigan, A.; Figg, N.; Pung, Y.F.; Logan, A.; et al. Mitochondrial respiration is reduced in atherosclerosis, promoting necrotic core formation and reducing relative fibrous cap thickness. Arterioscler. Thromb. Vasc. Biol. 2017, 37, 2322–2332. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, M.; Torres, G.; Wu, S.; Ouyang, C.; Xie, Z.; Zou, M.-H. Metformin Suppresses Diabetes-Accelerated Atherosclerosis via the Inhibition of Drp1-Mediated Mitochondrial Fission. Diabetes 2017, 66, 193–205. [Google Scholar] [CrossRef]
- Hong, S.; Zhang, X.; Zhang, X.; Liu, W.; Fu, Y.; Liu, Y.; Shi, Z.; Chi, J.; Zhao, M.; Yin, X. Role of the calcium sensing receptor in cardiomyocyte apoptosis via mitochondrial dynamics in compensatory hypertrophied myocardium of spontaneously hypertensive rat. Biochem. Biophys. Res. Commun. 2017, 487, 728–733. [Google Scholar] [CrossRef] [PubMed]
- Rogers, M.; Maldonado, N.; Hutcheson, J.D.; Goettsch, C.; Goto, S.; Yamada, I.; Faits, T.; Sesaki, H.; Aikawa, M.; Aikawa, E. Dynamin-Related Protein 1 Inhibition Attenuates Cardiovascular Calcification in the Presence of Oxidative Stress. Circ. Res. 2017, 121, 220–233. [Google Scholar] [CrossRef]
- Zhang, J.; Yu, J.; Chen, Y.; Liu, L.; Xu, M.; Sun, L.; Luo, H.; Wang, Y.; Meng, G. Exogenous Hydrogen Sulfide Supplement Attenuates Isoproterenol-Induced Myocardial Hypertrophy in a Sirtuin 3-Dependent Manner. Oxidative Med. Cell. Longev. 2018, 2018, 9396089. [Google Scholar] [CrossRef] [PubMed]
- Moyzis, A.G.; Lally, N.S.; Liang, W.; Leon, L.J.; Najor, R.H.; Orogo, A.M.; Gustafsson, B. Mcl-1-mediated mitochondrial fission protects against stress but impairs cardiac adaptation to exercise. J. Mol. Cell. Cardiol. 2020, 146, 109–120. [Google Scholar] [CrossRef]
- Nakamura, M.; Sadoshima, J. Mechanisms of physiological and pathological cardiac hypertrophy. Nat. Rev. Cardiol. 2018, 15, 387–407. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.W.; Chang, Y.T.; Wang, Q.; Lin, J.J.C.; Chen, Y.J.; Chen, C.C. Quantitative phosphoproteomic study of pressure-overloaded mouse heart reveals dynamin-related protein 1 as a modulator of cardiac hypertrophy. Mol. Cell. Proteom. 2013, 12, 3094–3107. [Google Scholar] [CrossRef] [PubMed]
- Hasan, P.; Saotome, M.; Ikoma, T.; Iguchi, K.; Kawasaki, H.; Iwashita, T.; Hayashi, H.; Maekawa, Y. Mitochondrial fission protein, dynamin-related protein 1, contributes to the promotion of hypertensive cardiac hypertrophy and fibrosis in Dahl-salt sensitive rats. J. Mol. Cell. Cardiol. 2018, 121, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Shirakabe, A.; Zhai, P.; Ikeda, Y.; Saito, T.; Maejima, Y.; Hsu, C.-P.; Nomura, M.; Egashira, K.; Levine, B.; Sadoshima, J.; et al. Drp1-Dependent Mitochondrial Autophagy Plays a Protective Role Against Pressure Overload–Induced Mitochondrial Dysfunction and Heart Failure. Circulation 2016, 133, 1249–1263. [Google Scholar] [CrossRef] [PubMed]
- Cooper, H.A.; Eguchi, S. Inhibition of mitochondrial fission as a novel therapeutic strategy to reduce mortality upon myocardial infarction. Clin. Sci. 2018, 132, 2163–2167. [Google Scholar] [CrossRef]
- Givvimani, S.; Pushpakumar, S.; Metreveli, N.; Veeranki, S.; Kundu, S.; Tyagi, S. Role of mitochondrial fission and fusion in cardiomyocyte contractility. Int. J. Cardiol. 2015, 187, 325–333. [Google Scholar] [CrossRef]
- Ong, S.-B.; Kwek, X.-Y.; Katwadi, K.; Hernandez-Resendiz, S.; Crespo-Avilan, G.E.; Ismail, N.I.; Lin, Y.-H.; Yap, E.P.; Lim, S.-Y.; Ja, K.P.M.M.; et al. Targeting Mitochondrial Fission Using Mdivi-1 in A Clinically Relevant Large Animal Model of Acute Myocardial Infarction: A Pilot Study. Int. J. Mol. Sci. 2019, 20, 3972. [Google Scholar] [CrossRef]
- Ong, S.-B.; Subrayan, S.; Lim, S.Y.; Yellon, D.M.; Davidson, S.M.; Hausenloy, D.J. Inhibiting Mitochondrial Fission Protects the Heart Against Ischemia/Reperfusion Injury. Circulation 2010, 121, 2012–2022. [Google Scholar] [CrossRef]
- Zhou, H.; Wang, J.; Zhu, P.; Zhu, H.; Toan, S.; Hu, S.; Ren, J.; Chen, Y. NR4A1 aggravates the cardiac microvascular ischemia reperfusion injury through suppressing FUNDC1-mediated mitophagy and promoting Mff-required mitochondrial fission by CK2α. Basic Res. Cardiol. 2018, 113, 23. [Google Scholar] [CrossRef] [PubMed]
- Disatnik, M.H.; Ferreira, J.C.; Campos, J.C.; Gomes, K.S.; Dourado, P.M.; Qi, X.; Mochly-Rosen, D. Acute inhibition of excessive mitochondrial fission after myocardial infarction prevents long-term cardiac dysfunction. J. Am. Heart Assoc. 2013, 2, e000461. [Google Scholar] [CrossRef]
- Nishimura, A.; Shimauchi, T.; Tanaka, T.; Shimoda, K.; Toyama, T.; Kitajima, N.; Ishikawa, T.; Shindo, N.; Numaga-Tomita, T.; Yasuda, S.; et al. Hypoxia-induced interaction of filamin with Drp1 causes mitochondrial hyperfission–associated myocardial senescence. Sci. Signal. 2018, 11, eaat5185. [Google Scholar] [CrossRef]




| Name of Protein | Gene Name | Yeast Ortholog | Uniport ID | Human Homolog | Location | Molecular Functions | References |
|---|---|---|---|---|---|---|---|
| Dynamin 1 | DNM1 | DNM1, YLL001W, L1381 | P54861 | Drp1 or DNM1 | Cytoplasm and cytosol | GTPase activity, GTP binding, identical protein binding, microtubule binding, protein kinase binding, RNA binding | [29,30,31] |
| Dynamin 1-like protein | DNM1L | Dnm1p | O00429 | DNM1L, DLP1, DRP1 | Cytoplasm and cytosol | GTPase activator activity, GTPase activity, GTP binding, GTP-dependent protein binding, identical protein binding, lipid binding, microtubule binding, protein homodimerization activity, small GTPase binding, ubiquitin protein ligase binding | [32,33] |
| Dynamin 2 | DNM2 | Dnm2 | P50570 | DNM2, DYN2 | Cytoplasm and cytosol | D2 dopamine receptor binding, enzyme binding, GTPase activity, GTP binding, microtubule binding, nitric-oxide synthase binding, phosphatidylinositol 3-kinase regulatory subunit binding, protein-containing complex binding, protein kinase binding, SH3 domain binding, WW domain binding | [30,31] |
| Dynamin 3 | DNM3 | Dnm3 | Q9UQ16 | Drp3 or DNM3 | Cytoplasm and cytosol | GTPase activity, GTP binding, identical protein binding, microtubule binding, nitric-oxide synthase binding, structural constituent of postsynapse, type 1 metabotropic glutamate receptor binding, type 5 metabotropic glutamate receptor binding | [30,34] |
| Mitochondrial fission 1 protein | FIS1 | Fis1, MDV2, YIL065C | Q9Y3D6 | Fis1, TTC11, CGI-135 | OMM and peroxisome membrane | Identical protein binding | [35,36] |
| Ganglioside-induced differentiation-associated protein 1 | GDAP1 | - | Q8TB36 | GDAP1 | OMM | Regulates the mitochondrial network by promoting mitochondrial fission | [37,38] |
| Putative gametogenetin-binding protein 1 | GGNBP1 | - | Q5YKI7 | GGNBP1 | Cytoplasm and cytosol | Involved in spermatogenesis | [39] |
| Vascular endothelial growth factor receptor 2 | KDR | - | P35968 | KDR, FLK1, VEGFR2 | ER and plasma membrane | ATP binding, cadherin binding, growth factor binding, heat shock protein 90 (HSP90) binding, identical protein binding, integrin binding, protein serine/threonine/tyrosine kinase activity, transmembrane receptor protein tyrosine kinase activity, vascular endothelial growth factor-activated receptor activity, vascular endothelial growth factor binding | [40,41,42,43,44] |
| Calcium uniporter protein, mitochondrial | MCU | - | Q8NE86 | MCU, C10orf42, CCDC109A | Inner mitochondrial membrane (IMM) | Calcium channel activity, identical protein binding, uniporter activity | [45,46] |
| Mitochondrial fission factor | MFF | - | Q9GZY8 | Mff, C2orf33, AD030, AD033, GL004 | OMM and peroxisome | Identical protein binding, protein homodimerization activity | [47,48] |
| Mitochondrial dynamics protein MiD51 | MIEF1 | - | Q9NQG6 | MIEF1, Mid51, SMCR7L | OMM | ADP binding, GDP binding, identical protein binding | [49,50] |
| Mitochondrial dynamics protein MiD49 | MIEF2 | - | Q96C03 | MIEF2, Mid49, SMCR7 | OMM | Regulation of mitochondrial organization, recruitment and association of the fission mediator dynamin-related protein 1 (DNM1L) | [51,52,53] |
| Mitochondrial fission process protein 1 | MTFP1 | - | Q9UDX5 | MTFP1, MTP18, HSPC242, My022 | IMM | Apoptotic process, mitochondrial fission, response to muscle activity | [54,55] |
| Mitochondrial division protein 1 | Mdv1p and Caf4p | MDV1, FIS2, GAG3, NET2, YJL112W, J0802 | P47025 | - | OMM | Ubiquitin binding | [56] |
| Group | Inhibitors | Mechanisms | Function | References |
|---|---|---|---|---|
| Peptide | P110 | Inhibiting Fis1/Drp1 PPI, enzyme activity and Fis1 TPR-binding peptide | Inhibition of mitochondrial fission | [136] |
| P259 | Drp1/Mff PPI specific inhibitor | Inhibitory effect on physiological fission | [138] | |
| Small molecule | Mdivi-1 | Inhibiting Drp1 activity | Inhibition of mitochondrial fission | [139] |
| Ginkgolide K | Reducing Drp1 recruitment | Inhibition of mitochondrial fission | [73,140] | |
| siRNA | Drp1 siRNA | Suppressing of Drp1 expression | Inhibition of mitochondrial fission | [141] |
| PPAR agonist | Supporting of Drp1 phosphorylation at Ser616 | Inhibition of mitochondrial fission | [142,143] | |
| Protein | AKAP1 | Increase the Drp1 phosphorylation at Ser637 | Inhibition of mitochondrial fission | [144] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zerihun, M.; Sukumaran, S.; Qvit, N. The Drp1-Mediated Mitochondrial Fission Protein Interactome as an Emerging Core Player in Mitochondrial Dynamics and Cardiovascular Disease Therapy. Int. J. Mol. Sci. 2023, 24, 5785. https://doi.org/10.3390/ijms24065785
Zerihun M, Sukumaran S, Qvit N. The Drp1-Mediated Mitochondrial Fission Protein Interactome as an Emerging Core Player in Mitochondrial Dynamics and Cardiovascular Disease Therapy. International Journal of Molecular Sciences. 2023; 24(6):5785. https://doi.org/10.3390/ijms24065785
Chicago/Turabian StyleZerihun, Mulate, Surya Sukumaran, and Nir Qvit. 2023. "The Drp1-Mediated Mitochondrial Fission Protein Interactome as an Emerging Core Player in Mitochondrial Dynamics and Cardiovascular Disease Therapy" International Journal of Molecular Sciences 24, no. 6: 5785. https://doi.org/10.3390/ijms24065785
APA StyleZerihun, M., Sukumaran, S., & Qvit, N. (2023). The Drp1-Mediated Mitochondrial Fission Protein Interactome as an Emerging Core Player in Mitochondrial Dynamics and Cardiovascular Disease Therapy. International Journal of Molecular Sciences, 24(6), 5785. https://doi.org/10.3390/ijms24065785






