Fishing the Targets of Bioactive Compounds from Psidium guajava L. Leaves in the Context of Diabetes
Abstract
1. Introduction
2. Results
2.1. Identification of the Phenolic Compositions
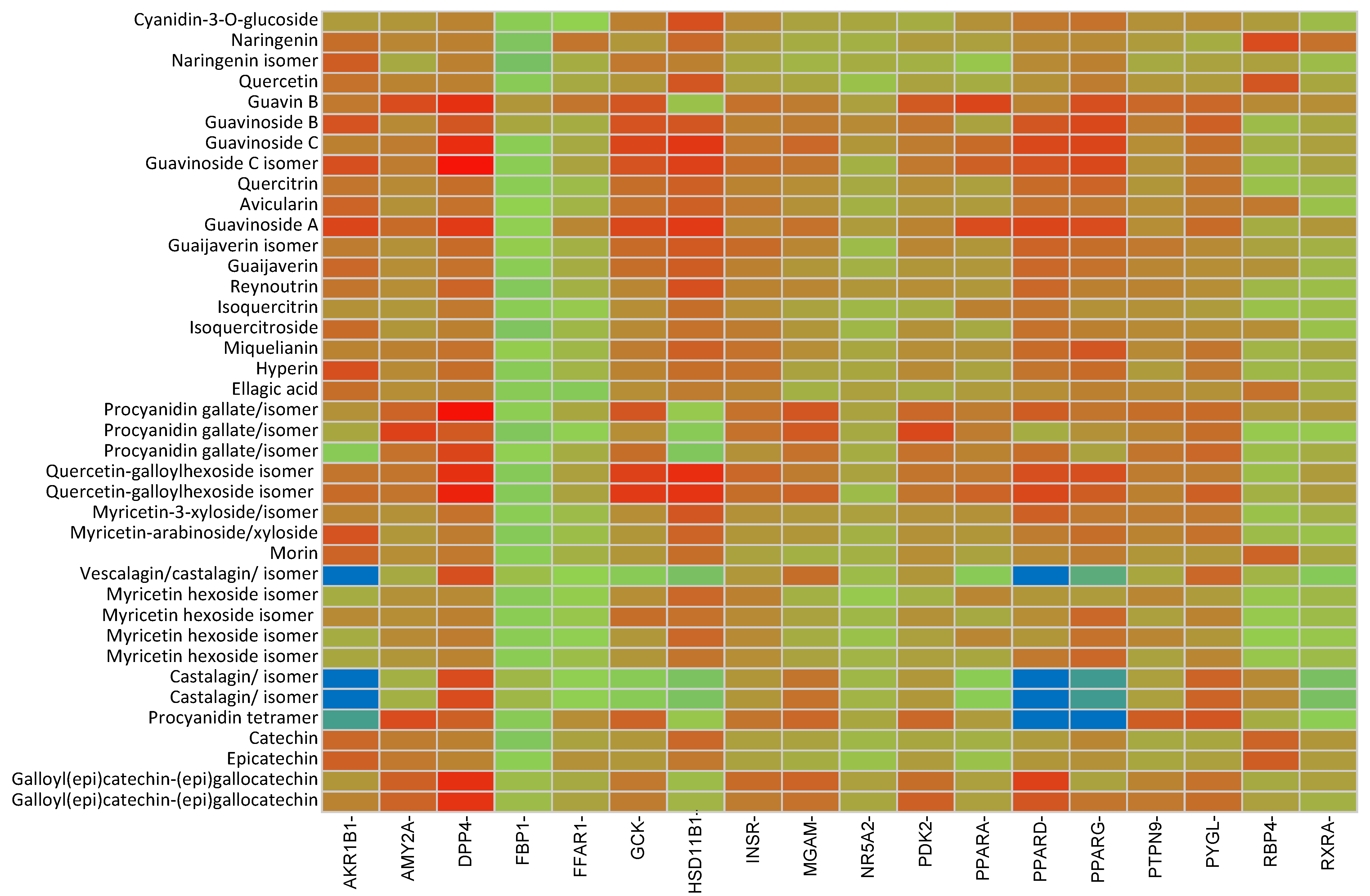
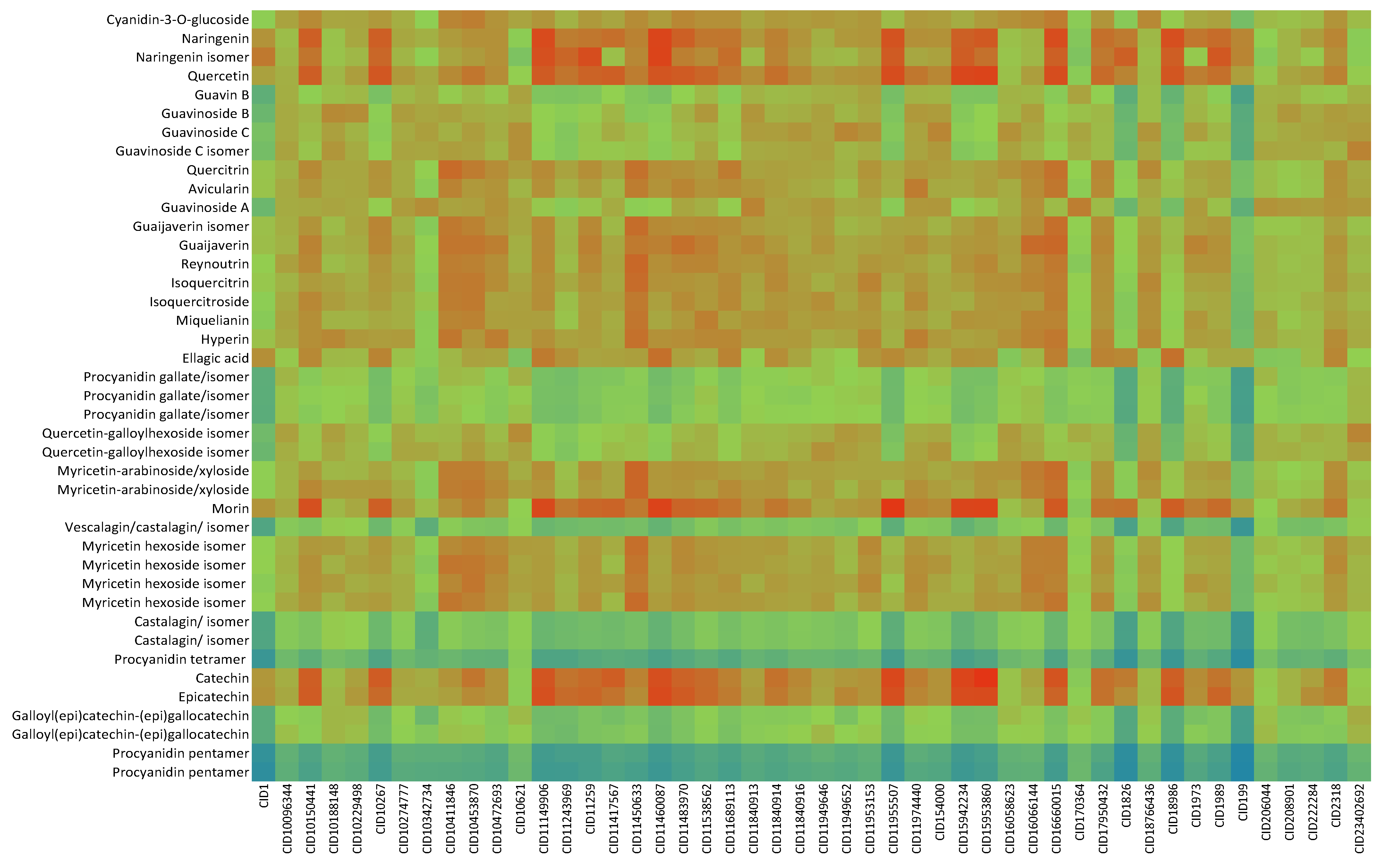
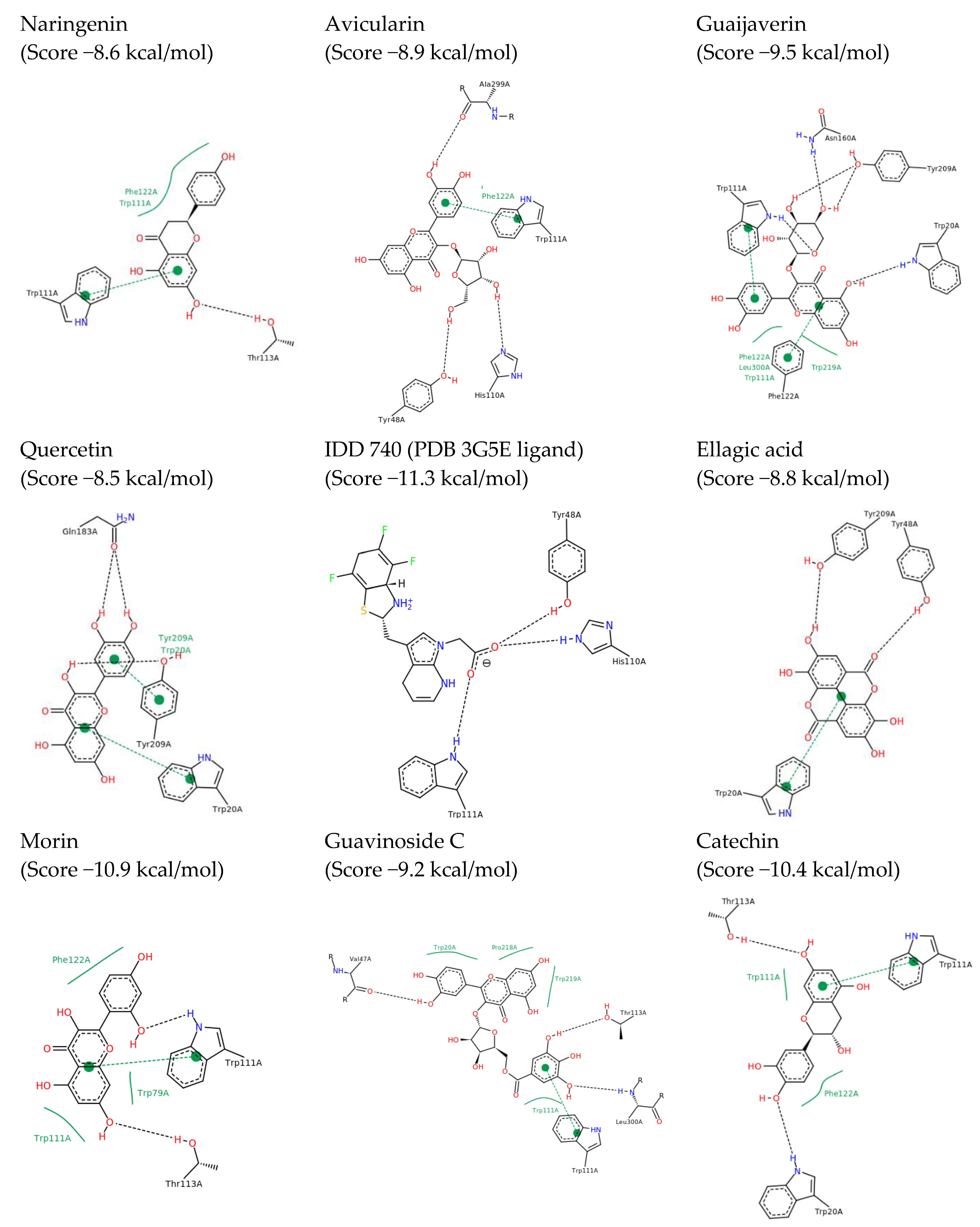
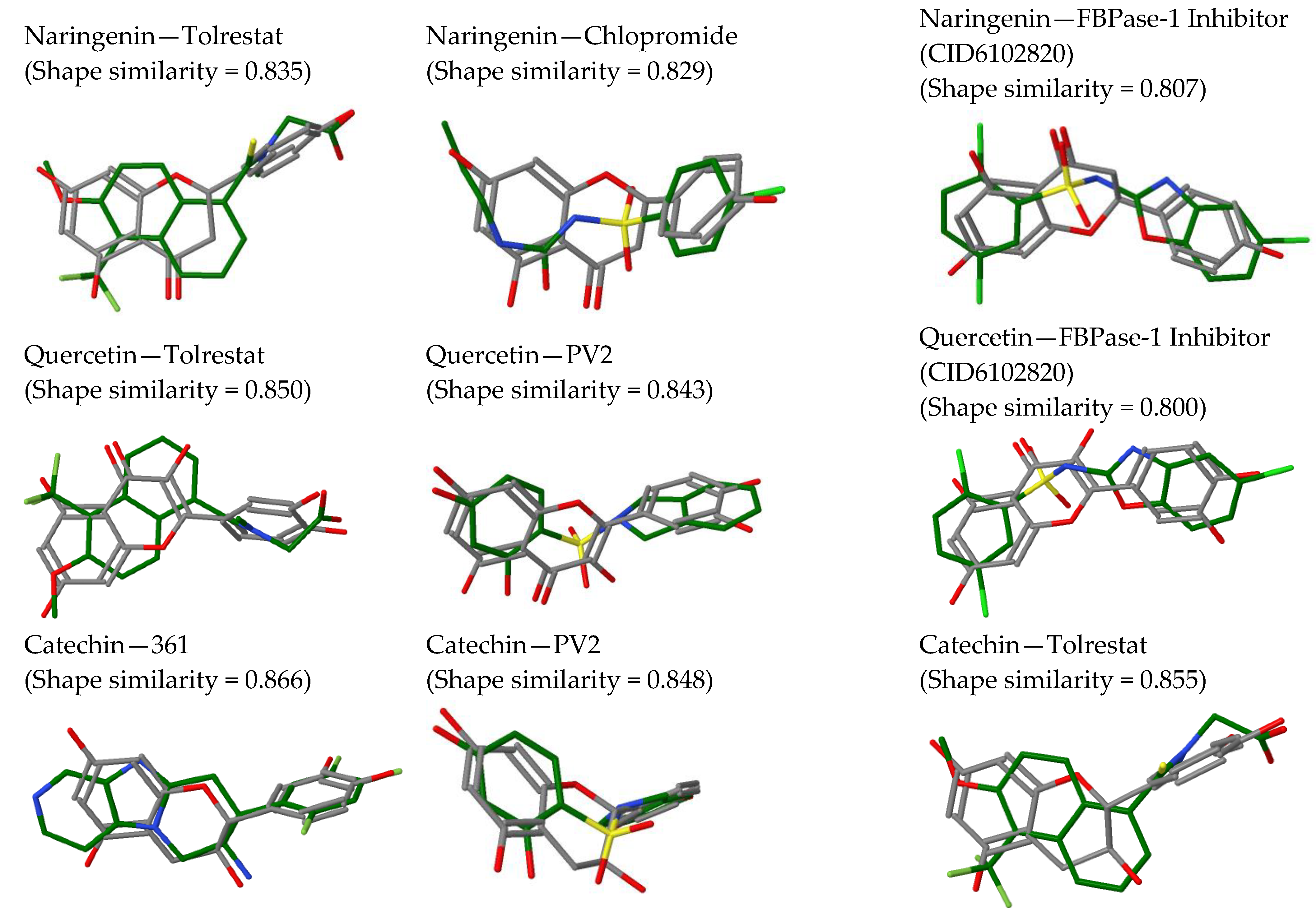
2.2. In Silico Results and Bibliography Searches
3. Discussion
4. Materials and Methods
4.1. Plant Material and Sample Preparation
4.2. HPLC-ESI-QTOF-MS Analyses
4.3. In Silico Approaches
4.4. Bibliography Searches
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Guariguata, L.; Whiting, D.R.; Hambleton, I.; Beagley, J.; Linnenkamp, U.; Shaw, J.E. Global estimates of diabetes prevalence for 2013 and projections for 2035 for the IDF Diabetes Atlas. Diabetes Res. Clin. Pract. 2013, 103, 137–149. [Google Scholar] [CrossRef]
- Soltani, N. Prevention of Diabetes Complications. In Type 1 Diabetes Complications; Wagner, D., Ed.; InTech: Rijeka, Croatia, 2011; pp. 353–366. [Google Scholar]
- Tan, C.; Wang, Q.; Luo, C.; Chen, S.; Li, Q.; Li, P. Yeast α-glucosidase inhibitory phenolic compounds isolated from Gynura medica leaf. Int. J. Mol. Sci. 2013, 14, 2551–2558. [Google Scholar] [CrossRef]
- Singh, R.; Kaur, N.; Kishore, L.; Gupta, G.K. Management of diabetic complications: A chemical constituents based approach. J. Ethnopharmacol. 2013, 150, 51–70. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, R.M.P.; Mitchell, S.; Solis, R.V. Psidium guajava: A review of its traditional uses, phytochemistry and pharmacology. J. Ethnopharmacol. 2008, 117, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Díaz-de-Cerio, E.; Gómez-Caravaca, A.M.; Verardo, V.; Fernández-Gutiérrez, A.; Segura-Carretero, A. Determination of guava (Psidium guajava L.) leaf phenolic compounds using HPLC-DAD-QTOF-MA. J. Funct. Foods. 2016, 22, 376–388. [Google Scholar] [CrossRef]
- Díaz-De-Cerio, E.; Verardo, V.; Gómez-Caravaca, A.M.; Fernández-Gutiérrez, A.; Segura-Carretero, A. Exploratory characterization of phenolic compounds with demonstrated anti-diabetic activity in guava leaves at different Oxidation States. Int. J. Mol. Sci. 2016, 17, 699. [Google Scholar] [CrossRef]
- Gambari, R. Predictive analyses of biological effects of natural products: From plant extracts to biomolecular laboratory and computer modeling. Evid. Based Complement. Altern. Med. 2011, 2011, 383290. [Google Scholar] [CrossRef]
- Prathipati, P.; Ngai, L.M.; Manjunatha, U.H.; Bender, A. Fishing the target of antitubercular compounds: In silico target deconvolution model development and validation. J. Proteome Res. 2009, 8, 2788–2798. [Google Scholar] [CrossRef] [PubMed]
- de Rijke, E.; Out, P.; Niessen, W.M.A.; Ariese, F.; Gooijer, C.; Brinkman, U.A.T. Analytical separation and detection methods for flavonoids. J. Chromatogr. A 2006, 1112, 31–63. [Google Scholar] [CrossRef] [PubMed]
- Wolfender, J.L. HPLC in natural product analysis: The detection issue. Planta Med. 2009, 75, 719–734. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Anand, S.; Arasakumari, M.; Prabu, P.; Amalraj, A.J. Anti-diabetic and aldose reductase inhibitory potential of Psidium guajava by in vitro analysis. Int. J. Pharm. Pharm. Sci. 2016, 8, 271–276. [Google Scholar] [CrossRef]
- Ortiz-Andrade, R.R.; Sánchez-Salgado, J.C.; Navarrete-Vázquez, G.; Webster, S.P.; Binnie, M.; García-Jiménez, S.; León-Rivera, I.; Cigarroa-Vázquez, P.; Villalobos-Molina, R.; Estrada-Soto, S. Antidiabetic and toxicological evaluations of naringenin in normoglycaemic and NIDDM rat models and its implications on extra-pancreatic glucose regulation. Diabetes Obes. Metab. 2008, 10, 1097–1104. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.S.; Qais, F.A.; Rehman, M.T.; Ismail, M.H.; Alokail, M.S.; Altwaijry, N.; Alafaleq, N.O.; AlAjmi, M.F.; Salem, N.; Alqhatani, R. Mechanistic inhibition of non-enzymatic glycation and aldose reductase activity by naringenin: Binding, enzyme kinetics and molecular docking analysis. Int. J. Biol. Macromol. 2020, 159, 87–97. [Google Scholar] [CrossRef]
- Fan, J.; Johnson, M.H.; Lila, M.A.; Yousef, G.; De Mejia, E.G. Berry and citrus phenolic compounds inhibit dipeptidyl peptidase IV: Implications in diabetes management. Evid. Based Complement. Altern. Med. 2013, 2013, 479505. [Google Scholar] [CrossRef]
- Goldwasser, J.; Cohen, P.Y.; Yang, E.; Balaguer, P.; Yarmush, M.L.; Nahmias, Y. Transcriptional regulation of human and rat hepatic lipid metabolism by the grapefruit flavonoid naringenin: Role of PPARα, PPARγ and LXRα. PLoS ONE 2010, 5, e12399. [Google Scholar] [CrossRef] [PubMed]
- Chethan, S.; Dharmesh, S.M.; Malleshi, N.G. Inhibition of aldose reductase from cataracted eye lenses by finger millet (Eleusine coracana) polyphenols. Bioorg. Med. Chem. 2008, 16, 10085–10090. [Google Scholar] [CrossRef]
- De La Fuente, J.Á.; Manzanaro, S.; Martín, M.J.; De Quesada, T.G.; Reymundo, I.; Luengo, S.M.; Gago, F. Synthesis, activity, and molecular modeling studies of novel human aldose reductase inhibitors based on a marine natural product. J. Med. Chem. 2003, 46, 5208–5221. [Google Scholar] [CrossRef]
- Ueda, H.; Tachibana, Y.; Moriyasu, M.; Kawanishi, K.; Alves, S.M. Aldose reductase inhibitors from the fruits of Caesalpinia ferrea Mart. Phytomedicine 2001, 8, 377–381. [Google Scholar] [CrossRef]
- Torres-Piedra, M.; Ortiz-Andrade, R.; Villalobos-Molina, R.; Singh, N.; Medina-Franco, J.L.; Webster, S.P.; Binnie, M.; Navarrete-Vázquez, G.; Estrada-Soto, S. A comparative study of flavonoid analogues on streptozotocin–nicotinamide induced diabetic rats: Quercetin as a potential antidiabetic agent acting via 11β-Hydroxysteroid dehydrogenase type 1 inhibition. Eur. J. Med. Chem. 2010, 45, 2606–2612. [Google Scholar] [CrossRef]
- Miranda, M.A.; Okamoto, A.K.; Ferreira, C.V.; Silva, T.L.; Granjeiro, J.M.; Aoyama, H. Differential effects of flavonoids on bovine kidney low molecular mass protein tyrosine phosphatase. J. Enzyme Inhib. Med. Chem. 2006, 21, 419–425. [Google Scholar] [CrossRef] [PubMed]
- Chuang, C.; Martinez, K.; Xie, G.; Kennedy, A.; Bumrungpert, A.; Overman, A.; Jia, W.; McIntosh, M.M. Quercetin is equally or more effective than resveratrol in attenuating tumor necrosis factor-α-mediated inflammation and insulin resistance in primary human adipocytes. Am. J. Clin. Nutr. 2010, 92, 1511–1521. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, M.; Nishida, N.; Shimoda, H.; Takada, M.; Kawahara, Y.; Matsuda, H. Polyphenol constituents from Salacia species: Quantitative analysis of mangiferin with alpha-glucosidase and aldose reductase inhibitory activities. Yakugaku Zasshi 2001, 121, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Dhagat, U.; Endo, S.; Hara, A.; El-Kabbani, O. Inhibition of 3(17)α-hydroxysteroid dehydrogenase (AKR1C21) by aldose reductase inhibitors. Bioorg. Med. Chem. 2008, 16, 3245–3254. [Google Scholar] [CrossRef]
- Kim, H.M.; Lee, D.G.; Lee, S. Plant-derived molecules from Saussurea grandifolia as inhibitors of aldose reductase. J. Korean Soc. Appl. Biol. Chem. 2015, 58, 365–371. [Google Scholar] [CrossRef]
- Jung, H.A.; Islam, M.D.N.; Kwon, Y.S.; Jin, S.E.; Son, Y.K.; Park, J.J.; Sohn, H.S.; Choi, J.S. Extraction and identification of three major aldose reductase inhibitors from Artemisia montana. Food Chem. Toxicol. 2011, 49, 376–384. [Google Scholar] [CrossRef]
- Yoshikawa, M.; Shimada, H.; Nishida, N.; Li, Y.; Toguchida, I.; Yamahara, J.; Matsuda, H. Antidiabetic principles of natural medicines. II. Aldose reductase and α-glucosidase inhibitors from Brazilian natural medicine, the leaves of Myrcia multiflora DC. (Myrtaceae): Structures of myrciacitrins I and II and myrciaphenones A and B. Chem. Pharm. Bull. 1998, 46, 113–119. [Google Scholar] [CrossRef]
- Choi, J.-S.; Bae, J.-Y.; Kim, D.S.; Li, J.; Kim, J.-L.; Lee, Y.-J.; Kang, Y.-H. Dietary Compound Quercitrin Dampens VEGF Induction and PPARγ Activation in Oxidized LDL-Exposed Murine Macrophages: Association with Scavenger Receptor CD36. J. Agric. Food Chem. 2010, 58, 1333–1341. [Google Scholar] [CrossRef]
- Brindis, F.; González-Trujano, M.E.; González-Andrade, M.; Aguirre-Hernández, E.; Villalobos-Molina, R. Aqueous extract of Annona macroprophyllata: A potential α-glucosidase inhibitor. BioMed Res. Int. 2013, 2013, 1–6. [Google Scholar] [CrossRef]
- Akileshwari, C.; Raghu, G.; Muthenna, P.; Mueller, N.H.; Suryanaryana, P.; Petrash, J.M.; Reddy, G.B. Bioflavonoid ellagic acid inhibits aldose reductase: Implications for prevention of diabetic complications. J. Funct. Foods 2014, 6, 374–383. [Google Scholar] [CrossRef]
- Hundsdörfer, C.; Hemmerling, H.J.; Götz, C.; Totzke, F.; Bednarski, P.; Le Borgne, M.; Jose, J. Indeno[1,2-b]indole derivatives as a novel class of potent human protein kinase CK2 inhibitors. Bioorg. Med. Chem. 2012, 20, 2282–2289. [Google Scholar] [CrossRef] [PubMed]
- Naeem, S.; Hylands, P.; Barlow, D. Construction of an Indonesian herbal constituents database and its use in Random Forest modelling in a search for inhibitors of aldose reductase. Bioorg. Med. Chem. 2012, 20, 1251–1258. [Google Scholar] [CrossRef]
- Sawant, L.; Singh, V.K.; Dethe, S.; Bhaskar, A.; Balachandran, J.; Mundkinajeddu, D.; Agarwal, A. Aldose reductase and protein tyrosine phosphatase 1B inhibitory active compounds from Syzygium cumini seeds. Pharm. Biol. 2015, 53, 1176–1182. [Google Scholar] [CrossRef]
- Kawanishi, K.; Moriyasu, M.; Kuroiwa, E.; Tachibana, Y.; Ayala, F.; Ueda, H. Aldose reductase inhibitors from the leaves of Myrciaria dubia (H. B. & K.) McVaugh. Phytomedicine 2004, 11, 652–656. [Google Scholar]
- Chatzileontiadou, D.S.M.; Skamnaki, V.T.; Kyriakis, E.; Kantsadi, A.L.; Stravodimos, G.A.; Leonidas, D.D. Natural flavonoids as antidiabetic agents. The binding of gallic and ellagic acids to glycogen phosphorylase b. FEBS Lett. 2015, 589, 1787–1794. [Google Scholar]
- Zoechling, A.; Liebner, F.; Jungbauer, A. Red wine: A source of potent ligands for peroxisome proliferator-activated receptor γ. Food Funct. 2011, 2, 28–38. [Google Scholar] [CrossRef]
- Adisakwattana, S.; Chanathong, B. α-Glucosidase inhibitory activity and lipid-lowering mechanisms of Moringa oleifera leaf extract. Eur. Rev. Med. Pharmacol. Sci. 2011, 15, 803–808. [Google Scholar]
- Toma, A.; Makonnen, E.; Mekonnen, Y.; Debella, A.; Addisakwattana, S. Intestinal α-glucosidase and some pancreatic enzymes inhibitory effect of hydroalcholic extract of Moringa stenopetala leaves. BMC Complement. Altern. Med. 2014, 14, 180. [Google Scholar] [CrossRef]
- Palanisamy, U.D.; Ling, L.T.; Manaharan, T.; Appleton, D. Rapid isolation of geraniin from Nephelium lappaceum rind waste and its anti-hyperglycemic activity. Food Chem. 2011, 127, 21–27. [Google Scholar] [CrossRef]
- Abbas, G.; Hussain, H.; Hamaed, A.; Supuran, C.T. The management of diabetes mellitus-imperative role of natural products against dipeptidyl peptidase-4, α-glucosidase and sodium-dependent glucose co-transporter 2 (SGLT2). Bioorg. Chem. 2019, 86, 305–315. [Google Scholar] [CrossRef]
- Zhu, Q.; Ge, F.; Dong, Y.; Sun, W.; Wang, Z.; Shan, Y.; Chen, R.; Sun, J.; Ge, R.-S. Comparison of flavonoids and isoflavonoids to inhibit rat and human 11β-hydroxysteroid dehydrogenase 1 and 2. Steroids 2018, 132, 25–32. [Google Scholar] [CrossRef]
- Shen, S.C.; Cheng, F.C.; Wu, N.J. Effect of guava (Psidium guajava Linn.) leaf soluble solids on glucose metabolism in type 2 diabetic rats. Phytother. Res. 2008, 22, 1458–1464. [Google Scholar] [CrossRef]
- Lee, Y.; Lim, Y.; Kwon, O. Selected phytochemicals and culinary plant extracts inhibit fructose uptake in caco-2 cells. Molecules 2015, 20, 17393–17404. [Google Scholar] [CrossRef]
- Wu, W.J.; Yan, W.L.; Yu, S.C.; Gunawan, G.; Lin, C.Y.; Huang, C.Y.; Chang, C.T.; Chen, H.W.; Lii, C.K.; Yu, A.L.; et al. Inactivation of Protein Tyrosine Phosphatase 1B (PTP1B) Activity by the Aqueous Partition of Guava Leaf Extract. J. Pharm. Pharmacol. 2018, 6, 890–906. [Google Scholar]
- Cheng, F.C.; Shen, S.C.; Wu, J.S.B. Effect of guava (Psidium guajava L.) leaf extract on glucose uptake in rat hepatocytes. J. Food Sci. 2009, 74, H132–H138. [Google Scholar] [CrossRef]
- Yoon, S.-Y.; Lee, J.H.; Kwon, S.J.; Kang, H.J.; Chung, S.J. Ginkgolic acid as a dual-targeting inhibitor for protein tyrosine phosphatases relevant to insulin resistance. Bioorg. Chem. 2018, 81, 264–269. [Google Scholar] [CrossRef]
- Qiang, G.; Xue, S.; Yang, J.J.; Du, G.; Pang, X.; Li, X.; Goswami, D.; Griffin, P.R.; Ortlund, E.A.; Chan, C.B. Identification of a small molecular insulin receptor agonist with potent antidiabetes activity. Diabetes 2014, 63, 1394–1409. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.W.; Wang, Y.C.; Lu, H.C.; Chiang, W.D. Optimization of ultrasound-assisted extraction conditions for total phenols with anti-hyperglycemic activity from Psidium guajava leaves. Process. Biochem. 2014, 49, 1601–1605. [Google Scholar] [CrossRef]
- Oghogho, O.O.; Nimenibo-Uadia, R. Phytochemical screening, GC-MS analysis and in vitro inhibition of alpha-amylase and alpha-glucosidase activities by methanol extract of Psidium guajava leaves and fractions. J. Pharmacogn. Phytochem. 2019, 8, 634–640. [Google Scholar]
- Wang, H.; Du, Y.J.; Song, H.C. α-Glucosidase and α-amylase inhibitory activities of guava leaves. Food Chem. 2010, 123, 6–13. [Google Scholar] [CrossRef]
- Banu, M.; Sujatha, K.; Kamala, M.; Kumar, K.V. Hypoglycaemic and hypolipidaemic potentials of isolated fraction of Psidium guajava leaf in alloxan-induced diabetic rats. Int. J. Pharm. Innov. 2012, 2, 16–22. [Google Scholar]
- Tella, T.; Masola, B.; Mukaratirwa, S. The effect of Psidium guajava aqueous leaf extract on liver glycogen enzymes, hormone sensitive lipase and serum lipid profile in diabetic rats. Biomed. Pharmacother. 2019, 109, 2441–2446. [Google Scholar] [CrossRef] [PubMed]
- Vinayagam, R.; Jayachandran, M.; Chung, S.S.M.; Xu, B. Guava leaf inhibits hepatic gluconeogenesis and increases glycogen synthesis via AMPK/ACC signaling pathways in streptozotocin-induced diabetic rats. Biomed. Pharmacother. 2018, 103, 1012–1017. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Caravaca, A.M.; Verardo, V.; Toselli, M.; Segura-Carretero, A.; Fernández-Gutiérrez, A.; Caboni, M.F. Determination of the major phenolic compounds in pomegranate juices by HPLC-DAD-ESI-MS. J. Agric. Food Chem. 2013, 61, 5328–5337. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Pérez, A.; Muñoz, A.; Peña-García, J.; Den-Haan, H.; Bekas, N.; Katsikoudi, A.; Tzakos, A.G.; Péréz-Sánchez, H. DIA-DB: A Web-Accessible Database for the Prediction of Diabetes Drugs. In Bioinformatics and Biomedical Engineering, Proceedings of the Third International Conference, IWBBIO 2015, Granada, Spain, 15–17 April 2015; Ortuño, F., Rojas, I., Eds.; Springer International Publishing: Berlin/Heidelberg, Germany, 2015. [Google Scholar]
- Ge, H.; Wang, Y.; Zhao, W.; Lin, W.; Yan, X.; Xu, J. Scaffold hopping of potential anti-tumor agents by WEGA: A shape-based approach. MedChemComm 2014, 5, 737–741. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A. Autodock vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
| No. | Compound | rt (min) | m/z Exp | m/z Calc | Molecular Formula | Score | Error (ppm) |
|---|---|---|---|---|---|---|---|
| Negative mode | |||||||
| 1 | Galloyl-Hexahydroxydiphenoyl (HHDP) glucose isomer 1 | 1.93 | 481.06 | 481.34 | C20H18O14 | 96.51 | −2.55 |
| 2 | HHDP glucose isomer 2 | 2.14 | 481.06 | 481.34 | C20H18O14 | 99.09 | −0.19 |
| 3 | HHDP glucose isomer 3 | 2.52 | 481.06 | 481.34 | C20H18O14 | 97.21 | −2.24 |
| 4 | Prodelphinidin B isomer | 3.85 | 609.13 | 609.51 | C30H26O14 | 97.84 | −1.7 |
| 5 | Gallic acid | 4.02 | 169.01 | 169.11 | C7H6O5 | 99.27 | 0.37 |
| 6 | Pedunculagin/Casuariin isomer 1 | 5.87 | 783.07 | 783.53 | C34H24O22 | 98.57 | −1.29 |
| 7 | Prodelphinidin Dimer isomer 1 | 7.27 | 593.13 | 593.51 | C30H26O13 | 96.51 | −2.35 |
| 8 | (epi)-gallocatechin isomer 1 | 7.81 | 305.07 | 305.26 | C15H14O7 | 95.55 | −3.32 |
| 9 | Vescalagin/castalagin isomer | 7.95 | 933.07 | 933.62 | C41H26O26 | 99.19 | −0.79 |
| 10 | Prodelphinidin Dimer isomer 2 | 8.12 | 593.13 | 593.51 | C30H26O13 | 96.51 | −2.35 |
| 11 | Uralenneoside | 9.39 | 285.06 | 285.23 | C12H14O8 | 97.80 | −2.69 |
| 12 | Geraniin isomer 1 | 9.50 | 951.08 | 951.64 | C41H28O27 | 99.56 | −0.20 |
| 13 | Pedunculagin/Casuariin isomer 2 | 9.54 | 783.07 | 783.53 | C34H24O22 | 98.39 | −1.36 |
| 14 | Geraniin isomer 2 | 9.65 | 951.08 | 951.64 | C41H28O27 | 99.56 | −0.20 |
| 15 | Procyanidin B isomer 1 | 10.02 | 577.14 | 577.51 | C30H26O12 | 95.68 | −2.55 |
| 16 | Galloyl(epi)catechin-(epi)gallocatechin | 10.35 | 745.14 | 745.62 | C37H30O17 | 96.90 | −0.62 |
| 17 | Procyanidin B isomer 2 | 10.36 | 577.14 | 577.51 | C30H26O13 | 99.41 | −0.61 |
| 18 | Tellimagrandin I isomer | 10.74 | 785.09 | 785.55 | C34H26O22 | 99.13 | −0.96 |
| 19 | Pterocarinin A isomer 1 | 11.00 | 1067.12 | 1067.75 | C46H36O30 | 99.82 | −0.11 |
| 20 | Pterocarinin A isomer 2 | 11.21 | 1067.12 | 1067.75 | C46H36O30 | 98.39 | −1.26 |
| 21 | Stenophyllanin A | 11.25 | 1207.15 | 1207.89 | C56H40O31 | 98.64 | −1.08 |
| 22 | Procyanidin trimer isomer 1 | 11.25 | 865.20 | 865.77 | C45H38O18 | 97.53 | −1.59 |
| 23 | (epi)-catechin | 11.26 | 289.07 | 289.26 | C15H14O6 | 96.76 | −3.18 |
| 24 | Procyanidin tetramer | 11.34 | 1153.26 | 1153.03 | C60H50O24 | 99.60 | −0.50 |
| 25 | Procyanidin trimer isomer 2 | 11.41 | 865.20 | 865.77 | C45H38O18 | 97.53 | −1.59 |
| 26 | Guavin A | 11.50 | 1223.14 | 1223.89 | C56H40O32 | 99.05 | 0.85 |
| 27 | Casuarinin/Casuarictin isomer | 11.90 | 935.08 | 935.64 | C41H28O26 | 97.67 | −1.43 |
| 28 | Galloyl(epi)catechin-(epi)gallocatechin | 12.10 | 745.14 | 745.62 | C37H30O17 | 96.90 | −0.62 |
| 29 | Procyanidin pentamer | 12.14 | 1441.32 | 1441.27 | C75H62O30 | 95.66 | 1.97 |
| 30 | Galloyl-(epi)catechin trimer isomer 1 | 12.17 | 1017.21 | 1017.87 | C52H42O22 | 99.72 | −0.01 |
| 31 | (epi)-gallocatechin isomer 2 | 12.33 | 305.07 | 305.26 | C15H14O7 | 95.55 | −3.32 |
| 32 | Tellimagrandin I isomer | 12.50 | 785.09 | 785.55 | C34H26O22 | 98.44 | −1.38 |
| 33 | Vescalagin | 12.76 | 933.07 | 933.62 | C41H26O26 | 96.33 | −0.80 |
| 34 | Stenophyllanin A isomer | 12.93 | 1207.15 | 1207.89 | C56H40O31 | 98.37 | 0.89 |
| 35 | Galloyl-(epi)catechin trimer isomer 2 | 12.99 | 1017.21 | 1017.87 | C52H42O22 | 98.17 | −1.35 |
| 36 | Myricetin hexoside isomer 1 | 13.28 | 479.08 | 479.37 | C21H20O13 | 98.36 | −0.92 |
| 37 | Stachyuranin A | 13.41 | 1225.16 | 1225.91 | C56H42O32 | 95.54 | 1.35 |
| 38 | Procyanidin gallate isomer | 13.52 | 729.15 | 729.62 | C37H30O16 | 96.89 | −1.91 |
| 39 | Myricetin hexoside isomer 2 | 13.68 | 479.08 | 479.37 | C21H20O13 | 97.89 | −0.08 |
| 40 | Vescalagin/castalagin isomer | 13.84 | 933.07 | 933.62 | C41H26O26 | 88.32 | −1.57 |
| 41 | Myricetin-arabinoside/xylopyranoside isomer 1 | 13.99 | 449.07 | 449.34 | C20H18O12 | 98.39 | −1.65 |
| 42 | Myricetin-arabinoside/xylopyranoside isomer 2 | 14.21 | 449.07 | 449.34 | C20H18O12 | 98.02 | −1.65 |
| 43 | Procyanidin gallate isomer | 14.56 | 729.64 | 577.51 | C30H26O12 | 98.17 | −1.73 |
| 44 | Myricetin-arabinoside/xylopyranoside isomer 3 | 14.99 | 449.07 | 449.34 | C20H18O12 | 98.66 | −1.65 |
| 45 | Myricetin hexoside isomer 3 | 15.03 | 479.08 | 479.37 | C21H20O13 | 97.08 | −1.92 |
| 46 | Myricetin hexoside isomer 4 | 15.22 | 479.08 | 479.37 | C21H20O13 | 97.08 | −1.92 |
| 47 | Myricetin-arabinoside/xylopyranoside Isomer 4 | 15.60 | 449.07 | 449.34 | C20H18O12 | 98.39 | −1.65 |
| 48 | Quercetin-galloylhexoside isomer | 15.63 | 615.10 | 615.47 | C28H24O16 | 99.16 | −0.98 |
| 49 | Ellagic acid deoxyhexoside | 15.84 | 447.06 | 447.33 | C20H16O12 | 91.25 | −3.19 |
| 50 | Quercetin-galloylhexoside isomer | 16.04 | 615.10 | 615.47 | C28H24O16 | 99.16 | −0.98 |
| 51 | Myricetin-arabinoside/xylopyranoside isomer 5 | 16.19 | 449.07 | 449.34 | C20H18O12 | 98.39 | −1.65 |
| 52 | Morin | 16.28 | 301.04 | 301.23 | C15H10O7 | 97.46 | −2.50 |
| 53 | Myricetin-arabinoside/xylopyranoside isomer 6 | 16.46 | 449.07 | 449.34 | C20H18O12 | 98.39 | −1.65 |
| 54 | Ellagic acid | 16.51 | 301.00 | 301.19 | C14H6O8 | 98.88 | −1.71 |
| 55 | Hyperin | 16.62 | 463.09 | 463.37 | C21H20O12 | 96.41 | −2.65 |
| 56 | Quercetin glucoronide | 16.72 | 477.07 | 477.35 | C21H18O13 | 98.10 | −1.83 |
| 57 | Isoquercitrin | 16.95 | 463.09 | 463.37 | C21H20O12 | 97.04 | −2.33 |
| 58 | Procyanidin gallate isomer | 17.04 | 729.15 | 729.62 | C37H30O16 | 96.89 | −1.91 |
| 59 | Reynoutrin | 17.50 | 433.08 | 433.34 | C20H18O11 | 95.94 | −2.90 |
| 60 | Guajaverin | 17.80 | 433.08 | 433.34 | C20H18O11 | 97.99 | −1.91 |
| 61 | Guavinoside A isomer 1 | 17.96 | 543.12 | 544.46 | C26H24O13 | 98.10 | −1.77 |
| 62 | Avicularin | 18.21 | 433.08 | 433.34 | C20H18O11 | 96.70 | −2.20 |
| 63 | Quercitrin | 19.19 | 447.10 | 447.37 | C21H20O11 | 95.23 | −3.02 |
| 64 | Myrciaphenone B | 19.21 | 481.10 | 481.38 | C21H22O13 | 97.20 | −2.23 |
| 65 | Guavinoside C | 19.77 | 585.09 | 585.45 | C27H22O15 | 97.19 | −1.92 |
| 66 | Guavinoside B isomer 1 | 20.77 | 571.15 | 571.51 | C28H28O13 | 97.26 | −2.05 |
| 67 | Guavinoside A isomer 2 | 20.70 | 543.12 | 543.45 | C26H24O13 | 98.10 | −1.77 |
| 68 | Guavinoside B isomer 2 | 21.67 | 571.15 | 571.51 | C28H28O13 | 97.26 | −2.05 |
| 69 | 2,6-dihydroxy-3-methyl-4-O-(6″-O-galloyl-β-D-glucopyranosyl)-benzophenone | 21.97 | 557.13 | 557.48 | C27H26O13 | 96.93 | −2.12 |
| 70 | Guavin B | 22.24 | 693.11 | 693.54 | C33H26O17 | 97.82 | −1.67 |
| 71 | Quercetin | 22.31 | 301.04 | 301.23 | C15H10O7 | 98.90 | −1.34 |
| 72 | Naringenin isomer | 26.74 | 271.06 | 271.25 | C15H12O5 | 96.09 | −3.67 |
| Positive mode | |||||||
| 73 | Cyanidin-3-O-glucoside | 3.66 | 449.11 | 449.39 | C21H21O11 | 96.97 | −2.34 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Díaz-de-Cerio, E.; Girón, F.; Pérez-Garrido, A.; Pereira, A.S.P.; Gabaldón-Hernández, J.A.; Verardo, V.; Segura Carretero, A.; Pérez-Sánchez, H. Fishing the Targets of Bioactive Compounds from Psidium guajava L. Leaves in the Context of Diabetes. Int. J. Mol. Sci. 2023, 24, 5761. https://doi.org/10.3390/ijms24065761
Díaz-de-Cerio E, Girón F, Pérez-Garrido A, Pereira ASP, Gabaldón-Hernández JA, Verardo V, Segura Carretero A, Pérez-Sánchez H. Fishing the Targets of Bioactive Compounds from Psidium guajava L. Leaves in the Context of Diabetes. International Journal of Molecular Sciences. 2023; 24(6):5761. https://doi.org/10.3390/ijms24065761
Chicago/Turabian StyleDíaz-de-Cerio, Elixabet, Francisco Girón, Alfonso Pérez-Garrido, Andreia S. P. Pereira, José Antonio Gabaldón-Hernández, Vito Verardo, Antonio Segura Carretero, and Horacio Pérez-Sánchez. 2023. "Fishing the Targets of Bioactive Compounds from Psidium guajava L. Leaves in the Context of Diabetes" International Journal of Molecular Sciences 24, no. 6: 5761. https://doi.org/10.3390/ijms24065761
APA StyleDíaz-de-Cerio, E., Girón, F., Pérez-Garrido, A., Pereira, A. S. P., Gabaldón-Hernández, J. A., Verardo, V., Segura Carretero, A., & Pérez-Sánchez, H. (2023). Fishing the Targets of Bioactive Compounds from Psidium guajava L. Leaves in the Context of Diabetes. International Journal of Molecular Sciences, 24(6), 5761. https://doi.org/10.3390/ijms24065761








