MiRNAs in Hematopoiesis and Acute Lymphoblastic Leukemia
Abstract
1. Introduction
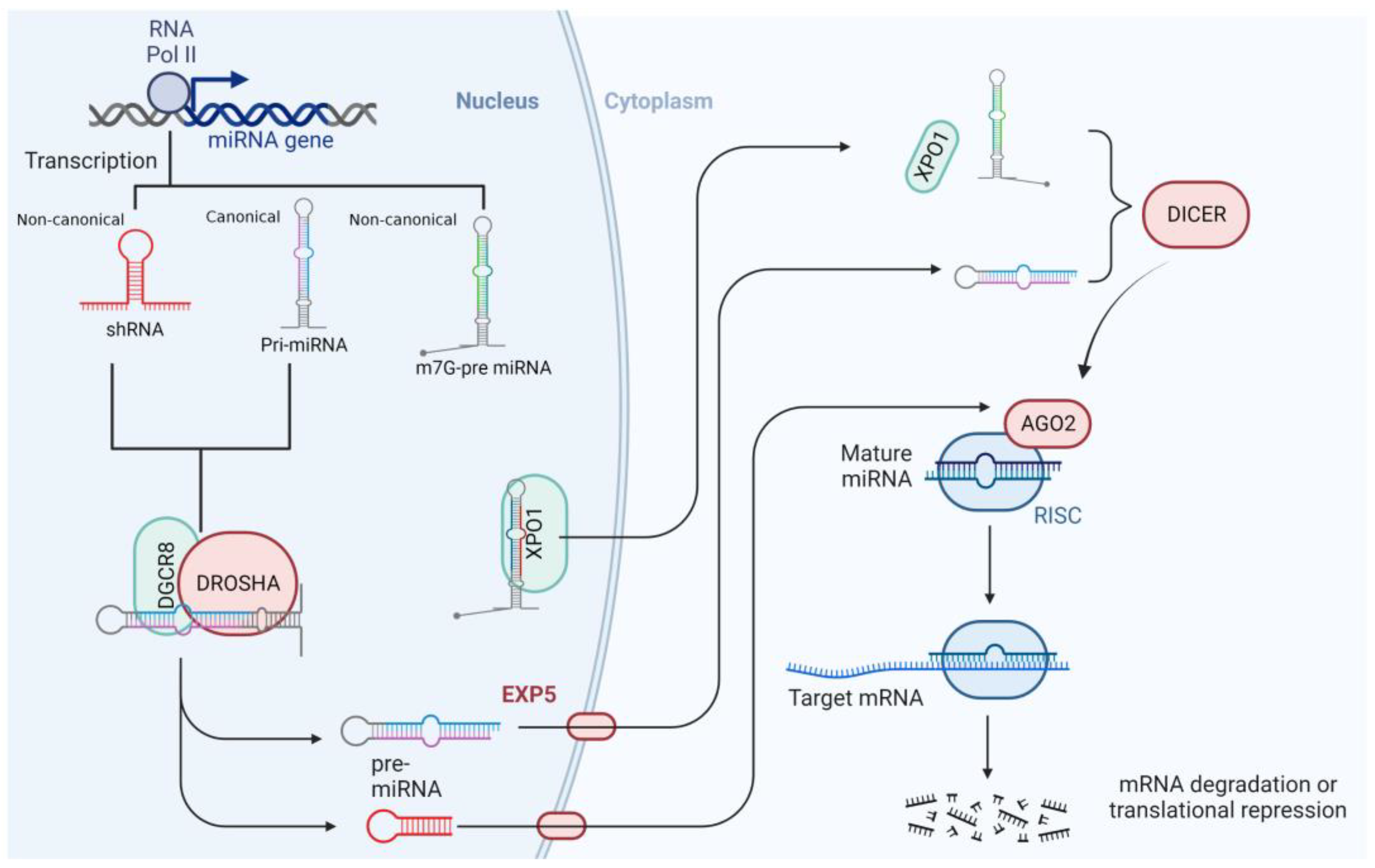
2. MiRNAs’ Features and Biogenesis
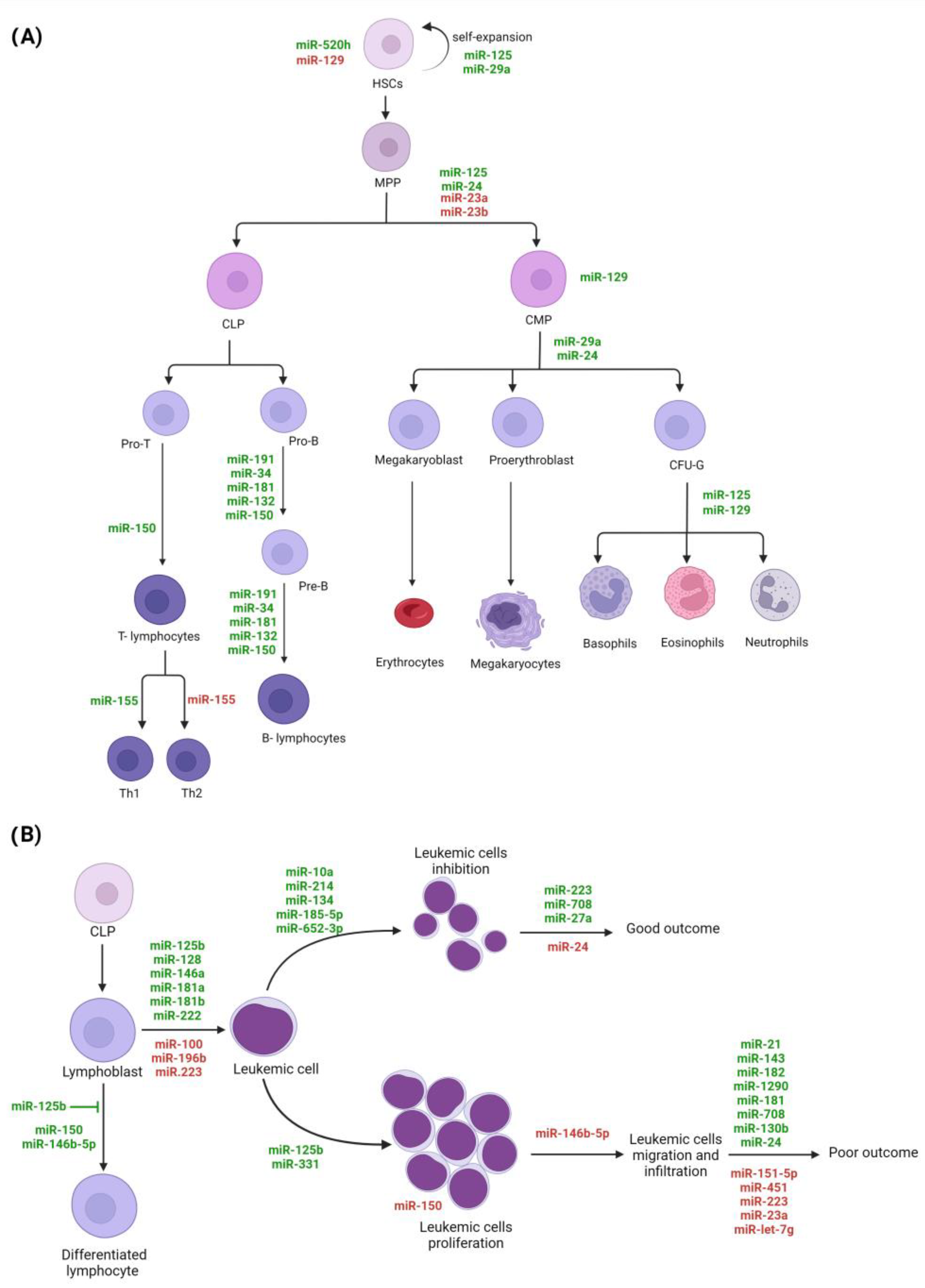
3. MiRNAs and Hematopoiesis
4. MiRNAs’ Role in Acute Lymphoblastic Leukemia
4.1. MiRNAs Polymorphisms as Risk Factors
4.2. MiRNAs’ SNPs and Drug Metabolism
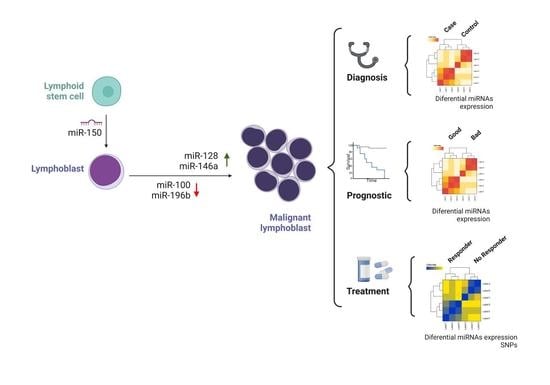
4.3. MiRNAs’ Expression Profiles and Their Role as Biomarkers in ALL
4.3.1. Diagnostic Biomarkers
4.3.2. Immunophenotype Classification
4.3.3. Molecular Subtype Identification
4.3.4. Disease-Free Survival and Overall Survival
5. Molecular Mechanisms of miRNAs in ALL
6. Closing Remarks and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fernandes, M.R.; Souza Vinagre, L.W.M.; Rodrigues, J.C.G.; Wanderley, A.V.; Fernandes, S.M.; Gellen, L.P.A.; Alcantara, A.L.; Sousa, B.B.; Burbano, R.M.R.; Assumpcao, P.P.; et al. Correlation of Genetic Variants and the Incidence, Prevalence and Mortality Rates of Acute Lymphoblastic Leukemia. J. Pers. Med. 2022, 12, 370. [Google Scholar]
- Jammal, N.; Chew, S.; Jabbour, E.; Kantarjian, H. Antibody based therapy in relapsed acute lymphoblastic leukemia. Best Pract. Res. Clin. Haematol. 2020, 33, 101225. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, J.C.; Brassesco, M.S.; Scrideli, C.A.; Tone, L.G.; Narendran, A. MicroRNA expression and activity in pediatric acute lymphoblastic leukemia (ALL). Pediatr. Blood Cancer 2012, 59, 599–604. [Google Scholar] [CrossRef]
- Jaime-Perez, J.C.; Jimenez-Castillo, R.A.; Pinzon-Uresti, M.A.; Cantu-Rodriguez, O.G.; Herrera-Garza, J.L.; Marfil-Rivera, L.J.; Gomez-Almaguer, D. Real-world outcomes of treatment for acute lymphoblastic leukemia during adolescence in a financially restricted environment: Results at a single center in Latin America. Pediatr. Blood Cancer 2017, 64, e26396. [Google Scholar]
- Nunez-Enriquez, J.C.; Barcenas-Lopez, D.A.; Hidalgo-Miranda, A.; Jimenez-Hernandez, E.; Bekker-Mendez, V.C.; Flores-Lujano, J.; Solis-Labastida, K.A.; Martinez-Morales, G.B.; Sanchez-Munoz, F.; Espinoza-Hernandez, L.E.; et al. Gene Expression Profiling of Acute Lymphoblastic Leukemia in Children with Very Early Relapse. Arch. Med. Res. 2016, 47, 644–655. [Google Scholar] [PubMed]
- Jimenez-Hernandez, E.; Jaimes-Reyes, E.Z.; Arellano-Galindo, J.; Garcia-Jimenez, X.; Tiznado-Garcia, H.M.; Duenas-Gonzalez, M.T.; Martinez Villegas, O.; Sanchez-Jara, B.; Bekker-Mendez, V.C.; Ortiz-Torres, M.G.; et al. Survival of Mexican Children with Acute Lymphoblastic Leukaemia under Treatment with the Protocol from the Dana-Farber Cancer Institute 00-01. Biomed. Res. Int. 2015, 2015, 576950. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Hernandez, E.; Duenas-Gonzalez, M.T.; Arellano-Galindo, J.; Medrano-Ortiz-De-Zarate, M.E.; Bekker-Mendez, V.C.; Berges-Garcia, A.; Solis-Labastida, K.; Sanchez-Jara, B.; Tiznado-Garcia, H.M.; Jaimes-Reyes, E.Z.; et al. Survival of Mexican children with acute myeloid leukaemia who received early intensification chemotherapy and an autologous transplant. Biomed. Res. Int. 2015, 2015, 940278. [Google Scholar]
- Ko, R.H.; Ji, L.; Barnette, P.; Bostrom, B.; Hutchinson, R.; Raetz, E.; Seibel, N.L.; Twist, C.J.; Eckroth, E.; Sposto, R.; et al. Outcome of patients treated for relapsed or refractory acute lymphoblastic leukemia: A Therapeutic Advances in Childhood Leukemia Consortium study. J. Clin. Oncol. 2010, 28, 648–654. [Google Scholar] [CrossRef]
- Wu, C.; Li, W. Genomics and pharmacogenomics of pediatric acute lymphoblastic leukemia. Crit. Rev. Oncol. Hematol. 2018, 126, 100–111. [Google Scholar]
- Nana-Sinkam, S.P.; Croce, C.M. MicroRNAs as therapeutic targets in cancer. Transl. Res. 2011, 157, 216–225. [Google Scholar] [CrossRef]
- Palazzo, A.F.; Lee, E.S. Non-coding RNA: What is functional and what is junk? Front. Genet. 2015, 6, 2. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Wu, W.; Chen, Q.; Chen, M. Non-Coding RNAs and their Integrated Networks. J. Integr. Bioinform. 2019, 16. [Google Scholar] [CrossRef] [PubMed]
- Bhatti, G.K.; Khullar, N.; Sidhu, I.S.; Navik, U.S.; Reddy, A.P.; Reddy, P.H.; Bhatti, J.S. Emerging role of non-coding RNA in health and disease. Metab. Brain Dis. 2021, 36, 1119–1134. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef] [PubMed]
- Croce, C.M. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 2009, 10, 704–714. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Lee, H.; Han, S.; Kwon, C.S.; Lee, D. Biogenesis and regulation of the let-7 miRNAs and their functional implications. Protein Cell 2016, 7, 100–113. [Google Scholar] [CrossRef]
- Elshazli, R.M.; Toraih, E.A.; Hussein, M.H.; Ruiz, E.M.; Kandil, E.; Fawzy, M.S. Pan-Cancer Study on Variants of Canonical miRNA Biogenesis Pathway Components: A Pooled Analysis. Cancers 2023, 15, 338. [Google Scholar] [CrossRef]
- Lee, Y.S.; Nakahara, K.; Pham, J.W.; Kim, K.; He, Z.; Sontheimer, E.J.; Carthew, R.W. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 2004, 117, 69–81. [Google Scholar] [CrossRef]
- Yi, R.; Qin, Y.; Macara, I.G.; Cullen, B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes. Dev. 2003, 17, 3011–3016. [Google Scholar] [CrossRef]
- Makarova, J.A.; Shkurnikov, M.U.; Wicklein, D.; Lange, T.; Samatov, T.R.; Turchinovich, A.A.; Tonevitsky, A.G. Intracellular and extracellular microRNA: An update on localization and biological role. Prog. Histochem. Cytochem. 2016, 51, 33–49. [Google Scholar] [CrossRef] [PubMed]
- De Rie, D.; Abugessaisa, I.; Alam, T.; Arner, E.; Arner, P.; Ashoor, H.; Astrom, G.; Babina, M.; Bertin, N.; Burroughs, A.M.; et al. An integrated expression atlas of miRNAs and their promoters in human and mouse. Nat. Biotechnol. 2017, 35, 872–878. [Google Scholar] [CrossRef]
- Tanzer, A.; Stadler, P.F. Molecular evolution of a microRNA cluster. J. Mol. Biol. 2004, 339, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Broughton, J.P.; Lovci, M.T.; Huang, J.L.; Yeo, G.W.; Pasquinelli, A.E. Pairing beyond the Seed Supports MicroRNA Targeting Specificity. Mol. Cell 2016, 64, 320–333. [Google Scholar] [CrossRef] [PubMed]
- Huang, V.; Qin, Y.; Wang, J.; Wang, X.; Place, R.F.; Lin, G.; Lue, T.F.; Li, L.C. RNAa is conserved in mammalian cells. PLoS ONE 2010, 5, e8848. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Jiang, Q.; Chang, N.; Wang, X.; Liu, C.; Xiong, J.; Cao, H.; Liang, Z. Small activating RNA binds to the genomic target site in a seed-region-dependent manner. Nucleic Acids Res. 2016, 44, 2274–2282. [Google Scholar] [CrossRef]
- Portnoy, V.; Lin, S.H.; Li, K.H.; Burlingame, A.; Hu, Z.H.; Li, H.; Li, L.C. saRNA-guided Ago2 targets the RITA complex to promoters to stimulate transcription. Cell Res. 2016, 26, 320–335. [Google Scholar] [CrossRef]
- Vasudevan, S. Posttranscriptional upregulation by microRNAs. Wiley Interdiscip. Rev. 2012, 3, 311–330. [Google Scholar] [CrossRef]
- Comazzetto, S.; Shen, B.; Morrison, S.J. Niches that regulate stem cells and hematopoiesis in adult bone marrow. Dev. Cell 2021, 56, 1848–1860. [Google Scholar] [CrossRef]
- Man, Y.; Yao, X.; Yang, T.; Wang, Y. Hematopoietic Stem Cell Niche During Homeostasis, Malignancy, and Bone Marrow Transplantation. Front. Cell Dev. Biol. 2021, 9, 621214. [Google Scholar] [CrossRef]
- Kim, M.; Civin, C.I.; Kingsbury, T.J. MicroRNAs as regulators and effectors of hematopoietic transcription factors. Wiley Interdiscip. Rev. 2019, 10, e1537. [Google Scholar] [CrossRef] [PubMed]
- Levesque, J.P.; Winkler, I.G. Hierarchy of immature hematopoietic cells related to blood flow and niche. Curr. Opin. Hematol. 2011, 18, 220–225. [Google Scholar] [CrossRef] [PubMed]
- Kovtonyuk, L.V.; Fritsch, K.; Feng, X.; Manz, M.G.; Takizawa, H. Inflamm-Aging of Hematopoiesis, Hematopoietic Stem Cells, and the Bone Marrow Microenvironment. Front. Immunol. 2016, 7, 502. [Google Scholar] [CrossRef] [PubMed]
- Frobel, J.; Landspersky, T.; Percin, G.; Schreck, C.; Rahmig, S.; Ori, A.; Nowak, D.; Essers, M.; Waskow, C.; Oostendorp, R.A.J.; et al. The Hematopoietic Bone Marrow Niche Ecosystem. Front. Cell Dev. Biol. 2021, 9, 705410. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri-Fard, S.; Niazi, V.; Taheri, M. Role of miRNAs and lncRNAs in hematopoietic stem cell differentiation. Noncoding RNA Res. 2021, 6, 8–14. [Google Scholar] [CrossRef]
- Bissels, U.; Bosio, A.; Wagner, W. MicroRNAs are shaping the hematopoietic landscape. Haematologica 2012, 97, 160–167. [Google Scholar] [CrossRef]
- Luinenburg, D.G.; de Haan, G. MicroRNAs in hematopoietic stem cell aging. Mech. Ageing Dev. 2020, 189, 111281. [Google Scholar] [CrossRef]
- Edginton-White, B.; Bonifer, C. The transcriptional regulation of normal and malignant blood cell development. FEBS J. 2022, 289, 1240–1255. [Google Scholar] [CrossRef]
- Neaga, A.; Bagacean, C.; Tempescul, A.; Jimbu, L.; Mesaros, O.; Blag, C.; Tomuleasa, C.; Bocsan, C.; Gaman, M.; Zdrenghea, M.; et al. MicroRNAs Associated with a Good Prognosis of Acute Myeloid Leukemia and Their Effect on Macrophage Polarization. Front. Immunol. 2020, 11, 582915. [Google Scholar] [CrossRef]
- Alemdehy, M.F.; Erkeland, S.J. MicroRNAs: Key players of normal and malignant myelopoiesis. Curr. Opin. Hematol. 2012, 19, 261–267. [Google Scholar] [CrossRef] [PubMed]
- Luan, C.; Yang, Z.; Chen, B. The functional role of microRNA in acute lymphoblastic leukemia: Relevance for diagnosis, differential diagnosis, prognosis, and therapy. Onco Targets Ther. 2015, 8, 2903–2914. [Google Scholar] [PubMed]
- Attaway, M.; Chwat-Edelstein, T.; Vuong, B.Q. Regulatory Non-Coding RNAs Modulate Transcriptional Activation During B Cell Development. Front. Genet. 2021, 12, 678084. [Google Scholar] [CrossRef] [PubMed]
- Wallaert, A.; Durinck, K.; Taghon, T.; van Vlierberghe, P.; Speleman, F. T-ALL and thymocytes: A message of noncoding RNAs. J. Hematol. Oncol. 2017, 10, 66. [Google Scholar] [CrossRef]
- Winter, S.J.; Krueger, A. Development of Unconventional T Cells Controlled by MicroRNA. Front. Immunol. 2019, 10, 2520. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; So, A.Y.; Sookram, R.; Wong, S.; Wang, J.K.; Ouyang, Y.; He, P.; Su, Y.; Casellas, R.; Baltimore, D. Epigenetic silencing of miR-125b is required for normal B-cell development. Blood 2018, 131, 1920–1930. [Google Scholar] [CrossRef]
- Kretov, D.A.; Shafik, A.M.; Cifuentes, D. Assessing miR-451 Activity and Its Role in Erythropoiesis. Methods Mol. Biol. 2018, 1680, 179–190. [Google Scholar]
- Renou, L.; Boelle, P.Y.; Deswarte, C.; Spicuglia, S.; Benyoucef, A.; Calvo, J.; Uzan, B.; Belhocine, M.; Cieslak, A.; Landman-Parker, J.; et al. Homeobox protein TLX3 activates miR-125b expression to promote T-cell acute lymphoblastic leukemia. Blood Adv. 2017, 1, 733–747. [Google Scholar] [CrossRef]
- Liu, Z.; Smith, K.R.; Khong, H.T.; Huang, J.; Ahn, E.E.; Zhou, M.; Tan, M. miR-125b regulates differentiation and metabolic reprogramming of T cell acute lymphoblastic leukemia by directly targeting A20. Oncotarget 2016, 7, 78667–78679. [Google Scholar] [CrossRef]
- Podshivalova, K.; Wang, E.A.; Hart, T.; Salomon, D.R. Expression of the miR-150 tumor suppressor is restored by and synergizes with rapamycin in a human leukemia T-cell line. Leuk. Res. 2018, 74, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Correia, N.C.; Fragoso, R.; Carvalho, T.; Enguita, F.J.; Barata, J.T. MiR-146b negatively regulates migration and delays progression of T-cell acute lymphoblastic leukemia. Sci. Rep. 2016, 6, 31894. [Google Scholar] [CrossRef] [PubMed]
- Weiss, C.N.; Ito, K. A Macro View of MicroRNAs: The Discovery of MicroRNAs and Their Role in Hematopoiesis and Hematologic Disease. Int. Rev. Cell Mol. Biol. 2017, 334, 99–175. [Google Scholar] [PubMed]
- Sole, C.; Larrea, E.; Manterola, L.; Goicoechea, I.; Armesto, M.; Arestin, M.; Caffarel, M.; Araujo, A.; Fernandez, M.; Araiz, M.; et al. Aberrant expression of MicroRNAs in B-cell lymphomas. Microrna 2016, 5, 87–105. [Google Scholar] [CrossRef]
- Guo, J.R.; Li, W.; Wu, Y.; Wu, L.Q.; Li, X.; Guo, Y.F.; Zheng, X.H.; Lian, X.L.; Huang, H.F.; Chen, Y.Z.; et al. Hepatocyte growth factor promotes proliferation, invasion, and metastasis of myeloid leukemia cells through PI3K-AKT and MAPK/ERK signaling pathway. Am. J. Transl. Res. 2016, 8, 3630–3644. [Google Scholar] [PubMed]
- Tapeh, B.E.G.; Alivand, M.R.; Solali, S. The role of microRNAs in acute lymphoblastic leukaemia: From biology to applications. Cell Biochem. Funct. 2020, 38, 334–346. [Google Scholar] [CrossRef] [PubMed]
- Turk, A.; Calin, G.A.; Kunej, T. MicroRNAs in Leukemias: A Clinically Annotated Compendium. Int. J. Mol. Sci. 2022, 23, 3469. [Google Scholar] [CrossRef] [PubMed]
- Mardani, R.; Jafari Najaf Abadi, M.H.; Motieian, M.; Taghizadeh-Boroujeni, S.; Bayat, A.; Farsinezhad, A.; Gheibi Hayat, S.M.; Motieian, M.; Pourghadamyari, H. MicroRNA in leukemia: Tumor suppressors and oncogenes with prognostic potential. J. Cell Physiol. 2019, 234, 8465–8486. [Google Scholar] [CrossRef]
- Rawoof, A.; Swaminathan, G.; Tiwari, S.; Nair, R.A.; Dinesh Kumar, L. LeukmiR: A database for miRNAs and their targets in acute lymphoblastic leukemia. Database 2020, 2020, baz151. [Google Scholar] [CrossRef]
- Xie, Y.; Xu, Z.; Wang, K.; Wang, N.; Wang, Y.; Bao, J. Networks of micrornas and genes in acute lymphoblastic leukemia. Mol. Med. Rep. 2015, 12, 5361–5368. [Google Scholar] [CrossRef]
- Gutierrez-Camino, A.; Lopez-Lopez, E.; Martin-Guerrero, I.; Pinan, M.A.; Garcia-Miguel, P.; Sanchez-Toledo, J.; Carbone Baneres, A.; Uriz, J.; Navajas, A.; Garcia-Orad, A.; et al. Noncoding RNA-related polymorphisms in pediatric acute lymphoblastic leukemia susceptibility. Pediatr. Res. 2014, 75, 767–773. [Google Scholar] [CrossRef]
- Jemimah Devanandan, H.; Venkatesan, V.; Scott, J.X.; Magatha, L.S.; Durairaj Paul, S.F.; Koshy, T. MicroRNA 146a Polymorphisms and Expression in Indian Children with Acute Lymphoblastic Leukemia. Lab. Med. 2019, 50, 249–253. [Google Scholar] [CrossRef]
- Pei, J.S.; Chang, W.S.; Hsu, P.C.; Chen, C.C.; Chin, Y.T.; Huang, T.L.; Hsu, Y.N.; Kuo, C.C.; Wang, Y.C.; Tsai, C.W.; et al. Significant Association Between the MiR146a Genotypes and Susceptibility to Childhood Acute Lymphoblastic Leukemia in Taiwan. Cancer Genom. Proteom. 2020, 17, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Zou, D.; Yin, J.; Ye, Z.; Zeng, Q.; Tian, C.; Wang, Y.; Chen, Q.; Chen, R. Association Between the miR-146a Rs2910164 Polymorphism and Childhood Acute Lymphoblastic Leukemia Susceptibility in an Asian Population. Front. Genet. 2020, 11, 886. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Zhu, Y.; Wei, Q.K.; Yuan, Y.; Zhou, F.; Ge, Y.Y.; Yang, J.R.; Su, H.; Zhuang, S.M. A functional polymorphism in the miR-146a gene is associated with the risk for hepatocellular carcinoma. Carcinogenesis 2008, 29, 2126–2131. [Google Scholar] [CrossRef] [PubMed]
- Hasani, S.S.; Hashemi, M.; Eskandari-Nasab, E.; Naderi, M.; Omrani, M.; Sheybani-Nasab, M. A functional polymorphism in the miR-146a gene is associated with the risk of childhood acute lymphoblastic leukemia: A preliminary report. Tumour Biol. 2014, 35, 219–225. [Google Scholar] [CrossRef]
- Jimenez-Morales, S.; Nunez-Enriquez, J.C.; Cruz-Islas, J.; Bekker-Mendez, V.C.; Jimenez-Hernandez, E.; Medina-Sanson, A.; Olarte-Carrillo, I.; Martinez-Tovar, A.; Flores-Lujano, J.; Ramirez-Bello, J.; et al. Association Analysis Between the Functional Single Nucleotide Variants in miR-146a, miR-196a-2, miR-499a, and miR-612 with Acute Lymphoblastic Leukemia. Front. Oncol. 2021, 11, 762063. [Google Scholar] [CrossRef]
- Chansing, K.; Pakakasama, S.; Hongeng, S.; Thongmee, A.; Pongstaporn, W. Lack of Association between the MiR146a Polymorphism and Susceptibility to Thai Childhood Acute Lymphoblastic Leukemia. Asian Pac. J. Cancer Prev. 2016, 17, 2435–2438. [Google Scholar]
- Xue, Y.; Yang, X.; Hu, S.; Kang, M.; Chen, J.; Fang, Y. A genetic variant in miR-100 is a protective factor of childhood acute lymphoblastic leukemia. Cancer Med. 2019, 8, 2553–2560. [Google Scholar] [CrossRef]
- Hoffman, A.E.; Zheng, T.; Yi, C.; Leaderer, D.; Weidhaas, J.; Slack, F.; Zhang, Y.; Paranjape, T.; Zhu, Y. microRNA miR-196a-2 and breast cancer: A genetic and epigenetic association study and functional analysis. Cancer Res. 2009, 69, 5970–5977. [Google Scholar] [CrossRef]
- Xu, W.; Xu, J.; Liu, S.; Chen, B.; Wang, X.; Li, Y.; Qian, Y.; Zhao, W.; Wu, J. Effects of common polymorphisms rs11614913 in miR-196a2 and rs2910164 in miR-146a on cancer susceptibility: A meta-analysis. PLoS ONE 2011, 6, e20471. [Google Scholar] [CrossRef]
- Tian, T.; Xu, Y.; Dai, J.; Wu, J.; Shen, H.; Hu, Z. Functional polymorphisms in two pre-microRNAs and cancer risk: A meta-analysis. Int. J. Mol. Epidemiol. Genet. 2010, 1, 358–366. [Google Scholar] [PubMed]
- Hu, Z.; Chen, J.; Tian, T.; Zhou, X.; Gu, H.; Xu, L.; Zeng, Y.; Miao, R.; Jin, G.; Ma, H.; et al. Genetic variants of miRNA sequences and non-small cell lung cancer survival. J. Clin. Invest. 2008, 118, 2600–2608. [Google Scholar] [CrossRef] [PubMed]
- Tong, N.; Xu, B.; Shi, D.; Du, M.; Li, X.; Sheng, X.; Wang, M.; Chu, H.; Fang, Y.; Li, J.; et al. Hsa-miR-196a2 polymorphism increases the risk of acute lymphoblastic leukemia in Chinese children. Mutat. Res. 2014, 759, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Rakmanee, S.; Pakakasama, S.; Hongeng, S.; Sanguansin, S.; Thongmee, A.; Pongstaporn, W. Increased Risk of Thai Childhood Acute Lymphoblastic Leukemia with the MiR196a2 T>C Polymorphism. Asian Pac. J. Cancer Prev. 2017, 18, 1117–1120. [Google Scholar] [PubMed]
- Chen, C.C.; Hsu, P.C.; Shih, L.C.; Hsu, Y.N.; Kuo, C.C.; Chao, C.Y.; Chang, W.S.; Tsai, C.W.; Bau, D.T.; Pei, J.S.; et al. MiR-196a-2 Genotypes Determine the Susceptibility and Early Onset of Childhood Acute Lymphoblastic Leukemia. Anticancer Res. 2020, 40, 4465–4469. [Google Scholar] [CrossRef]
- Liu, Z.; Li, G.; Wei, S.; Niu, J.; El-Naggar, A.K.; Sturgis, E.M.; Wei, Q. Genetic variants in selected pre-microRNA genes and the risk of squamous cell carcinoma of the head and neck. Cancer 2010, 116, 4753–4760. [Google Scholar] [CrossRef]
- Hu, Z.; Liang, J.; Wang, Z.; Tian, T.; Zhou, X.; Chen, J.; Miao, R.; Wang, Y.; Wang, X.; Shen, H.; et al. Common genetic variants in pre-microRNAs were associated with increased risk of breast cancer in Chinese women. Hum. Mutat. 2009, 30, 79–84. [Google Scholar] [CrossRef]
- Xiang, Y.; Fan, S.; Cao, J.; Huang, S.; Zhang, L.P. Association of the microRNA-499 variants with susceptibility to hepatocellular carcinoma in a Chinese population. Mol. Biol. Rep. 2012, 39, 7019–7023. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, B.; Ren, X. Hsa-miR-499 polymorphism (rs3746444) and cancer risk: A meta-analysis of 17 case-control studies. Gene 2012, 509, 267–272. [Google Scholar] [CrossRef]
- Wang, L.; Qian, S.; Zhi, H.; Zhang, Y.; Wang, B.; Lu, Z. The association between hsa-miR-499 T>C polymorphism and cancer risk: A meta-analysis. Gene 2012, 508, 9–14. [Google Scholar] [CrossRef]
- de Souza, T.P.; de Carvalho, D.C.; Wanderley, A.V.; Fernandes, S.M.; Rodrigues, J.C.G.; Cohen-Paes, A.; Fernandes, M.R.; Mello Junior, F.A.R.; Pastana, L.F.; Vinagre, L.; et al. Influence of variants of the drosha, mir499a, and mir938 genes on susceptibility to acute lymphoblastic leukemia in an admixed population from the brazilian amazon. Am. J. Transl. Res. 2020, 12, 8216–8224. [Google Scholar] [PubMed]
- Kim, H.K.; Prokunina-Olsson, L.; Chanock, S.J. Common genetic variants in miR-1206 (8q24.2) and miR-612 (11q13.3) affect biogenesis of mature miRNA forms. PLoS ONE 2012, 7, e47454. [Google Scholar] [CrossRef] [PubMed]
- Siyadat, P.; Ayatollahi, H.; Barati, M.; Sheikhi, M.; Shahidi, M. High Resolution Melting Analysis for Evaluation of mir-612 (Rs12803915) Genetic Variant with Susceptibility to Pediatric Acute Lymphoblastic Leukemia. Rep. Biochem. Mol. Biol. 2021, 9, 385–393. [Google Scholar] [CrossRef]
- Trevino, L.R.; Yang, W.; French, D.; Hunger, S.P.; Carroll, W.L.; Devidas, M.; Willman, C.; Neale, G.; Downing, J.; Raimondi, S.C.; et al. Germline genomic variants associated with childhood acute lymphoblastic leukemia. Nat. Genet. 2009, 41, 1001–1005. [Google Scholar] [CrossRef] [PubMed]
- Papaemmanuil, E.; Hosking, F.J.; Vijayakrishnan, J.; Price, A.; Olver, B.; Sheridan, E.; Kinsey, S.E.; Lightfoot, T.; Roman, E.; Irving, J.A.; et al. Loci on 7p12.2, 10q21.2 and 14q11.2 are associated with risk of childhood acute lymphoblastic leukemia. Nat. Genet. 2009, 41, 1006–1010. [Google Scholar] [CrossRef] [PubMed]
- Son, M.S.; Jang, M.J.; Jeon, Y.J.; Kim, W.H.; Kwon, C.I.; Ko, K.H.; Park, P.W.; Hong, S.P.; Rim, K.S.; Kwon, S.W.; et al. Promoter polymorphisms of pri-miR-34b/c are associated with hepatocellular carcinoma. Gene 2013, 524, 156–160. [Google Scholar] [CrossRef]
- Ji, T.X.; Zhi, C.; Guo, X.G.; Zhou, Q.; Wang, G.Q.; Chen, B.; Ma, F.F. MiR-34b/c rs4938723 Polymorphism Significantly Decreases the Risk of Digestive Tract Cancer: Meta-analysis. Asian Pac. J. Cancer Prev. 2015, 16, 6099–6104. [Google Scholar] [CrossRef]
- Yuan, F.; Sun, R.; Chen, P.; Liang, Y.; Ni, S.; Quan, Y.; Huang, J.; Zhang, L.; Gao, L. Combined analysis of pri-miR-34b/c rs4938723 and TP53 Arg72Pro with cervical cancer risk. Tumor Biol. 2016, 37, 6267–6273. [Google Scholar] [CrossRef]
- Sanaei, S.; Hashemi, M.; Rezaei, M.; Hashemi, S.M.; Bahari, G.; Ghavami, S. Evaluation of the pri-miR-34b/c rs4938723 polymorphism and its association with breast cancer risk. Biomed. Rep. 2016, 5, 125–129. [Google Scholar] [CrossRef]
- Hashemi, M.; Moazeni-Roodi, A.; Bahari, G.; Taheri, M.; Ghavami, S. Association between miR-34b/c rs4938723 polymorphism and risk of cancer: An updated meta-analysis of 27 case-control studies. J. Cell Biochem. 2019, 120, 3306–3314. [Google Scholar] [CrossRef]
- Tong, N.; Chu, H.; Wang, M.; Xue, Y.; Du, M.; Lu, L.; Zhang, H.; Wang, F.; Fang, Y.; Li, J.; et al. Pri-miR-34b/c rs4938723 polymorphism contributes to acute lymphoblastic leukemia susceptibility in Chinese children. Leuk. Lymphoma 2016, 57, 1436–1441. [Google Scholar] [CrossRef] [PubMed]
- Hashemi, M.; Bahari, G.; Naderi, M.; Sadeghi-Bojd, S.; Taheri, M. Pri-miR-34b/c rs4938723 polymorphism is associated with the risk of childhood acute lymphoblastic leukemia. Cancer Genet. 2016, 209, 493–496. [Google Scholar] [CrossRef] [PubMed]
- Amstutz, U.; Offer, S.M.; Sistonen, J.; Joerger, M.; Diasio, R.B.; Largiader, C.R. Polymorphisms in MIR27A Associated with Early-Onset Toxicity in Fluoropyrimidine-Based Chemotherapy. Clin. Cancer Res. 2015, 21, 2038–2044. [Google Scholar] [CrossRef] [PubMed]
- Meulendijks, D.; Henricks, L.M.; Amstutz, U.; Froehlich, T.K.; Largiader, C.R.; Beijnen, J.H.; de Boer, A.; Deenen, M.J.; Cats, A.; Schellens, J.H.; et al. Rs895819 in MIR27A improves the predictive value of DPYD variants to identify patients at risk of severe fluoropyrimidine-associated toxicity. Int. J. Cancer 2016, 138, 2752–2761. [Google Scholar] [CrossRef] [PubMed]
- Zhan, X.; Wu, W.; Han, B.; Gao, G.; Qiao, R.; Lv, J.; Zhang, S.; Zhang, W.; Fan, W.; Chen, H.; et al. Hsa-miR-196a2 functional SNP is associated with severe toxicity after platinum-based chemotherapy of advanced nonsmall cell lung cancer patients in a Chinese population. J. Clin. Lab. Anal. 2012, 26, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Qiu, A.; Min, S.H.; Jansen, M.; Malhotra, U.; Tsai, E.; Cabelof, D.C.; Matherly, L.H.; Zhao, R.; Akabas, M.H.; Goldman, I.D.; et al. Rodent intestinal folate transporters (SLC46A1): Secondary structure, functional properties, and response to dietary folate restriction. Am. J. Physiol. Cell Physiol. 2007, 293, C1669–C1678. [Google Scholar] [CrossRef]
- Zhao, R.; Qiu, A.; Tsai, E.; Jansen, M.; Akabas, M.H.; Goldman, I.D. The proton-coupled folate transporter: Impact on pemetrexed transport and on antifolates activities compared with the reduced folate carrier. Mol. Pharmacol. 2008, 74, 854–862. [Google Scholar] [CrossRef]
- Desmoulin, S.K.; Hou, Z.; Gangjee, A.; Matherly, L.H. The human proton-coupled folate transporter: Biology and therapeutic applications to cancer. Cancer Biol. Ther. 2012, 13, 1355–1373. [Google Scholar] [CrossRef]
- Iparraguirre, L.; Gutierrez-Camino, A.; Umerez, M.; Martin-Guerrero, I.; Astigarraga, I.; Navajas, A.; Sastre, A.; Garcia de Andoin, N.; Garcia-Orad, A. MiR-pharmacogenetics of methotrexate in childhood B-cell acute lymphoblastic leukemia. Pharmacogenet. Genomics 2016, 26, 517–525. [Google Scholar] [CrossRef]
- Yokoi, T.; Nakajima, M. microRNAs as mediators of drug toxicity. Annu. Rev. Pharmacol. Toxicol. 2013, 53, 377–400. [Google Scholar] [CrossRef]
- Lopez-Lopez, E.; Gutierrez-Camino, A.; Pinan, M.A.; Sanchez-Toledo, J.; Uriz, J.J.; Ballesteros, J.; Garcia-Miguel, P.; Navajas, A.; Garcia-Orad, A. Pharmacogenetics of microRNAs and microRNAs biogenesis machinery in pediatric acute lymphoblastic leukemia. PLoS ONE 2014, 9, e91261. [Google Scholar] [CrossRef]
- Ward, L.D.; Kellis, M. HaploReg: A resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012, 40, D930–D934. [Google Scholar] [CrossRef] [PubMed]
- Paugh, S.W.; Bonten, E.J.; Savic, D.; Ramsey, L.B.; Thierfelder, W.E.; Gurung, P.; Malireddi, R.K.; Actis, M.; Mayasundari, A.; Min, J.; et al. NALP3 inflammasome upregulation and CASP1 cleavage of the glucocorticoid receptor cause glucocorticoid resistance in leukemia cells. Nat. Genet. 2015, 47, 607–614. [Google Scholar] [CrossRef] [PubMed]
- Pui, C.H.; Schrappe, M.; Ribeiro, R.C.; Niemeyer, C.M. Childhood and adolescent lymphoid and myeloid leukemia. Hematol. Am. Soc. Hematol. Educ. Program 2004, 2004, 118–145. [Google Scholar] [CrossRef] [PubMed]
- Randolph, T.R. Advances in acute lymphoblastic leukemia. Clin. Lab. Sci. 2004, 17, 235–245. [Google Scholar] [PubMed]
- Ultimo, S.; Martelli, A.M.; Zauli, G.; Vitale, M.; Calin, G.A.; Neri, L.M. Roles and clinical implications of microRNAs in acute lymphoblastic leukemia. J. Cell Physiol. 2018, 233, 5642–5654. [Google Scholar] [CrossRef] [PubMed]
- Longjohn, M.N.; Squires, W.R.B.; Christian, S.L. Meta-analysis of microRNA profiling data does not reveal a consensus signature for B cell acute lymphoblastic leukemia. Gene 2022, 821, 146211. [Google Scholar] [CrossRef]
- Zanette, D.L.; Rivadavia, F.; Molfetta, G.A.; Barbuzano, F.G.; Proto-Siqueira, R.; Silva, W.A., Jr.; Falcao, R.P.; Zago, M.A. miRNA expression profiles in chronic lymphocytic and acute lymphocytic leukemia. Braz. J. Med. Biol. Res. 2007, 40, 1435–1440. [Google Scholar] [CrossRef]
- Schotte, D.; Chau, J.C.; Sylvester, G.; Liu, G.; Chen, C.; van der Velden, V.H.; Broekhuis, M.J.; Peters, T.C.; Pieters, R.; den Boer, M.L.; et al. Identification of new microRNA genes and aberrant microRNA profiles in childhood acute lymphoblastic leukemia. Leukemia 2009, 23, 313–322. [Google Scholar] [CrossRef]
- Schotte, D.; de Menezes, R.X.; Akbari Moqadam, F.; Khankahdani, L.M.; Lange-Turenhout, E.; Chen, C.; Pieters, R.; Den Boer, M.L. MicroRNA characterize genetic diversity and drug resistance in pediatric acute lymphoblastic leukemia. Haematologica 2011, 96, 703–711. [Google Scholar] [CrossRef]
- Zhang, H.; Yang, J.H.; Zheng, Y.S.; Zhang, P.; Chen, X.; Wu, J.; Xu, L.; Luo, X.Q.; Ke, Z.Y.; Zhou, H.; et al. Genome-wide analysis of small RNA and novel MicroRNA discovery in human acute lymphoblastic leukemia based on extensive sequencing approach. PLoS ONE 2009, 4, e6849. [Google Scholar] [CrossRef] [PubMed]
- Ju, X.; Li, D.; Shi, Q.; Hou, H.; Sun, N.; Shen, B. Differential microRNA expression in childhood B-cell precursor acute lymphoblastic leukemia. Pediatr. Hematol. Oncol. 2009, 26, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Fayed, D.; Donia, T.; El-Shanshory, M.; Ali, E.M.M.; Mohamed, T.M. Evaluation of MicroRNA92, MicroRNA638 in Acute Lymphoblastic Leukemia of Egyptian Children. Asian Pac. J. Cancer Prev. 2021, 22, 1567–1572. [Google Scholar] [CrossRef]
- Swellam, M.; Hashim, M.; Mahmoud, M.S.; Ramadan, A.; Hassan, N.M. Aberrant Expression of Some Circulating miRNAs in Childhood Acute Lymphoblastic Leukemia. Biochem. Genet. 2018, 56, 283–294. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Song, L.; Wang, H.; Yang, W.; Hu, L.; Yang, Y. Extracellular vesicles carrying miRNA-181b-5p affects the malignant progression of acute lymphoblastic leukemia. J. Transl. Med. 2021, 19, 511. [Google Scholar] [CrossRef] [PubMed]
- Luna-Aguirre, C.M.; de la Luz Martinez-Fierro, M.; Mar-Aguilar, F.; Garza-Veloz, I.; Trevino-Alvarado, V.; Rojas-Martinez, A.; Jaime-Perez, J.C.; Malagon-Santiago, G.I.; Gutierrez-Aguirre, C.H.; Gonzalez-Llano, O.; et al. Circulating microRNA expression profile in B-cell acute lymphoblastic leukemia. Cancer Biomark 2015, 15, 299–310. [Google Scholar] [CrossRef]
- Shafik, R.E.; Abd El Wahab, N.; Mokhtar, M.M.; El Taweel, M.A.; Ebeid, E. Expression of microRNA-181a and microRNA-196b in Egyptian Pediatric acute Lymphoblastic Leukemia. Asian Pac. J. Cancer. Prev. 2020, 21, 3429–3434. [Google Scholar] [CrossRef]
- Dawidowska, M.; Jaksik, R.; Drobna, M.; Szarzynska-Zawadzka, B.; Kosmalska, M.; Sedek, L.; Machowska, L.; Lalik, A.; Lejman, M.; Ussowicz, M.; et al. Comprehensive Investigation of miRNome Identifies Novel Candidate miRNA-mRNA Interactions Implicated in T-Cell Acute Lymphoblastic Leukemia. Neoplasia 2019, 21, 294–310. [Google Scholar] [CrossRef]
- Swellam, M.; El-Khazragy, N. Clinical impact of circulating microRNAs as blood-based marker in childhood acute lymphoblastic leukemia. Tumor Biol. 2016, 37, 10571–10576. [Google Scholar] [CrossRef]
- Shafik, R.E.; Abd El Wahab, N.; Senoun, S.A.; Ebeid, E.; El Taweel, M.A. Expression of Micro-RNA 128 and Let-7b in Pediatric Acute Lymphoblastic Leukemia Cases. Asian Pac. J. Cancer Prev. 2018, 19, 2263–2267. [Google Scholar]
- De Oliveira, J.C.; Scrideli, C.A.; Brassesco, M.S.; Morales, A.G.; Pezuk, J.A.; Queiroz Rde, P.; Yunes, J.A.; Brandalise, S.R.; Tone, L.G. Differential miRNA expression in childhood acute lymphoblastic leukemia and association with clinical and biological features. Leuk. Res. 2012, 36, 293–298. [Google Scholar] [CrossRef] [PubMed]
- Shahid, S.; Shahid, W.; Shaheen, J.; Akhtar, M.W.; Sadaf, S. Circulating miR-146a expression as a non-invasive predictive biomarker for acute lymphoblastic leukemia. Sci. Rep. 2021, 11, 22783. [Google Scholar] [CrossRef] [PubMed]
- Hassan, N.M.; Refaat, L.A.; Ismail, G.N.; Abdellateif, M.; Fadel, S.A.; AbdelAziz, R.S. Diagnostic, prognostic and predictive values of miR-100 and miR-210 in pediatric acute lymphoblastic Leukemia. Hematology 2020, 25, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Mosakhani, N.; Missiry, M.E.; Vakkila, E.; Knuutila, S.; Vakkila, J. Low Expression of miR-18a as a Characteristic of Pediatric Acute Lymphoblastic Leukemia. J. Pediatr. Hematol. Oncol. 2017, 39, 585–588. [Google Scholar] [CrossRef] [PubMed]
- Ghodousi, E.S.; Rahgozar, S. MicroRNA-326 and microRNA-200c: Two novel biomarkers for diagnosis and prognosis of pediatric acute lymphoblastic leukemia. J. Cell Biochem. 2018, 119, 6024–6032. [Google Scholar] [CrossRef] [PubMed]
- Almeida, R.S.; Costa, E.S.M.; Coutinho, L.L.; Garcia Gomes, R.; Pedrosa, F.; Massaro, J.D.; Donadi, E.A.; Lucena-Silva, N. MicroRNA expression profiles discriminate childhood T- from B-acute lymphoblastic leukemia. Hematol. Oncol. 2019, 37, 103–112. [Google Scholar] [CrossRef]
- Fulci, V.; Colombo, T.; Chiaretti, S.; Messina, M.; Citarella, F.; Tavolaro, S.; Guarini, A.; Foa, R.; Macino, G. Characterization of B- and T-lineage acute lymphoblastic leukemia by integrated analysis of MicroRNA and mRNA expression profiles. Genes Chromosomes Cancer 2009, 48, 1069–1082. [Google Scholar] [CrossRef]
- Gebarowska, K.; Mroczek, A.; Kowalczyk, J.R.; Lejman, M. MicroRNA as a Prognostic and Diagnostic Marker in T-Cell Acute Lymphoblastic Leukemia. Int. J. Mol. Sci. 2021, 22, 5317. [Google Scholar] [CrossRef]
- Nair, R.A.; Verma, V.K.; Beevi, S.S.; Rawoof, A.; Alexander, L.E.; Prasad, E.R.; Kumari, P.K.; Kumar, P.; Dinesh Kumar, L. MicroRNA Signatures in Blood or Bone Marrow Distinguish Subtypes of Pediatric Acute Lymphoblastic Leukemia. Transl. Oncol. 2020, 13, 100800. [Google Scholar] [CrossRef]
- Fan, S.J.; Li, H.B.; Cui, G.; Kong, X.L.; Sun, L.L.; Zhao, Y.Q.; Li, Y.H.; Zhou, J. miRNA-149* promotes cell proliferation and suppresses apoptosis by mediating JunB in T-cell acute lymphoblastic leukemia. Leuk. Res. 2016, 41, 62–70. [Google Scholar] [CrossRef]
- Krzanowski, J.; Madzio, J.; Pastorczak, A.; Tracz, A.; Braun, M.; Tabarkiewicz, J.; Pluta, A.; Mlynarski, W.; Zawlik, I. Selected miRNA levels are associated with IKZF1 microdeletions in pediatric acute lymphoblastic leukemia. Oncol. Lett. 2017, 14, 3853–3861. [Google Scholar] [CrossRef] [PubMed]
- Malouf, C.; Antunes, E.T.B.; O’Dwyer, M.; Jakobczyk, H.; Sahm, F.; Landua, S.L.; Anderson, R.A.; Soufi, A.; Halsey, C.; Ottersbach, K. miR-130b and miR-128a are essential lineage-specific codrivers of t(4;11) MLL-AF4 acute leukemia. Blood 2021, 138, 2066–2092. [Google Scholar] [CrossRef] [PubMed]
- Mousavian, Z.; Nowzari-Dalini, A.; Stam, R.W.; Rahmatallah, Y.; Masoudi-Nejad, A. Network-based expression analysis reveals key genes related to glucocorticoid resistance in infant acute lymphoblastic leukemia. Cell Oncol. 2017, 40, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Dou, L.; Li, J.; Zheng, D.; Li, Y.; Gao, X.; Xu, C.; Gao, L.; Wang, L.; Yu, L. MicroRNA-142-3p inhibits cell proliferation in human acute lymphoblastic leukemia by targeting the MLL-AF4 oncogene. Mol. Biol. Rep. 2013, 40, 6811–6819. [Google Scholar] [CrossRef] [PubMed]
- Vendramini, E.; Giordan, M.; Giarin, E.; Michielotto, B.; Fazio, G.; Cazzaniga, G.; Biondi, A.; Silvestri, D.; Valsecchi, M.G.; Muckenthaler, M.U.; et al. High expression of miR-125b-2 and SNORD116 noncoding RNA clusters characterize ERG-related B cell precursor acute lymphoblastic leukemia. Oncotarget 2017, 8, 42398–42413. [Google Scholar] [CrossRef] [PubMed]
- Hirano, D.; Hayakawa, F.; Yasuda, T.; Tange, N.; Yamamoto, H.; Kojima, Y.; Morishita, T.; Imoto, N.; Tsuzuki, S.; Mano, H.; et al. Chromosomal translocation-mediated evasion from miRNA induces strong MEF2D fusion protein expression, causing inhibition of PAX5 transcriptional activity. Oncogene 2019, 38, 2263–2274. [Google Scholar] [CrossRef] [PubMed]
- Han, B.W.; Feng, D.D.; Li, Z.G.; Luo, X.Q.; Zhang, H.; Li, X.J.; Zhang, X.J.; Zheng, L.L.; Zeng, C.W.; Lin, K.Y.; et al. A set of miRNAs that involve in the pathways of drug resistance and leukemic stem-cell differentiation is associated with the risk of relapse and glucocorticoid response in childhood ALL. Hum. Mol. Genet. 2011, 20, 4903–4915. [Google Scholar] [CrossRef]
- El-Khazragy, N.; Noshi, M.A.; Abdel-Malak, C.; Zahran, R.F.; Swellam, M. miRNA-155 and miRNA-181a as prognostic biomarkers for pediatric acute lymphoblastic leukemia. J. Cell. Biochem. 2019, 120, 6315–6321. [Google Scholar] [CrossRef]
- Labib, H.A.; Elantouny, N.G.; Ibrahim, N.F.; Alnagar, A.A. Upregulation of microRNA-21 is a poor prognostic marker in patients with childhood B cell acute lymphoblastic leukemia. Hematology 2017, 22, 392–397. [Google Scholar] [CrossRef]
- Piatopoulou, D.; Avgeris, M.; Marmarinos, A.; Xagorari, M.; Baka, M.; Doganis, D.; Kossiva, L.; Scorilas, A.; Gourgiotis, D. miR-125b predicts childhood acute lymphoblastic leukaemia poor response to BFM chemotherapy treatment. Br. J. Cancer 2017, 117, 801–812. [Google Scholar] [CrossRef]
- Piatopoulou, D.; Avgeris, M.; Drakaki, I.; Marmarinos, A.; Xagorari, M.; Baka, M.; Pourtsidis, A.; Kossiva, L.; Gourgiotis, D.; Scorilas, A.; et al. Clinical utility of miR-143/miR-182 levels in prognosis and risk stratification specificity of BFM-treated childhood acute lymphoblastic leukemia. Ann. Hematol. 2018, 97, 1169–1182. [Google Scholar] [CrossRef]
- Ovcharenko, D.; Kelnar, K.; Johnson, C.; Leng, N.; Brown, D. Genome-scale microRNA and small interfering RNA screens identify small RNA modulators of TRAIL-induced apoptosis pathway. Cancer Res. 2007, 67, 10782–10788. [Google Scholar] [CrossRef]
- Agirre, X.; Jimenez-Velasco, A.; San Jose-Eneriz, E.; Garate, L.; Bandres, E.; Cordeu, L.; Aparicio, O.; Saez, B.; Navarro, G.; Vilas-Zornoza, A.; et al. Down-regulation of hsa-miR-10a in chronic myeloid leukemia CD34+ cells increases USF2-mediated cell growth. Mol. Cancer. Res. 2008, 6, 1830–1840. [Google Scholar] [CrossRef]
- Hussenet, T.; Dali, S.; Exinger, J.; Monga, B.; Jost, B.; Dembele, D.; Martinet, N.; Thibault, C.; Huelsken, J.; Brambilla, E.; et al. SOX2 is an oncogene activated by recurrent 3q26.3 amplifications in human lung squamous cell carcinomas. PLoS ONE 2010, 5, e8960. [Google Scholar] [CrossRef] [PubMed]
- Stamatopoulos, B.; Meuleman, N.; Haibe-Kains, B.; Saussoy, P.; van den Neste, E.; Michaux, L.; Heimann, P.; Martiat, P.; Bron, D.; Lagneaux, L.; et al. microRNA-29c and microRNA-223 down-regulation has in vivo significance in chronic lymphocytic leukemia and improves disease risk stratification. Blood 2009, 113, 5237–5245. [Google Scholar] [CrossRef]
- Amankwah, E.K.; Devidas, M.; Teachey, D.T.; Rabin, K.R.; Brown, P.A. Six Candidate miRNAs Associated with Early Relapse in Pediatric B-Cell Acute Lymphoblastic Leukemia. Anticancer Res. 2020, 40, 3147–3153. [Google Scholar] [CrossRef] [PubMed]
- Avigad, S.; Verly, I.R.; Lebel, A.; Kordi, O.; Shichrur, K.; Ohali, A.; Hameiri-Grossman, M.; Kaspers, G.J.; Cloos, J.; Fronkova, E.; et al. miR expression profiling at diagnosis predicts relapse in pediatric precursor B-cell acute lymphoblastic leukemia. Genes Chromosomes Cancer 2016, 55, 328–339. [Google Scholar] [CrossRef] [PubMed]
- Organista-Nava, J.; Gomez-Gomez, Y.; Illades-Aguiar, B.; del Carmen Alarcon-Romero, L.; Saavedra-Herrera, M.V.; Rivera-Ramirez, A.B.; Garzon-Barrientos, V.H.; Leyva-Vazquez, M.A. High miR-24 expression is associated with risk of relapse and poor survival in acute leukemia. Oncol. Rep. 2015, 33, 1639–1649. [Google Scholar] [CrossRef] [PubMed]
- Rzepiel, A.; Kutszegi, N.; Gezsi, A.; Sagi, J.C.; Egyed, B.; Peter, G.; Butz, H.; Nyiro, G.; Muller, J.; Kovacs, G.T.; et al. Circulating microRNAs as minimal residual disease biomarkers in childhood acute lymphoblastic leukemia. J. Transl. Med. 2019, 17, 372. [Google Scholar] [CrossRef]
- Inaba, H.; Greaves, M.; Mullighan, C.G. Acute lymphoblastic leukaemia. Lancet 2013, 381, 1943–1955. [Google Scholar] [CrossRef]
- Zhang, H.; Luo, X.Q.; Zhang, P.; Huang, L.B.; Zheng, Y.S.; Wu, J.; Zhou, H.; Qu, L.H.; Xu, L.; Chen, Y.Q.; et al. MicroRNA patterns associated with clinical prognostic parameters and CNS relapse prediction in pediatric acute leukemia. PLoS ONE 2009, 4, e7826. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.N.; Tang, Y.L.; Ke, Z.Y.; Chen, Y.Q.; Luo, X.Q.; Zhang, H.; Huang, L.B. MiR-124 contributes to glucocorticoid resistance in acute lymphoblastic leukemia by promoting proliferation, inhibiting apoptosis and targeting the glucocorticoid receptor. J. Steroid Biochem. Mol. Biol. 2017, 172, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Lucafo, M.; Sicari, D.; Chicco, A.; Curci, D.; Bellazzo, A.; Di Silvestre, A.; Pegolo, C.; Autry, R.; Cecchin, E.; De Iudicibus, S.; et al. miR-331-3p is involved in glucocorticoid resistance reversion by rapamycin through suppression of the MAPK signaling pathway. Cancer Chemother. Pharmacol. 2020, 86, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Shen, T.; Wang, H.; Ke, Z.; Liang, Y.; Ouyang, J.; Jiang, T. MicroRNA-185-5p restores glucocorticoid sensitivity by suppressing the mammalian target of rapamycin complex (mTORC) signaling pathway to enhance glucocorticoid receptor autoregulation. Leuk. Lymphoma 2017, 58, 2657–2667. [Google Scholar] [CrossRef]
- Akbari Moqadam, F.; Lange-Turenhout, E.A.; Aries, I.M.; Pieters, R.; den Boer, M.L. MiR-125b, miR-100 and miR-99a co-regulate vincristine resistance in childhood acute lymphoblastic leukemia. Leuk. Res. 2013, 37, 1315–1321. [Google Scholar] [CrossRef]
- El-Khazragy, N.; Elshimy, A.A.; Hassan, S.S.; Matbouly, S.; Safwat, G.; Zannoun, M.; Riad, R.A. Dysregulation of miR-125b predicts poor response to therapy in pediatric acute lymphoblastic leukemia. J. Cell. Biochem. 2018, 120, 7428–7438. [Google Scholar] [CrossRef]
- Mei, Y.; Gao, C.; Wang, K.; Cui, L.; Li, W.; Zhao, X.; Liu, F.; Wu, M.; Deng, G.; Ding, W.; et al. Effect of microRNA-210 on prognosis and response to chemotherapeutic drugs in pediatric acute lymphoblastic leukemia. Cancer Sci. 2014, 105, 463–472. [Google Scholar] [CrossRef]
- Jiang, Q.; Lu, X.; Huang, P.; Gao, C.; Zhao, X.; Xing, T.; Li, G.; Bao, S.; Zheng, H. Expression of miR-652-3p and Effect on Apoptosis and Drug Sensitivity in Pediatric Acute Lymphoblastic Leukemia. Biomed. Res. Int. 2018, 2018, 5724686. [Google Scholar] [CrossRef]
- Chen, H.; Zhang, D.; Zhang, G.; Li, X.; Liang, Y.; Kasukurthi, M.V.; Li, S.; Borchert, G.M.; Huang, J. A semantics-oriented computational approach to investigate microRNA regulation on glucocorticoid resistance in pediatric acute lymphoblastic leukemia. BMC Med. Inform. Decis. Mak. 2018, 18, 57. [Google Scholar] [CrossRef]
- Moses, B.S.; Evans, R.; Slone, W.L.; Piktel, D.; Martinez, I.; Craig, M.D.; Gibson, L.F. Bone Marrow Microenvironment Niche Regulates miR-221/222 in Acute Lymphoblastic Leukemia. Mol. Cancer Res. 2016, 14, 909–919. [Google Scholar] [CrossRef]
- Cui, C.; Cui, Q. The relationship of human tissue microRNAs with those from body fluids. Sci. Rep. 2020, 10, 5644. [Google Scholar] [CrossRef]
- Su, F.; Gao, Z.; Liu, Y.; Zhou, G.; Cui, Y.; Deng, C.; Liu, Y.; Zhang, Y.; Ma, X.; Wang, Y.; et al. Integrated Tissue and Blood miRNA Expression Profiles Identify Novel Biomarkers for Accurate Non-Invasive Diagnosis of Breast Cancer: Preliminary Results and Future Clinical Implications. Genes 2022, 13, 1931. [Google Scholar] [CrossRef] [PubMed]
- Sharifi, Z.; Talkhabi, M.; Taleahmad, S. Identification of potential microRNA diagnostic panels and uncovering regulatory mechanisms in breast cancer pathogenesis. Sci. Rep. 2022, 12, 20135. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Li, J.; Cairns, M.J. Identifying miRNAs, targets and functions. Brief. Bioinform. 2014, 15, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, P.; Singh, M.; Singh, A.; Sharma, P.; Trehan, A.; Varma, N. Epigenetic analysis reveals significant differential expression of miR-378C and miR-128-2-5p in a cohort of relapsed pediatric B-acute lymphoblastic leukemia cases. Int. J. Lab. Hematol. 2021, 43, 1016–1023. [Google Scholar] [CrossRef] [PubMed]
- Drobna, M.; Szarzynska, B.; Jaksik, R.; Sedek, L.; Kuchmiy, A.; Taghon, T.; van Vlierberghe, P.; Szczepanski, T.; Witt, M.; Dawidowska, M.; et al. hsa-miR-20b-5p and hsa-miR-363-3p Affect Expression of PTEN and BIM Tumor Suppressor Genes and Modulate Survival of T-ALL Cells In Vitro. Cells 2020, 9, 1137. [Google Scholar] [CrossRef]
- Qian, L.; Zhang, W.; Lei, B.; He, A.; Ye, L.; Li, X.; Dong, X. MicroRNA-101 regulates T-cell acute lymphoblastic leukemia progression and chemotherapeutic sensitivity by targeting Notch1. Oncol. Rep. 2016, 36, 2511–2516. [Google Scholar] [CrossRef]
- Correia, N.C.; Melao, A.; Povoa, V.; Sarmento, L.; Gomez de Cedron, M.; Malumbres, M.; Enguita, F.J.; Barata, J.T. microRNAs regulate TAL1 expression in T-cell acute lymphoblastic leukemia. Oncotarget 2016, 7, 8268–8281. [Google Scholar] [CrossRef]
- Popovic, R.; Riesbeck, L.E.; Velu, C.S.; Chaubey, A.; Zhang, J.; Achille, N.J.; Erfurth, F.E.; Eaton, K.; Lu, J.; Grimes, H.L.; et al. Regulation of mir-196b by MLL and its overexpression by MLL fusions contributes to immortalization. Blood 2009, 113, 3314–3322. [Google Scholar] [CrossRef]
- Schotte, D.; Lange-Turenhout, E.A.; Stumpel, D.J.; Stam, R.W.; Buijs-Gladdines, J.G.; Meijerink, J.P.; Pieters, R.; Den Boer, M.L. Expression of miR-196b is not exclusively MLL-driven but is especially linked to activation of HOXA genes in pediatric acute lymphoblastic leukemia. Haematologica 2010, 95, 1675–1682. [Google Scholar] [CrossRef]
- Huang, Y.; Li, J.; Chen, Y.; Jiang, P.; Wang, L.; Hu, J. Identification of Early Recurrence Factors in Childhood and Adolescent B-Cell Acute Lymphoblastic Leukemia Based on Integrated Bioinformatics Analysis. Front. Oncol. 2020, 10, 565455. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Gregory, R.I. MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer 2015, 15, 321–333. [Google Scholar] [CrossRef] [PubMed]
- Cimmino, A.; Calin, G.A.; Fabbri, M.; Iorio, M.V.; Ferracin, M.; Shimizu, M.; Wojcik, S.E.; Aqeilan, R.I.; Zupo, S.; Dono, M.; et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. USA 2005, 102, 13944–13949. [Google Scholar] [CrossRef]
- Jackstadt, R.; Hermeking, H. MicroRNAs as regulators and mediators of c-MYC function. Biochim. Biophys. Acta 2015, 1849, 544–553. [Google Scholar] [CrossRef] [PubMed]
- Simonson, B.; Das, S. MicroRNA Therapeutics: The Next Magic Bullet? Mini Rev. Med. Chem. 2015, 15, 467–474. [Google Scholar] [CrossRef]
- Wang, Y.; Liang, H. When MicroRNAs Meet RNA Editing in Cancer: A Nucleotide Change Can Make a Difference. Bioessays 2018, 40, 1700188. [Google Scholar] [CrossRef]
- Moraes, F.C.; Pichon, C.; Letourneur, D.; Chaubet, F. miRNA Delivery by Nanosystems: State of the Art and Perspectives. Pharmaceutics 2021, 13, 1901. [Google Scholar] [CrossRef]
- Correia de Sousa, M.; Gjorgjieva, M.; Dolicka, D.; Sobolewski, C.; Foti, M. Deciphering miRNAs’ Action through miRNA Editing. Int. J. Mol. Sci. 2019, 20, 6249. [Google Scholar] [CrossRef]

| Upregulated | Downregulated |
|---|---|
| miR-7e [111], miR-9 [111], miR-9* [111], miR-34a [116], miR-92a [113], miR-100 [119], miR-125b-1 [114], miR-128 [111,120,121], miR-130b [111], miR-142-3p [112], miR-146a [115,119,122], miR-155 [115], miR-181 [121], miR-181a [111,117], miR-181b [111,115], miR-210 [123], miR-222 [112,116], miR-339 [112], miR-363 [111], miR-511 [116], miR-638 [113], miR-1943 [111], miR-1841 [111], miR-1931 [111], miR-1987 [111], miR-1890 [111], miR-1902 [111] | let-7e [121], miR-18a [124], miR-26a [116], miR-30a [111], miR-100 [121,123], miR-126 [111], miR-143 [111], miR-145 [115], miR-196a [119], miR-196b [117,121], miR-199b-3p [111], miR-200c [125], miR-203 [114], miR-221 [116], miR-223 [111,116], miR-326 [125], miR-373* [112], miR-451 [112], miR-582-5p [111], miR-1893 [111], miR-1971* [111], miR-1834 [111], miR-1842* [111], miR-1842 [111] |
| Upregulated | Downregulated |
|---|---|
| miR-29c-5p [126], miR-137 [129], miR-148a [127], miR-149* [130], miR-193b* [129], miR-424-5p [126], miR-424 [127], miR-450b-5p [126], miR-450a-5p [126], miR-506 [129], miR-509-5p [129], miR-510 [129], miR-542-5p [126], miR-629-5p [126] | miR-100 [128], miR-151 [127], miR-151a-5p [126], miR-151b [126], miR-195-5p [126], miR-371b-5p [126], miR-383 [129], miR-425-5p [126], miR-455-5p [126], miR-497-5p [126], miR-574-5p [126], miR-708 [128,129], miR-708-5p [126], miR-1266-5p [126] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mendiola-Soto, D.K.; Bárcenas-López, D.A.; Pérez-Amado, C.J.; Cruz-Miranda, G.M.; Mejía-Aranguré, J.M.; Ramírez-Bello, J.; Hidalgo-Miranda, A.; Jiménez-Morales, S. MiRNAs in Hematopoiesis and Acute Lymphoblastic Leukemia. Int. J. Mol. Sci. 2023, 24, 5436. https://doi.org/10.3390/ijms24065436
Mendiola-Soto DK, Bárcenas-López DA, Pérez-Amado CJ, Cruz-Miranda GM, Mejía-Aranguré JM, Ramírez-Bello J, Hidalgo-Miranda A, Jiménez-Morales S. MiRNAs in Hematopoiesis and Acute Lymphoblastic Leukemia. International Journal of Molecular Sciences. 2023; 24(6):5436. https://doi.org/10.3390/ijms24065436
Chicago/Turabian StyleMendiola-Soto, Diana Karen, Diego Alberto Bárcenas-López, Carlos Jhovani Pérez-Amado, Gabriela Marisol Cruz-Miranda, Juan Manuel Mejía-Aranguré, Julian Ramírez-Bello, Alfredo Hidalgo-Miranda, and Silvia Jiménez-Morales. 2023. "MiRNAs in Hematopoiesis and Acute Lymphoblastic Leukemia" International Journal of Molecular Sciences 24, no. 6: 5436. https://doi.org/10.3390/ijms24065436
APA StyleMendiola-Soto, D. K., Bárcenas-López, D. A., Pérez-Amado, C. J., Cruz-Miranda, G. M., Mejía-Aranguré, J. M., Ramírez-Bello, J., Hidalgo-Miranda, A., & Jiménez-Morales, S. (2023). MiRNAs in Hematopoiesis and Acute Lymphoblastic Leukemia. International Journal of Molecular Sciences, 24(6), 5436. https://doi.org/10.3390/ijms24065436