Dimeric Ankyrin with Inverted Module Promotes Bifunctional Property in Capturing Capsid to Impede HIV-1 Replication
Abstract
1. Introduction
2. Results
2.1. Inversion of AnkGAG1D4 Structure Does Not Alter Ankyrin Binding Surface
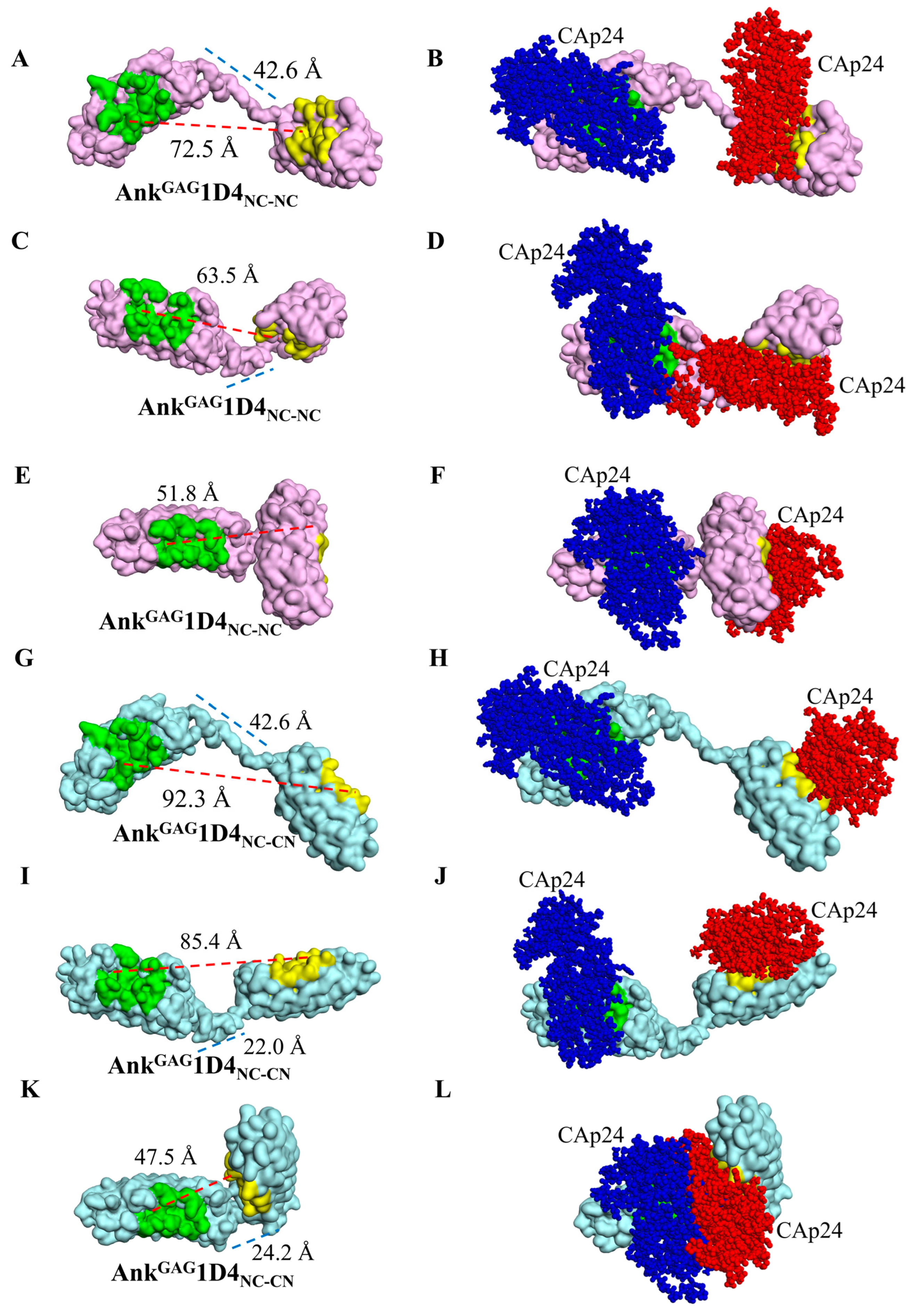
2.2. AnkGAG1D4 Dimers Exhibit Diverse Molecular Structures
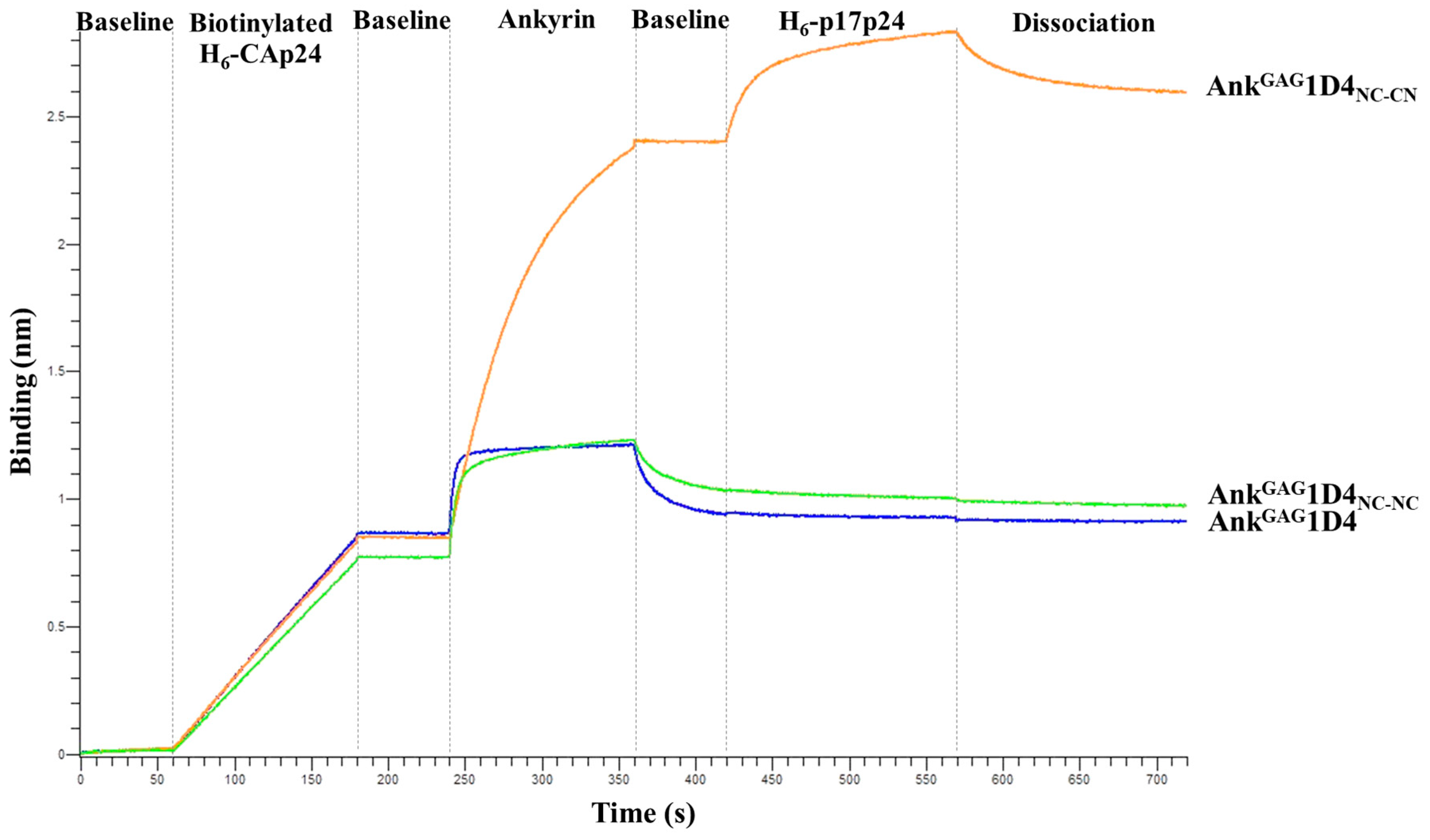
2.3. Structural Analysis Showed Functional Binding Property of AnkGAG1D4 Dimers
2.4. Bifunctional Module Relies on Dimeric AnkGAG1D4NC-CN
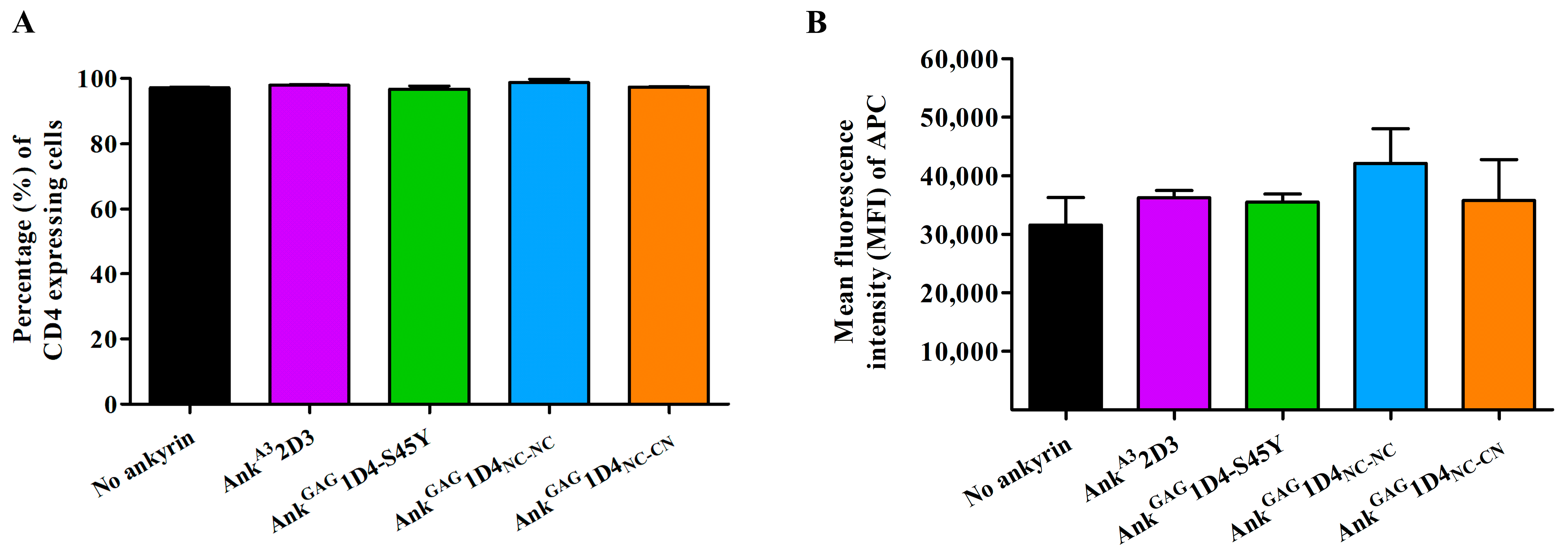
2.5. Ankyrin Expression Is Shunted to the Plasma Membrane and Does Not Interfere CD4 Expression in SupT1 Cells
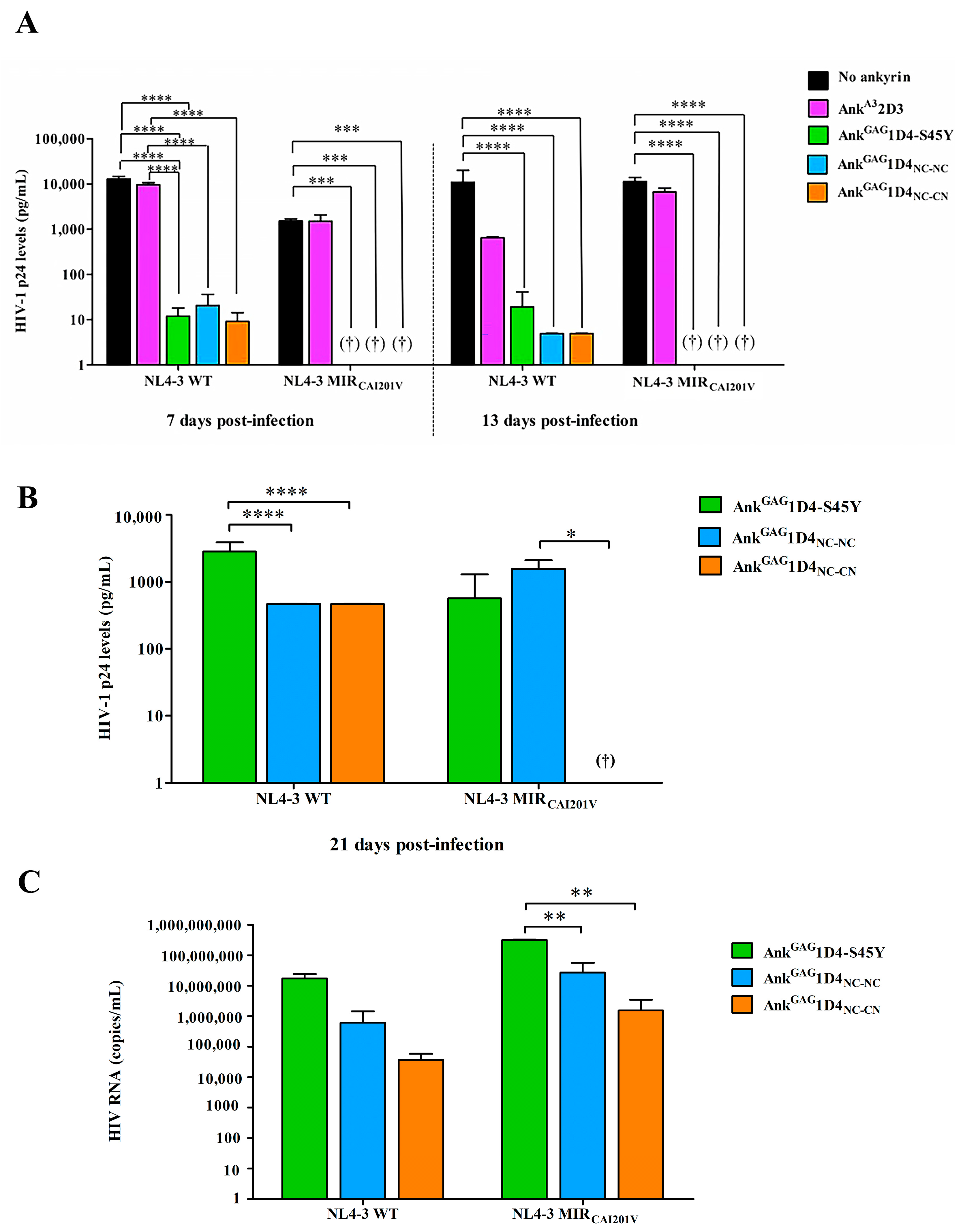
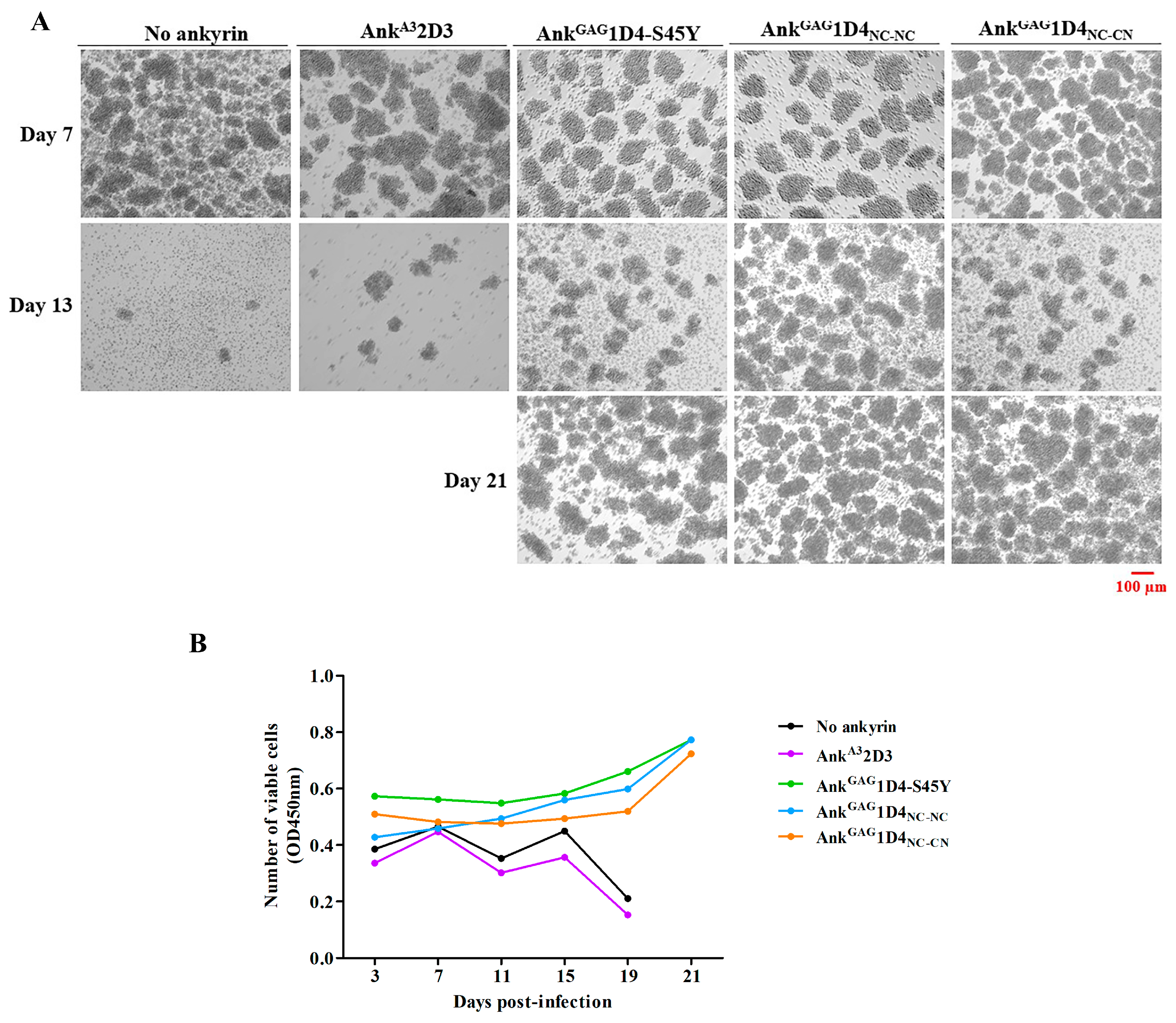
2.6. Dimeric Ankyrins Improves Antiviral Activity Than Monomeric Ankyrins against HIV-1 Replication
2.7. Dimeric Ankyrin Provides Superior Anti-Viral Activity against HIV-1 MIR Virus
3. Discussion
4. Materials and Methods
4.1. Construction of Inverted AnkGAG1D4
4.2. Structural Modeling of AnkGAG1D4 Dimer
4.3. Determination of the Distance of Binding Sites
4.4. Analysis of the Binding Ratio of Ankyrin Dimer to CAp24
4.5. Binding Activity of Dimeric AnkGAG1D4
4.6. Cell Lines and Plasmids
4.7. Lentivirus Production
4.8. Ankyrin-Expressing SupT1 Cells Establishment
4.9. Ankyrin Subcellular Localization
4.10. HIV-1 NL4-3 WT and HIV-1 NL4-3 MIRCAI201V Production
4.11. HIV-1 Infection
4.12. Viral Release Monitoring
4.13. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schweizer, A.; Rusert, P.; Berlinger, L.; Ruprecht, C.R.; Mann, A.; Corthésy, S.; Turville, S.G.; Aravantinou, M.; Fischer, M.; Robbiani, M.; et al. CD4-specific designed ankyrin repeat proteins are novel potent HIV entry inhibitors with unique characteristics. PLoS Pathog. 2008, 4, e1000109. [Google Scholar] [CrossRef] [PubMed]
- Mann, A.; Friedrich, N.; Krarup, A.; Weber, J.; Stiegeler, E.; Dreier, B.; Pugach, P.; Robbiani, M.; Riedel, T.; Moehle, K.; et al. Conformation-dependent recognition of HIV gp120 by designed ankyrin repeat proteins provides access to novel HIV entry inhibitors. J. Virol. 2013, 87, 5868–5881. [Google Scholar] [CrossRef] [PubMed]
- Sakkhachornphop, S.; Hadpech, S.; Wisitponchai, T.; Panto, C.; Kantamala, D.; Utaipat, U.; Praparattanapan, J.; Kotarathitithum, W.; Taejaroenkul, S.; Yasamut, U.; et al. Broad-Spectrum Antiviral Activity of an Ankyrin Repeat Protein on Viral Assembly against Chimeric NL4-3 Viruses Carrying Gag/PR Derived from Circulating Strains among Northern Thai Patients. Viruses 2018, 10, 625. [Google Scholar] [CrossRef] [PubMed]
- Nangola, S.; Urvoas, A.; Valerio-Lepiniec, M.; Khamaikawin, W.; Sakkhachornphop, S.; Hong, S.-S.; Boulanger, P.; Minard, P.; Tayapiwatana, C.J.R. Antiviral activity of recombinant ankyrin targeted to the capsid domain of HIV-1 Gag polyprotein. Retrovirology 2012, 9, 17. [Google Scholar] [CrossRef]
- Moonmuang, S.; Maniratanachote, R.; Chetprayoon, P.; Sornsuwan, K.; Thongkum, W.; Chupradit, K.; Tayapiwatana, C. Specific Interaction of DARPin with HIV-1 CA(NTD) Disturbs the Distribution of Gag, RNA Packaging, and Tetraspanin Remodelling in the Membrane. Viruses 2022, 14, 824. [Google Scholar] [CrossRef]
- Saoin, S.; Wisitponchai, T.; Intachai, K.; Chupradit, K.; Moonmuang, S.; Nangola, S.; Kitidee, K.; Fanhchaksai, K.; Lee, V.S.; Hong, S.S.; et al. Deciphering critical amino acid residues to modify and enhance the binding affinity of ankyrin scaffold specific to capsid protein of human immunodeficiency virus type 1. Asian Pac. J. Allergy Immunol. 2018, 36, 126–135. [Google Scholar] [CrossRef]
- Sornsuwan, K.; Thongkhum, W.; Pamonsupornwichit, T.; Carraway, T.S.; Soponpong, S.; Sakkhachornphop, S.; Tayapiwatana, C.; Yasamut, U. Performance of Affinity-Improved DARPin Targeting HIV Capsid Domain in Interference of Viral Progeny Production. Biomolecules 2021, 11, 1437. [Google Scholar] [CrossRef]
- Boersma, Y.L.; Chao, G.; Steiner, D.; Wittrup, K.D.; Plückthun, A. Bispecific designed ankyrin repeat proteins (DARPins) targeting epidermal growth factor receptor inhibit A431 cell proliferation and receptor recycling. J. Biol. Chem. 2011, 286, 41273–41285. [Google Scholar] [CrossRef]
- Chonira, V.; Kwon, Y.D.; Gorman, J.; Case, J.B.; Ku, Z.; Simeon, R.; Casner, R.G.; Harris, D.R.; Olia, A.S.; Stephens, T.; et al. A potent and broad neutralization of SARS-CoV-2 variants of concern by DARPins. Nat. Chem. Biol. 2022, 19, 284–291. [Google Scholar] [CrossRef]
- Rothenberger, S.; Hurdiss, D.L.; Walser, M.; Malvezzi, F.; Mayor, J.; Ryter, S.; Moreno, H.; Liechti, N.; Bosshart, A.; Iss, C.; et al. The trispecific DARPin ensovibep inhibits diverse SARS-CoV-2 variants. Nat. Biotechnol. 2022, 40, 1845–1854. [Google Scholar] [CrossRef]
- Juntit, O.A.; Yasamut, U.; Sakkhachornphop, S.; Chupradit, K.; Thongkum, W.; Srisawat, C.; Chokepaichitkool, T.; Kongtawelert, P.; Tayapiwatana, C. Biological properties of reverse ankyrin engineered for dimer construction to enhance HIV-1 capsid interaction. Asian Pac. J. Allergy Immunol. 2022. [Google Scholar] [CrossRef]
- Thenin-Houssier, S.; Valente, S.T. HIV-1 Capsid Inhibitors as Antiretroviral Agents. Curr. HIV Res. 2016, 14, 270–282. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Verheyen, J.; Rhee, S.-Y.; Voet, A.; Vandamme, A.-M.; Theys, K. Functional conservation of HIV-1 Gag: Implications for rational drug design. Retrovirology 2013, 10, 126. [Google Scholar] [CrossRef]
- Martin, D.E.; Salzwedel, K.; Allaway, G.P. Bevirimat: A Novel Maturation Inhibitor for the Treatment of HIV-1 Infection. Antivir. Chem. Chemother. 2008, 19, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Urano, E.; Ablan, S.D.; Mandt, R.; Pauly, G.T.; Sigano, D.M.; Schneider, J.P.; Martin, D.E.; Nitz, T.J.; Wild, C.T.; Freed, E.O. Alkyl Amine Bevirimat Derivatives Are Potent and Broadly Active HIV-1 Maturation Inhibitors. Antimicrob. Agents Chemother. 2016, 60, 190–197. [Google Scholar] [CrossRef]
- Waki, K.; Durell, S.R.; Soheilian, F.; Nagashima, K.; Butler, S.L.; Freed, E.O. Structural and functional insights into the HIV-1 maturation inhibitor binding pocket. PLoS Pathog. 2012, 8, e1002997. [Google Scholar] [CrossRef]
- Cevik, M.; Orkin, C. Insights into HIV-1 capsid inhibitors in preclinical and early clinical development as antiretroviral agents. Expert Opin. Investig. Drugs 2019, 28, 1021–1024. [Google Scholar] [CrossRef]
- Urano, E.; Timilsina, U.; Kaplan, J.A.; Ablan, S.; Ghimire, D.; Pham, P.; Kuruppu, N.; Mandt, R.; Durell, S.R.; Nitz, T.J.; et al. Resistance to Second-Generation HIV-1 Maturation Inhibitors. J. Virol. 2019, 93, e02017-18. [Google Scholar] [CrossRef]
- Welker, R.; Hohenberg, H.; Tessmer, U.; Huckhagel, C.; Kräusslich, H.G. Biochemical and structural analysis of isolated mature cores of human immunodeficiency virus type 1. J. Virol. 2000, 74, 1168–1177. [Google Scholar] [CrossRef]
- Vazquez-Lombardi, R.; Phan, T.G.; Zimmermann, C.; Lowe, D.; Jermutus, L.; Christ, D. Challenges and opportunities for non-antibody scaffold drugs. Drug Discov. Today 2015, 20, 1271–1283. [Google Scholar] [CrossRef]
- Barkauskas, C.; Mylonakis, E.; Poulakou, G.; Young, B.E.; Vock, D.M.; Siegel, L.; Engen, N.; Grandits, G.; Mosaly, N.R.; Vekstein, A.M.; et al. Efficacy and Safety of Ensovibep for Adults Hospitalized With COVID-19: A Randomized Controlled Trial. Ann. Intern. Med. 2022, 175, 1266–1274. [Google Scholar] [CrossRef] [PubMed]
- Léger, C.; Di Meo, T.; Aumont-Nicaise, M.; Velours, C.; Durand, D.; Li de la Sierra-Gallay, I.; van Tilbeurgh, H.; Hildebrandt, N.; Desmadril, M.; Urvoas, A.; et al. Ligand-induced conformational switch in an artificial bidomain protein scaffold. Sci. Rep. 2019, 9, 1178. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Zeng, W.; Ye, H.; Han, K.H.; Dharmarajan, V.; Novick, S.; Wilson, I.A.; Griffin, P.R.; Friedman, J.M.; Lerner, R.A. A General Method for Insertion of Functional Proteins within Proteins via Combinatorial Selection of Permissive Junctions. Chem. Biol. 2015, 22, 1134–1143. [Google Scholar] [CrossRef]
- Kim, T.Y.; Seo, H.D.; Lee, J.J.; Kang, J.A.; Kim, W.S.; Kim, H.M.; Song, H.Y.; Park, J.M.; Lee, D.E.; Kim, H.S. A dimeric form of a small-sized protein binder exhibits enhanced anti-tumor activity through prolonged blood circulation. J. Control. Release 2018, 279, 282–291. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.K.; Gregson, J.; Parkin, N.; Haile-Selassie, H.; Tanuri, A.; Andrade Forero, L.; Kaleebu, P.; Watera, C.; Aghokeng, A.; Mutenda, N.; et al. HIV-1 drug resistance before initiation or re-initiation of first-line antiretroviral therapy in low-income and middle-income countries: A systematic review and meta-regression analysis. Lancet Infect. Dis. 2018, 18, 346–355. [Google Scholar] [CrossRef]
- Praditwongwan, W.; Chuankhayan, P.; Saoin, S.; Wisitponchai, T.; Lee, V.S.; Nangola, S.; Hong, S.S.; Minard, P.; Boulanger, P.; Chen, C.J.; et al. Crystal structure of an antiviral ankyrin targeting the HIV-1 capsid and molecular modeling of the ankyrin-capsid complex. J. Comput.-Aided Mol. Des. 2014, 28, 869–884. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, I.C.; Hall, C.J.; Veverka, V.; Shi, J.Y.; Muskett, F.W.; Stephens, P.E.; Taylor, R.J.; Henry, A.J.; Carr, M.D. High Resolution NMR-based Model for the Structure of a scFv-IL-1β Complex. J. Biol. Chem. 2009, 284, 31928–31935. [Google Scholar] [CrossRef]
- MacKerell, A.D.; Bashford, D.; Bellott, M.; Dunbrack, R.L.; Evanseck, J.D.; Field, M.J.; Fischer, S.; Gao, J.; Guo, H.; Ha, S.; et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 1998, 102, 3586–3616. [Google Scholar] [CrossRef]
- Phillips, J.C.; Hardy, D.J.; Maia, J.D.C.; Stone, J.E.; Ribeiro, J.V.; Bernardi, R.C.; Buch, R.; Fiorin, G.; Hénin, J.; Jiang, W.; et al. Scalable molecular dynamics on CPU and GPU architectures with NAMD. J. Chem. Phys. 2020, 153, 044130. [Google Scholar] [CrossRef]





| Binding Ratio of Dimer to CAp24 (n = 403) | ||||||
|---|---|---|---|---|---|---|
| AnkGAG1D4NC-NC | AnkGAG1D4NC-CN | |||||
| 1:0 1 | 1:1 2 | 1:2 3 | 1:0 1 | 1:1 2 | 1:2 3 | |
| number | 6.0 | 181.0 | 216.0 | 0.0 | 46.0 | 357.0 |
| % | 1.5 | 44.9 | 53.6 | 0.0 | 11.4 | 88.6 |
| Variants | KD (M) | Kon (M·s−1) | Koff (s−1) |
|---|---|---|---|
| Monomeric AnkGAG1D4 | 3.3 × 10−8 | 6.8 × 105 | 2.2 × 10−2 |
| Dimeric AnkGAG1D4NC-NC | 1.9 × 10−8 | 1.3 × 105 | 9.9 × 10−3 |
| Dimeric AnkGAG1D4NC-CN | <1.0 × 10−12 | 1.1 × 105 | <1.0 × 10−7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Juntit, O.-a.; Sornsuwan, K.; Wisitponchai, T.; Sanghiran Lee, V.; Sakkhachornphop, S.; Yasamut, U.; Tayapiwatana, C. Dimeric Ankyrin with Inverted Module Promotes Bifunctional Property in Capturing Capsid to Impede HIV-1 Replication. Int. J. Mol. Sci. 2023, 24, 5266. https://doi.org/10.3390/ijms24065266
Juntit O-a, Sornsuwan K, Wisitponchai T, Sanghiran Lee V, Sakkhachornphop S, Yasamut U, Tayapiwatana C. Dimeric Ankyrin with Inverted Module Promotes Bifunctional Property in Capturing Capsid to Impede HIV-1 Replication. International Journal of Molecular Sciences. 2023; 24(6):5266. https://doi.org/10.3390/ijms24065266
Chicago/Turabian StyleJuntit, On-anong, Kanokporn Sornsuwan, Tanchanok Wisitponchai, Vannajan Sanghiran Lee, Supachai Sakkhachornphop, Umpa Yasamut, and Chatchai Tayapiwatana. 2023. "Dimeric Ankyrin with Inverted Module Promotes Bifunctional Property in Capturing Capsid to Impede HIV-1 Replication" International Journal of Molecular Sciences 24, no. 6: 5266. https://doi.org/10.3390/ijms24065266
APA StyleJuntit, O.-a., Sornsuwan, K., Wisitponchai, T., Sanghiran Lee, V., Sakkhachornphop, S., Yasamut, U., & Tayapiwatana, C. (2023). Dimeric Ankyrin with Inverted Module Promotes Bifunctional Property in Capturing Capsid to Impede HIV-1 Replication. International Journal of Molecular Sciences, 24(6), 5266. https://doi.org/10.3390/ijms24065266






