A Comprehensive Analysis Revealing FBXW9 as a Potential Prognostic and Immunological Biomarker in Breast Cancer
Abstract
1. Introduction
2. Results
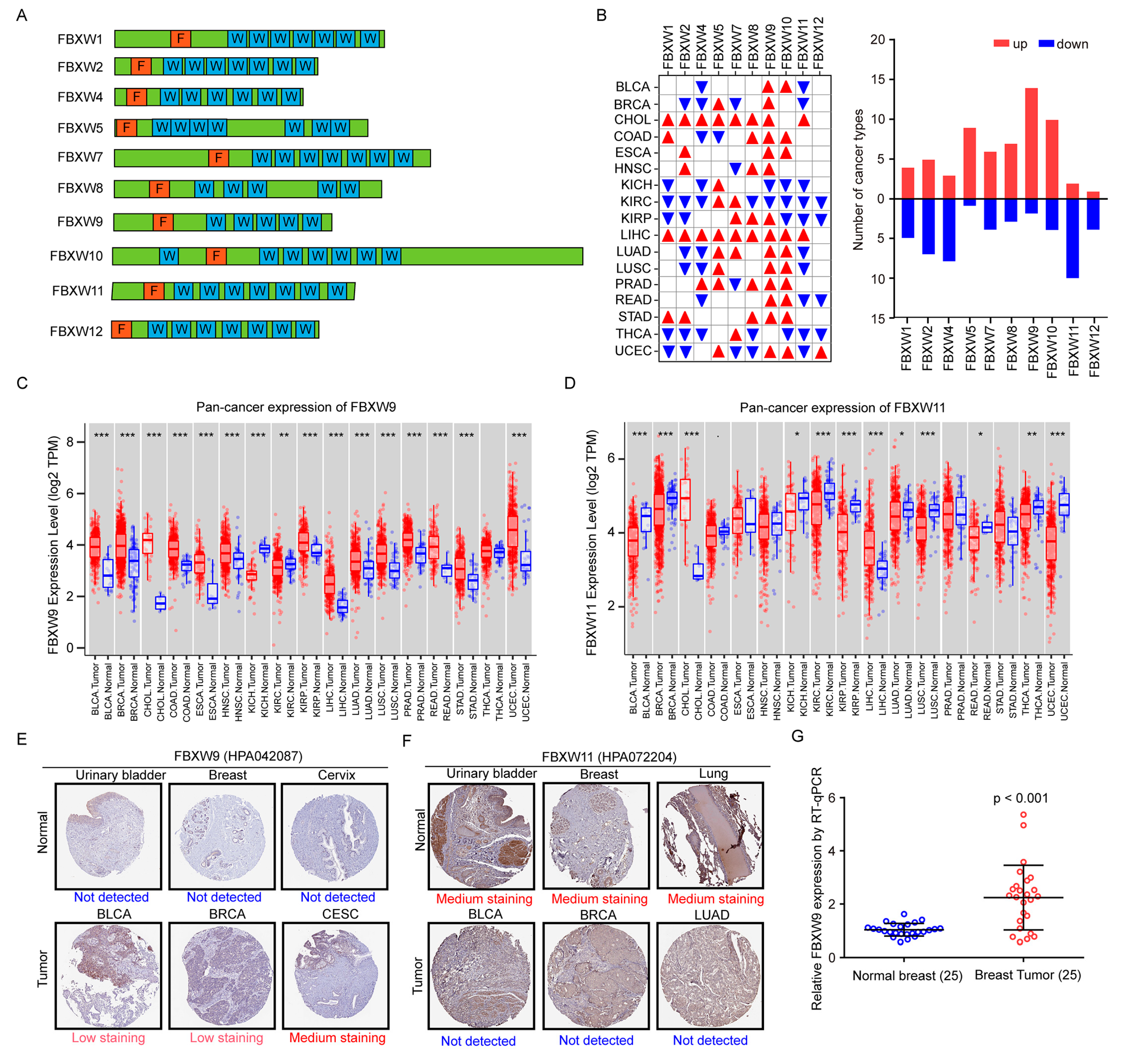
2.1. Analysis of mRNA Levels of FBXWs in Tumors of Various Cancer Types and Normal Tissues
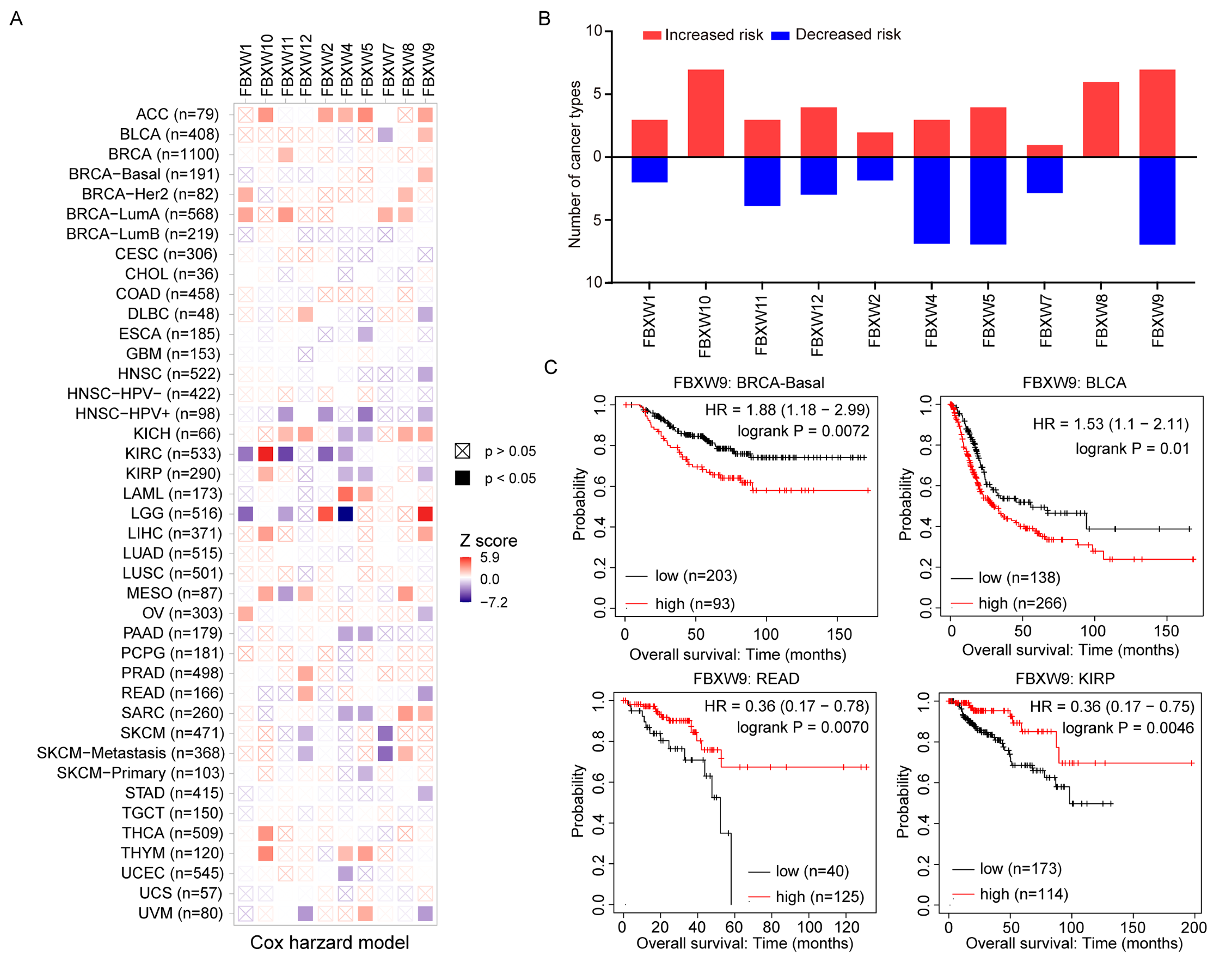
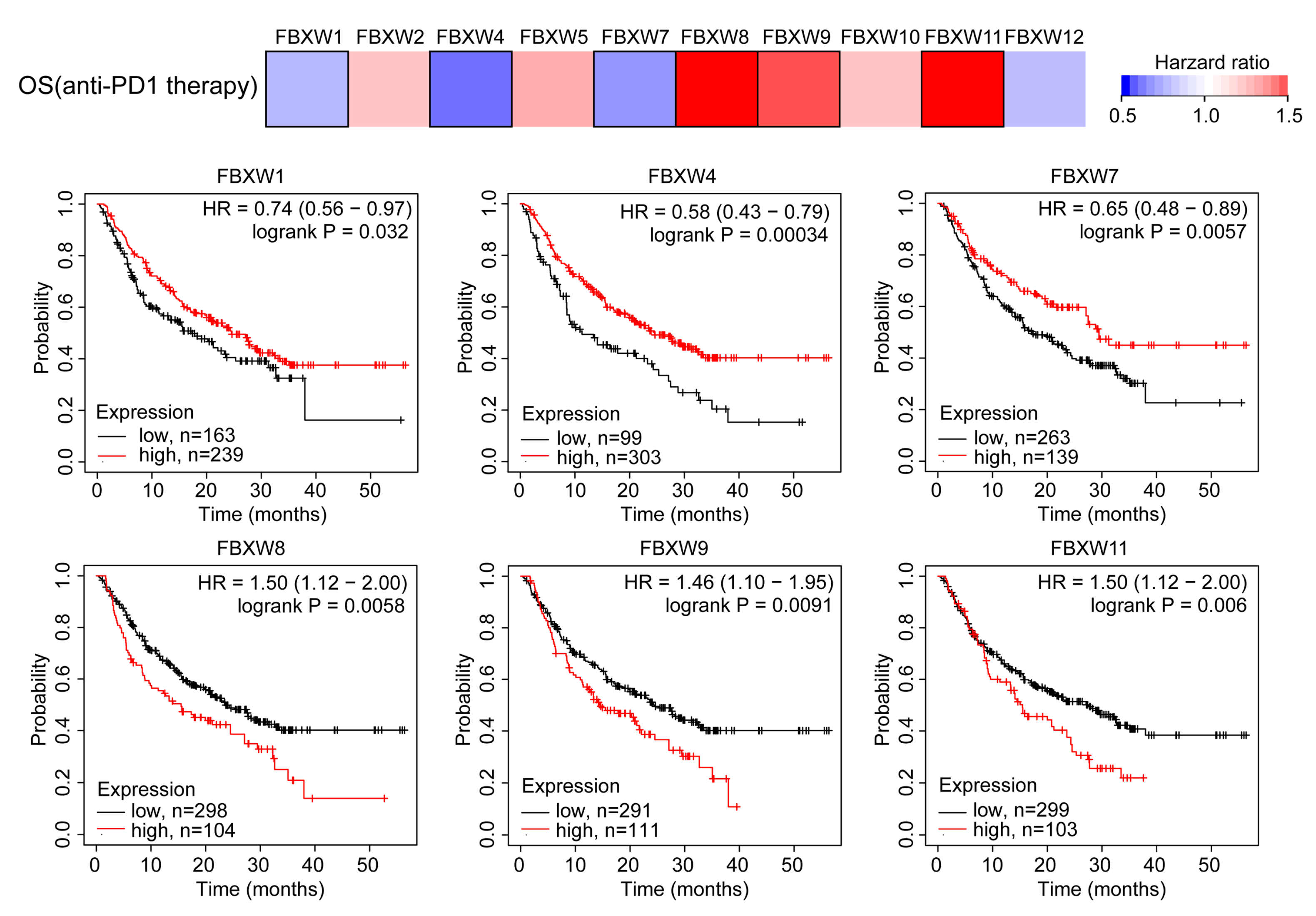
2.2. Analysis of the Association between mRNA Levels of FBXWs and the Prognosis of Patients with Cancer
2.3. Correlation of FBXW Expression and Immune Infiltration across Multiple Cancer Types
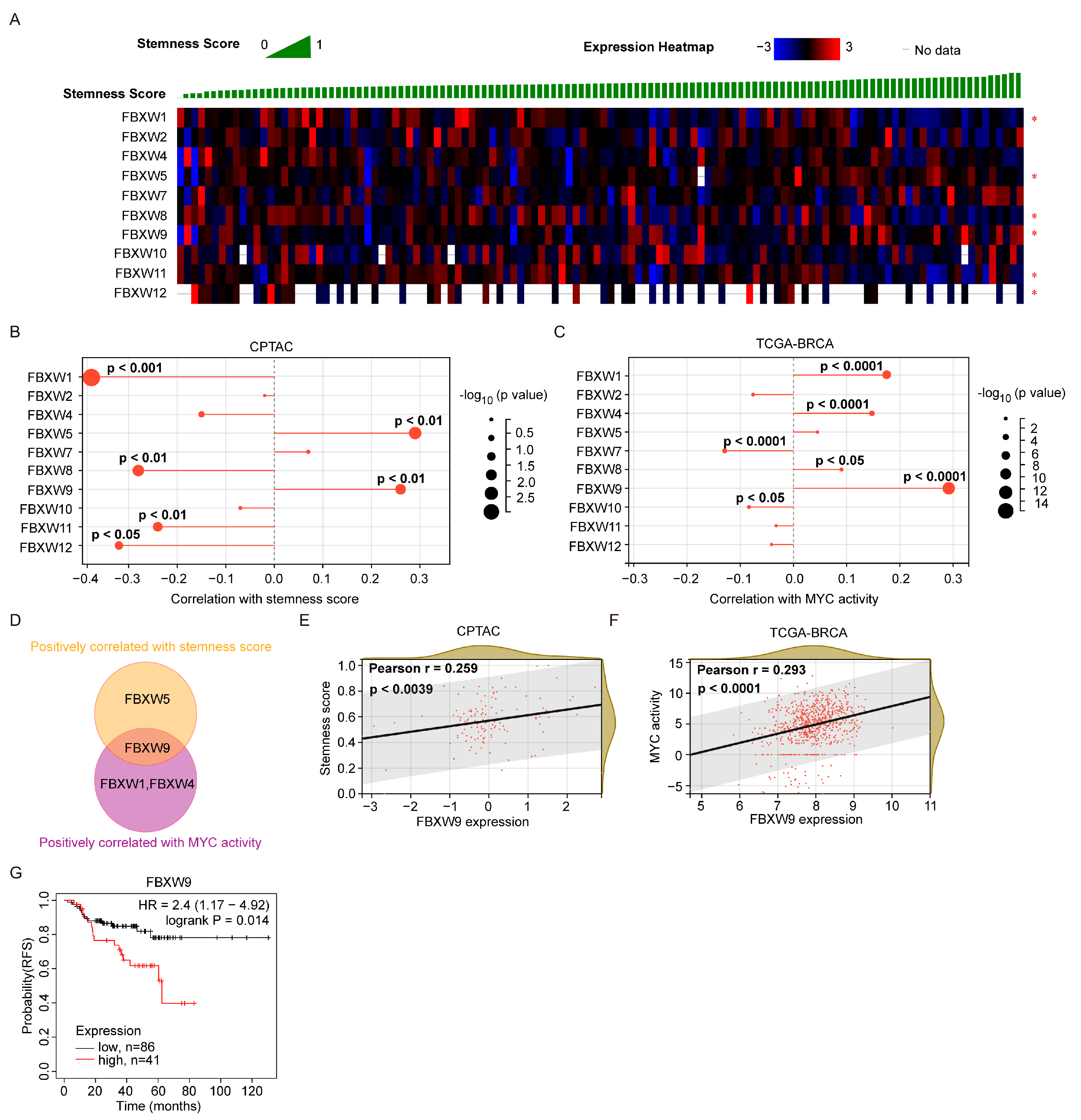
2.4. Analysis of Correlation between FBXW Expression and Stemness Score
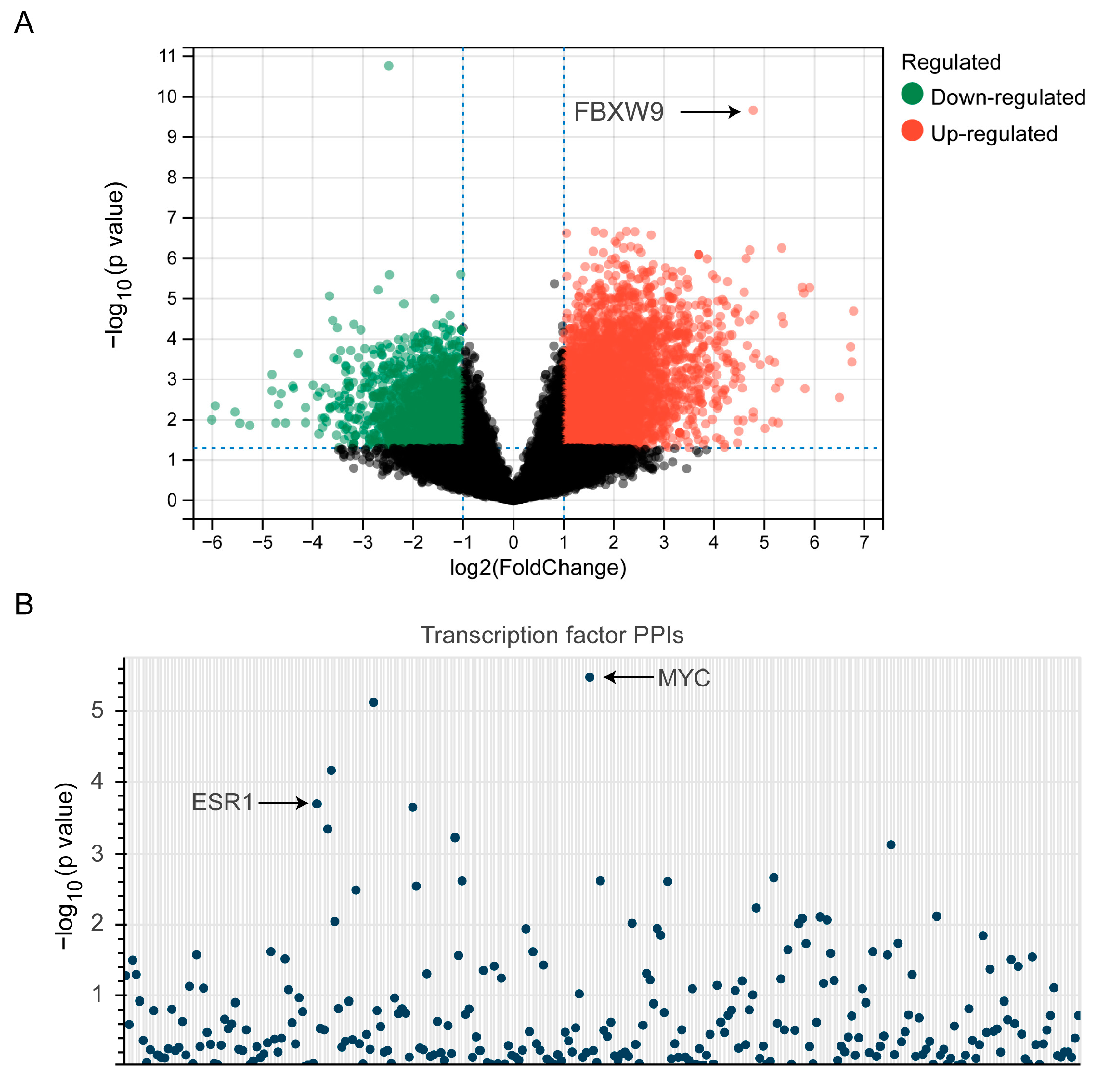
2.5. Analysis of Potential Substrates of FBXW9
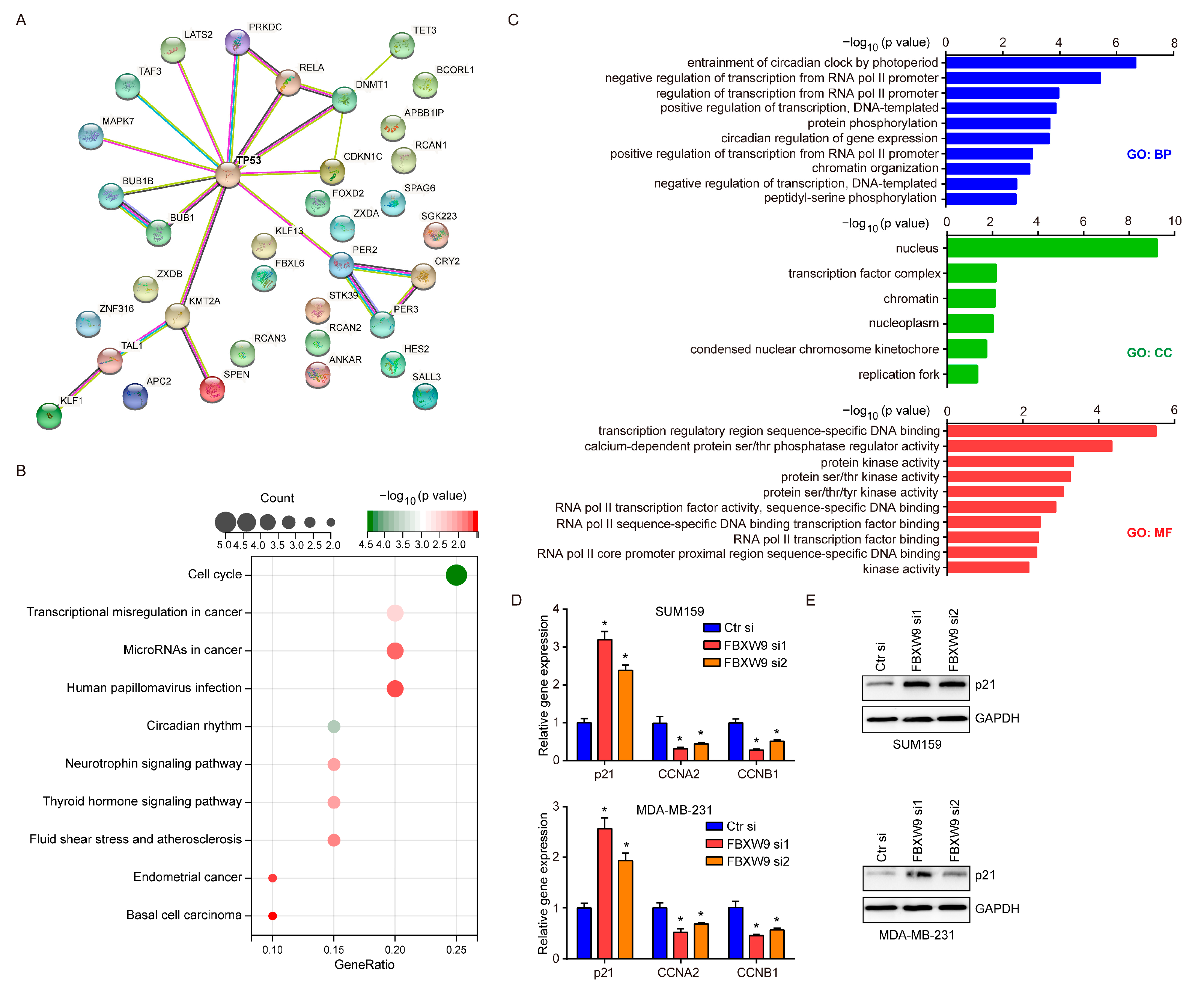
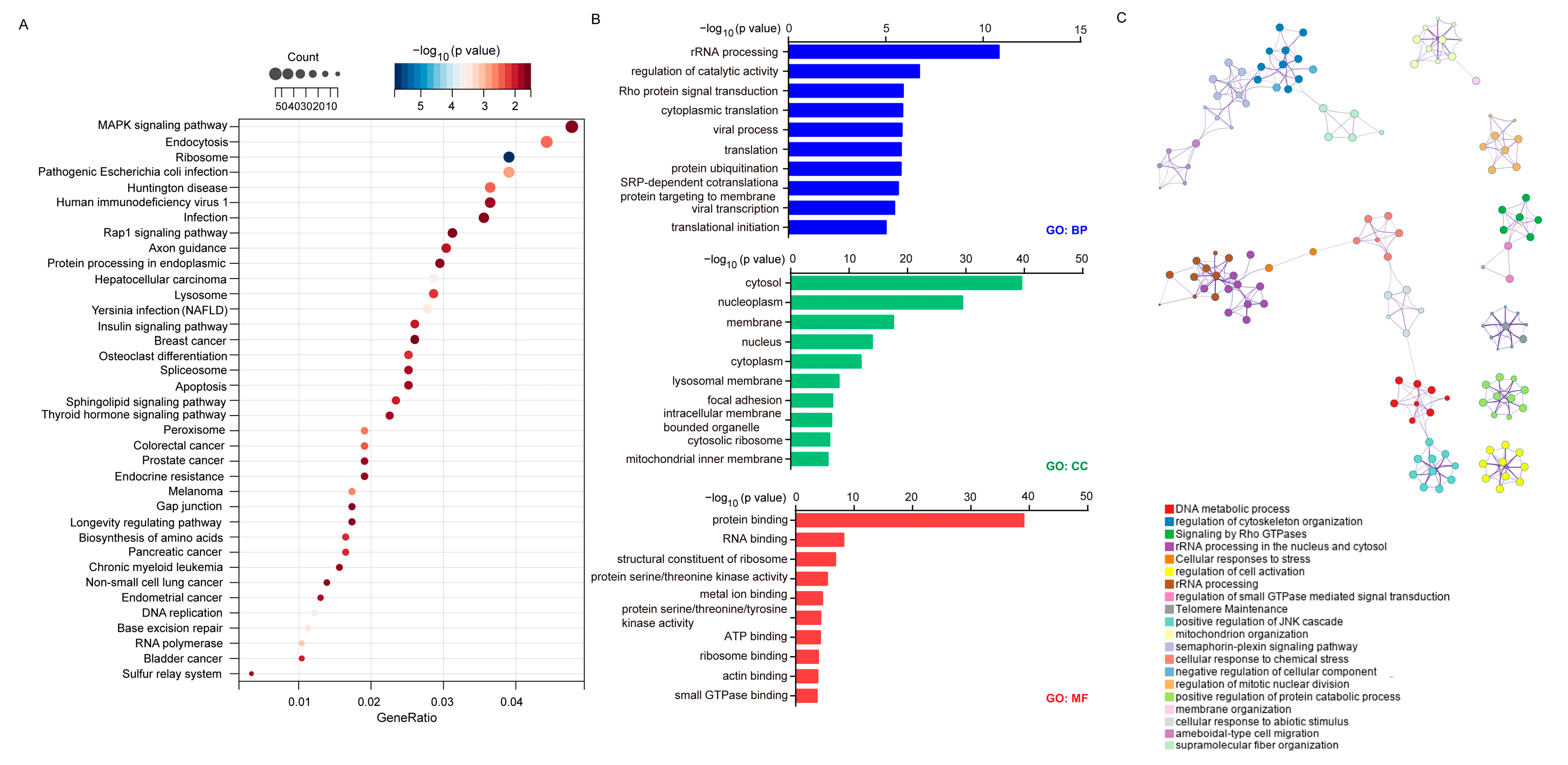
2.6. Establishment of Signaling Network with Co-Expressed Genes of FBXW9
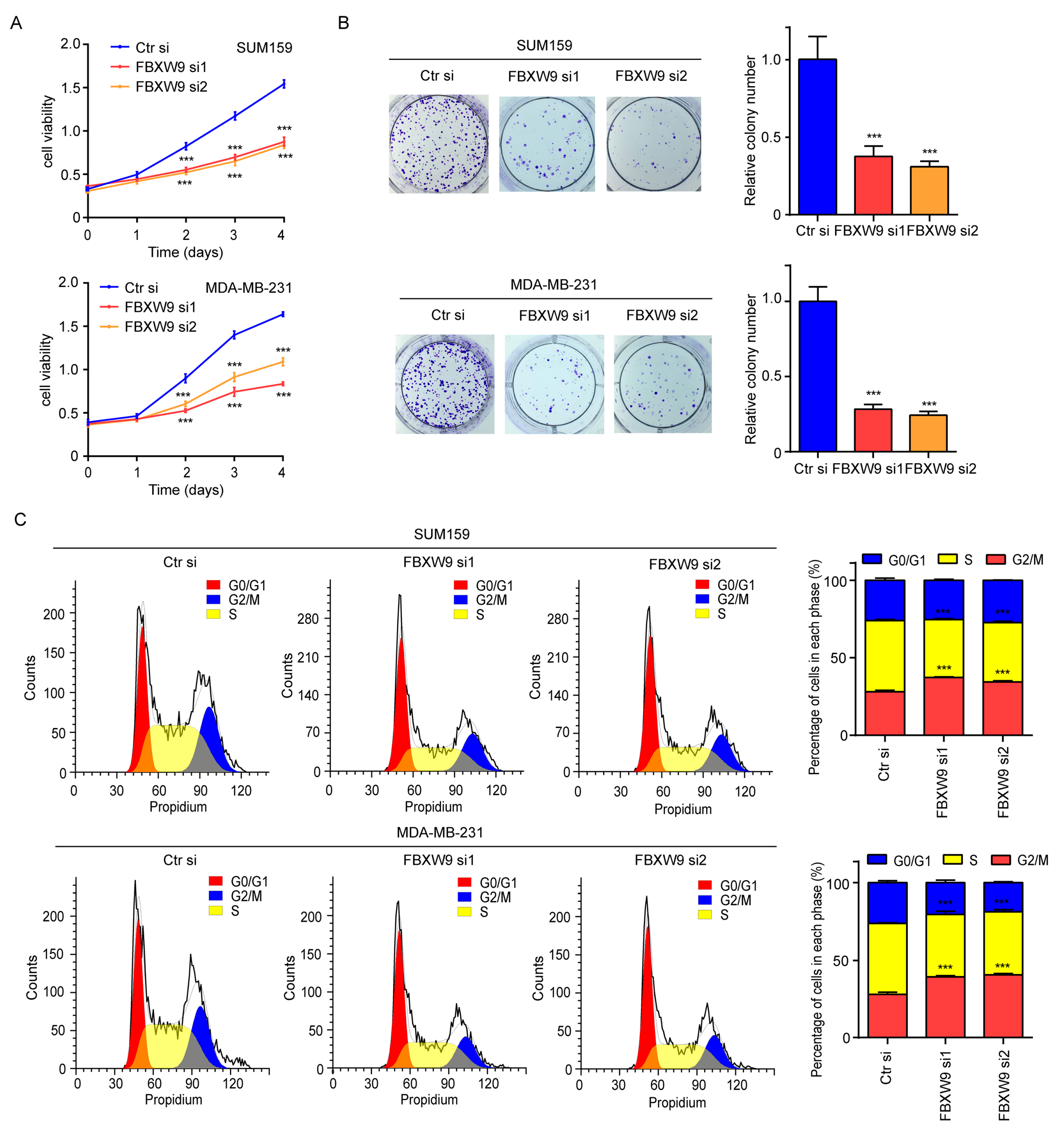
2.7. FBXW9 Silence Inhibited Cell Proliferation and Cell Cycle Progression in Breast Cancer Cells
3. Discussion
4. Materials and Methods
4.1. Pan-Cancer FBXW Expression Analysis
4.2. Pan-Cancer Analysis of the Association between FBXW Expression and Prognosis
4.3. Pan-Cancer Analysis of FBXW Expression and Cancer Immunology
4.4. Analysis of FBXWs’ mRNA Expression and Stemness of Breast Cancer
4.5. Prediction of Substrates of FBXW9
4.6. Pathway Enrichment Analysis of FBXW9-Correlated Genes in Breast Cancer
4.7. Cell Culture
4.8. siRNA-Mediated Gene Silencing
4.9. Cell Proliferation and Colony Forming Assays
4.10. Cell Cycle Analysis
4.11. Human Tissues Collection
4.12. RT-qPCR
4.13. Western Blotting
4.14. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ACC | Adrenocortical carcinoma |
| BLCA | Bladder urothelial carcinoma |
| BRCA | Breast invasive carcinoma |
| CESC | Cervical squamous cell carcinoma and endocervical adenocarcinoma |
| CHOL | Cholangiocarcinoma |
| COAD | Colon adenocarcinoma |
| DLBC | Lymphoid neoplasm diffuse large B-cell lymphoma |
| ESCA | Esophageal carcinoma |
| GBM | Glioblastoma multiforme |
| HNSC | Head and neck squamous cell carcinoma |
| KICH | Kidney chromophobe |
| KIRC | Kidney renal clear cell carcinoma |
| KIRP | Kidney renal papillary cell carcinoma |
| LAML | Acute myeloid leukemia |
| LGG | Brain lower-grade glioma |
| LIHC | Liver hepatocellular carcinoma |
| LUAD | Lung adenocarcinoma |
| LUSC | Lung squamous cell carcinoma |
| MESO | Mesothelioma |
| OV | Ovarian serous cystadenocarcinoma |
| PAAD | Pancreatic adenocarcinoma |
| PCPG | Pheochromocytoma and paraganglioma |
| PRAD | Prostate adenocarcinoma |
| READ | Rectum adenocarcinoma |
| SARC | Sarcoma |
| SKCM | Skin cutaneous melanoma |
| STAD | Stomach adenocarcinoma |
| TGCT | Testicular germ cell tumors |
| THCA | Thyroid carcinoma |
| THYM | Thymoma |
| UCEC | Uterine corpus endometrial carcinoma |
| UCS | Uterine carcinosarcoma |
| UVM | Uveal melanoma |
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Liu, W.; Sun, P.; Wang, F.; Feng, X. Pan-cancer analyses reveal regulation and clinical outcome association of the shelterin complex in cancer. Brief. Bioinform. 2021, 22, bbaa441. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Jiang, T.; Zhou, W.; Li, J.; Li, X.; Wang, Q.; Jin, X.; Yin, J.; Chen, L.; Zhang, Y.; et al. Pan-cancer characterization of immune-related lncRNAs identifies potential oncogenic biomarkers. Nat. Commun. 2020, 11, 1000. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Hershko, A.; Ciechanover, A. The ubiquitin system. Annu Rev. Biochem. 1998, 67, 425–479. [Google Scholar] [CrossRef]
- Bian, C.; Luan, Z.; Zhang, H.; Zhang, R.; Gao, J.; Wang, Y. miR-154-5p Affects the TGFbeta1/Smad3 Pathway on the Fibrosis of Diabetic Kidney Disease via Binding E3 Ubiquitin Ligase Smurf1. Oxid. Med. Cell. Longev. 2022, 2022, 7502632. [Google Scholar] [CrossRef]
- Senft, D.; Qi, J.; Ronai, Z. Ubiquitin ligases in oncogenic transformation and cancer therapy. Nat. Rev. Cancer 2017, 18, 69–88. [Google Scholar] [CrossRef]
- Xiong, B.; Huang, J.; Liu, Y.; Zou, M.; Zhao, Z.; Gong, J.; Wu, X.; Qiu, C. Ubiquitin-specific protease 2a promotes hepatocellular carcinoma progression via deubiquitination and stabilization of RAB1A. Cell. Oncol. 2020, 44, 329–343. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, P.; Inuzuka, H.; Wei, W. Roles of F-box proteins in cancer. Nat. Rev. Cancer 2014, 14, 233–247. [Google Scholar] [CrossRef]
- Frescas, D.; Pagano, M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: Tipping the scales of cancer. Nat. Rev. Cancer 2008, 8, 438–449. [Google Scholar] [CrossRef]
- Tekcham, D.S.; Chen, D.; Liu, Y.; Ling, T.; Zhang, Y.; Chen, H.; Wang, W.; Otkur, W.; Qi, H.; Xia, T.; et al. F-box proteins and cancer: An update from functional and regulatory mechanism to therapeutic clinical prospects. Theranostics 2020, 10, 4150–4167. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Mady, A.S.A.; Ma, Y.; Ryan, C.; Lawrence, T.S.; Nikolovska-Coleska, Z.; Sun, Y.; Morgan, M.A. The WD40 domain of FBXW7 is a poly(ADP-ribose)-binding domain that mediates the early DNA damage response. Nucleic Acids Res. 2019, 47, 4039–4053. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Xiang, Z.; Wu, H.; He, Q.; Dou, R.; Yang, C. The lncRNA SEMA3B-AS1/HMGB1/FBXW7 Axis Mediates the Peritoneal Metastasis of Gastric Cancer by Regulating BGN Protein Ubiquitination. Oxid. Med. Cell. Longev. 2022, 2022, 5055684. [Google Scholar] [CrossRef]
- Han, Q.; Zhang, Q.; Song, H.; Bamme, Y.; Song, C.; Ge, Z. FBXW4 Is Highly Expressed and Associated with Poor Survival in Acute Myeloid Leukemia. Front. Oncol. 2020, 10, 149. [Google Scholar] [CrossRef]
- Wei, P.; Jiang, J.; Xiao, M.; Zeng, M.; Liu, X.; Zhao, B.; Chen, F. The transcript ENST00000444125 of lncRNA LINC01503 promotes cancer stem cell properties of glioblastoma cells via reducing FBXW1 mediated GLI2 degradation. Exp. Cell Res. 2022, 412, 113009. [Google Scholar] [CrossRef] [PubMed]
- Lei, M.; Luo, C.; Zhang, J.; Cao, W.; Ge, J.; Zhao, M. A m6A methyltransferase-mediated immune signature determines prognosis, immune landscape and immunotherapy efficacy in patients with lung adenocarcinoma. Cell. Oncol. 2022, 45, 931–949. [Google Scholar] [CrossRef]
- Zou, Y.; Ye, F.; Kong, Y.; Hu, X.; Deng, X.; Xie, J.; Song, C.; Ou, X.; Wu, S.; Wu, L.; et al. The Single-Cell Landscape of Intratumoral Heterogeneity and The Immunosuppressive Microenvironment in Liver and Brain Metastases of Breast Cancer. Adv. Sci. 2022, 10, 2203699. [Google Scholar] [CrossRef]
- Yang, Z.; Zhao, N.; Cui, J.; Wu, H.; Xiong, J.; Peng, T. Exosomes derived from cancer stem cells of gemcitabine-resistant pancreatic cancer cells enhance drug resistance by delivering miR-210. Cell. Oncol. 2019, 43, 123–136. [Google Scholar] [CrossRef]
- Lawson, D.A.; Bhakta, N.R.; Kessenbrock, K.; Prummel, K.D.; Yu, Y.; Takai, K.; Zhou, A.; Eyob, H.; Balakrishnan, S.; Wang, C.-Y.; et al. Single-cell analysis reveals a stem-cell program in human metastatic breast cancer cells. Nature 2015, 526, 131–135. [Google Scholar] [CrossRef]
- Waldman, T.; Kinzler, K.W.; Vogelstein, B. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 1995, 55, 5187–5190. [Google Scholar]
- Olivero, C.E.; Martínez-Terroba, E.; Zimmer, J.; Liao, C.; Tesfaye, E.; Hooshdaran, N.; Schofield, J.; Bendor, J.; Fang, D.; Simon, M.D.; et al. p53 Activates the Long Noncoding RNA Pvt1b to Inhibit Myc and Suppress Tumorigenesis. Mol. Cell 2020, 77, 761–774.e8. [Google Scholar] [CrossRef] [PubMed]
- Popovic, D.; Vucic, D.; Dikic, I. Ubiquitination in disease pathogenesis and treatment. Nat. Med. 2014, 20, 1242–1253. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, S.Y.; Spiegelman, V.S.; Kumar, K.G. The many faces of beta-TrCP E3 ubiquitin ligases: Reflections in the magic mirror of cancer. Oncogene 2004, 23, 2028–2036. [Google Scholar] [CrossRef]
- Bi, Y.; Cui, D.; Xiong, X.; Zhao, Y. The characteristics and roles of β-TrCP1/2 in carcinogenesis. FEBS J. 2021, 288, 3351–3374. [Google Scholar] [CrossRef] [PubMed]
- Ren, C.; Han, X.; Lu, C.; Yang, T.; Qiao, P.; Sun, Y. Ubiquitination of NF-kappaB p65 by FBXW2 suppresses breast cancer stemness, tumorigenesis, and paclitaxel resistance. Cell Death Differ. 2022, 29, 381–392. [Google Scholar] [CrossRef]
- Lim, Y.X.; Lin, H.; Chu, T.; Lim, Y.P. WBP2 promotes BTRC mRNA stability to drive migration and invasion in triple-negative breast cancer via NF-kappaB activation. Mol. Oncol. 2022, 16, 422–446. [Google Scholar] [CrossRef]
- Lockwood, W.W.; Chandel, S.K.; Stewart, G.L.; Erdjument-Bromage, H.; Beverly, L.J. The Novel Ubiquitin Ligase Complex, SCFFbxw4, Interacts with the COP9 Signalosome in an F-Box Dependent Manner, Is Mutated, Lost and Under-Expressed in Human Cancers. PLoS ONE 2013, 8, e63610. [Google Scholar] [CrossRef]
- Sun, Y.; Hu, Z.; Goeb, Y.; Dreier, L. The F-Box Protein MEC-15 (FBXW9) Promotes Synaptic Transmission in GABAergic Motor Neurons in C. elegans. PLoS ONE 2013, 8, e59132. [Google Scholar] [CrossRef]
- Becht, E.; Giraldo, N.A.; Lacroix, L.; Buttard, B.; Elarouci, N.; Petitprez, F.; Selves, J.; Laurent-Puig, P.; Sautes-Fridman, C.; Fridman, W.H.; et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016, 17, 218. [Google Scholar] [CrossRef] [PubMed]
- Cordenonsi, M.; Zanconato, F.; Azzolin, L.; Forcato, M.; Rosato, A.; Frasson, C.; Inui, M.; Montagner, M.; Parenti, A.R.; Poletti, A.; et al. The Hippo Transducer TAZ Confers Cancer Stem Cell-Related Traits on Breast Cancer Cells. Cell 2011, 147, 759–772. [Google Scholar] [CrossRef]
- Ji, J.; Xu, R.; Zhang, X.; Han, M.; Xu, Y.; Wei, Y.; Ding, K.; Wang, S.; Huang, B.; Chen, A.; et al. Actin like-6A promotes glioma progression through stabilization of transcriptional regulators YAP/TAZ. Cell Death Dis. 2018, 9, 517. [Google Scholar] [CrossRef] [PubMed]
- Sui, Y.; Liu, Z.; Park, S.H.; Thatcher, S.E.; Zhu, B.; Fernandez, J.P. IKKbeta is a beta-catenin kinase that regulates mesenchymal stem cell differentiation. JCI Insight 2018, 3, e96660. [Google Scholar] [CrossRef] [PubMed]
- Aceto, N.; Sausgruber, N.; Brinkhaus, H.; Gaidatzis, D.; Martiny-Baron, G.; Mazzarol, G.; Confalonieri, S.; Quarto, M.; Hu, G.; Balwierz, P.J.; et al. Tyrosine phosphatase SHP2 promotes breast cancer progression and maintains tumor-initiating cells via activation of key transcription factors and a positive feedback signaling loop. Nat. Med. 2012, 18, 529–537. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef]
- Krug, K.; Jaehnig, E.J.; Satpathy, S.; Blumenberg, L.; Karpova, A.; Anurag, M.; Miles, G.; Mertins, P.; Geffen, Y.; Tang, L.C.; et al. Proteogenomic Landscape of Breast Cancer Tumorigenesis and Targeted Therapy. Cell 2020, 183, 1436–1456.e31. [Google Scholar] [CrossRef]
- Jiang, P.; Freedman, M.L.; Liu, J.S.; Liu, X.S. Inference of transcriptional regulation in cancers. Proc. Natl. Acad. Sci. USA 2015, 112, 7731–7736. [Google Scholar] [CrossRef]
- Wang, X.; Li, Y.; He, M.; Kong, X.; Jiang, P.; Liu, X.; Diao, L.; Zhang, X.; Li, H.; Ling, X.; et al. UbiBrowser 2.0: A comprehensive resource for proteome-wide known and predicted ubiquitin ligase/deubiquitinase–substrate interactions in eukaryotic species. Nucleic Acids Res. 2021, 50, D719–D728. [Google Scholar] [CrossRef]
- Sun, H.; Lin, D.-C.; Cao, Q.; Pang, B.; Gae, D.D.; Lee, V.K.M.; Lim, H.J.; Doan, N.; Said, J.W.; Gery, S.; et al. Identification of a Novel SYK/c-MYC/MALAT1 Signaling Pathway and Its Potential Therapeutic Value in Ewing Sarcoma. Clin. Cancer Res. 2017, 23, 4376–4387. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, S.; Liang, Z.; Fan, Z.; Cao, B.; Wang, N.; Wu, R.; Sun, H. A Comprehensive Analysis Revealing FBXW9 as a Potential Prognostic and Immunological Biomarker in Breast Cancer. Int. J. Mol. Sci. 2023, 24, 5262. https://doi.org/10.3390/ijms24065262
Yu S, Liang Z, Fan Z, Cao B, Wang N, Wu R, Sun H. A Comprehensive Analysis Revealing FBXW9 as a Potential Prognostic and Immunological Biomarker in Breast Cancer. International Journal of Molecular Sciences. 2023; 24(6):5262. https://doi.org/10.3390/ijms24065262
Chicago/Turabian StyleYu, Shiyi, Zhengyan Liang, Zhehao Fan, Binjie Cao, Ning Wang, Rui Wu, and Haibo Sun. 2023. "A Comprehensive Analysis Revealing FBXW9 as a Potential Prognostic and Immunological Biomarker in Breast Cancer" International Journal of Molecular Sciences 24, no. 6: 5262. https://doi.org/10.3390/ijms24065262
APA StyleYu, S., Liang, Z., Fan, Z., Cao, B., Wang, N., Wu, R., & Sun, H. (2023). A Comprehensive Analysis Revealing FBXW9 as a Potential Prognostic and Immunological Biomarker in Breast Cancer. International Journal of Molecular Sciences, 24(6), 5262. https://doi.org/10.3390/ijms24065262



