Genetic Diversity and Phylogeography of a Turf-Forming Cosmopolitan Marine Alga, Gelidium crinale (Gelidiales, Rhodo-Phyta)
Abstract
1. Introduction
“Long-range dispersal of seaweeds does exist, but it is an exception rather than the rule. If it were the rule, the world’s seaweed floras would show similar latitudinal gradients in species composition in the oceans and on both hemispheres. This is, however, not the case.”—van den Hoek [1].
2. Results
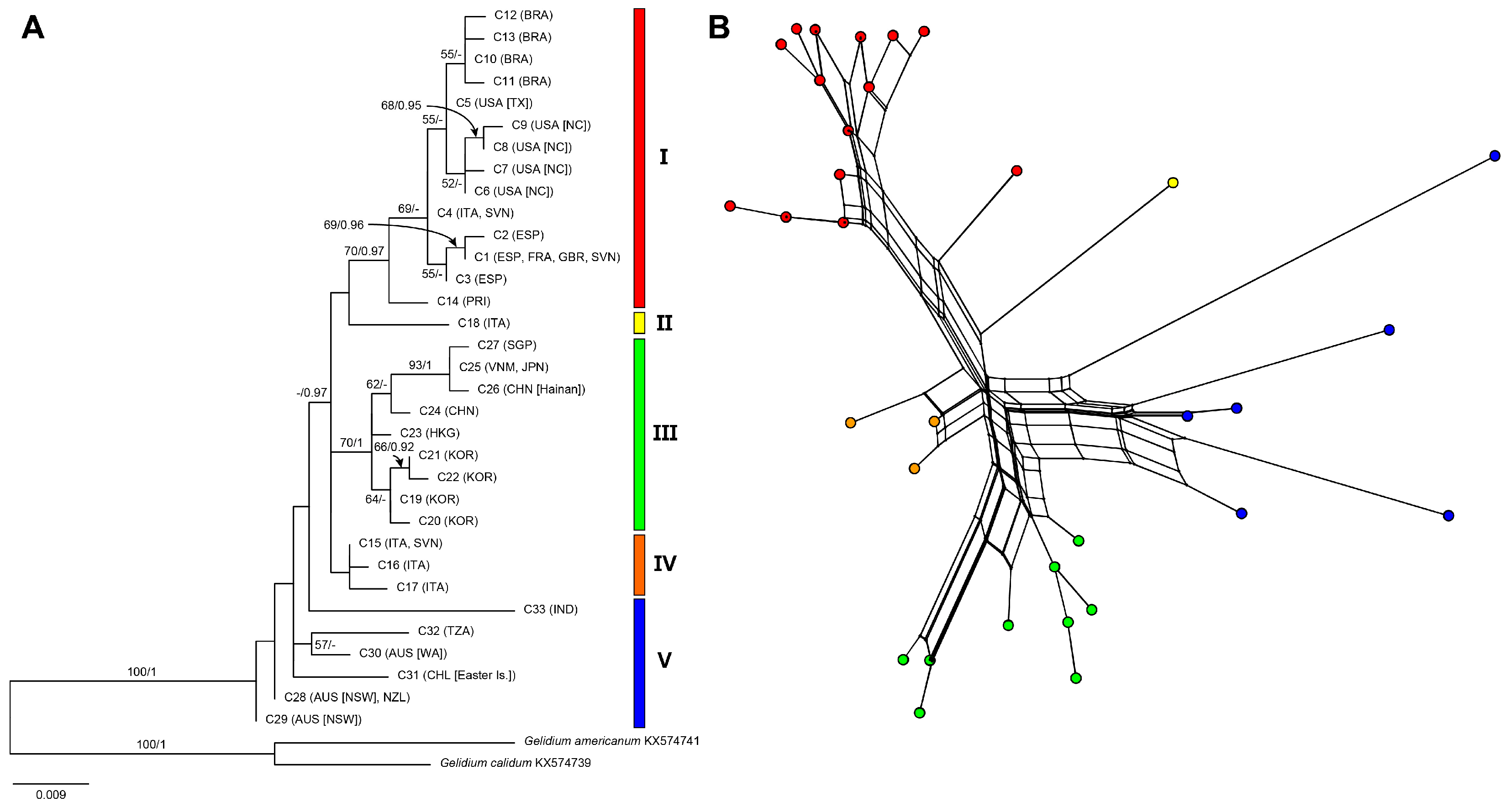
2.1. Phylogenies and Divergence Time
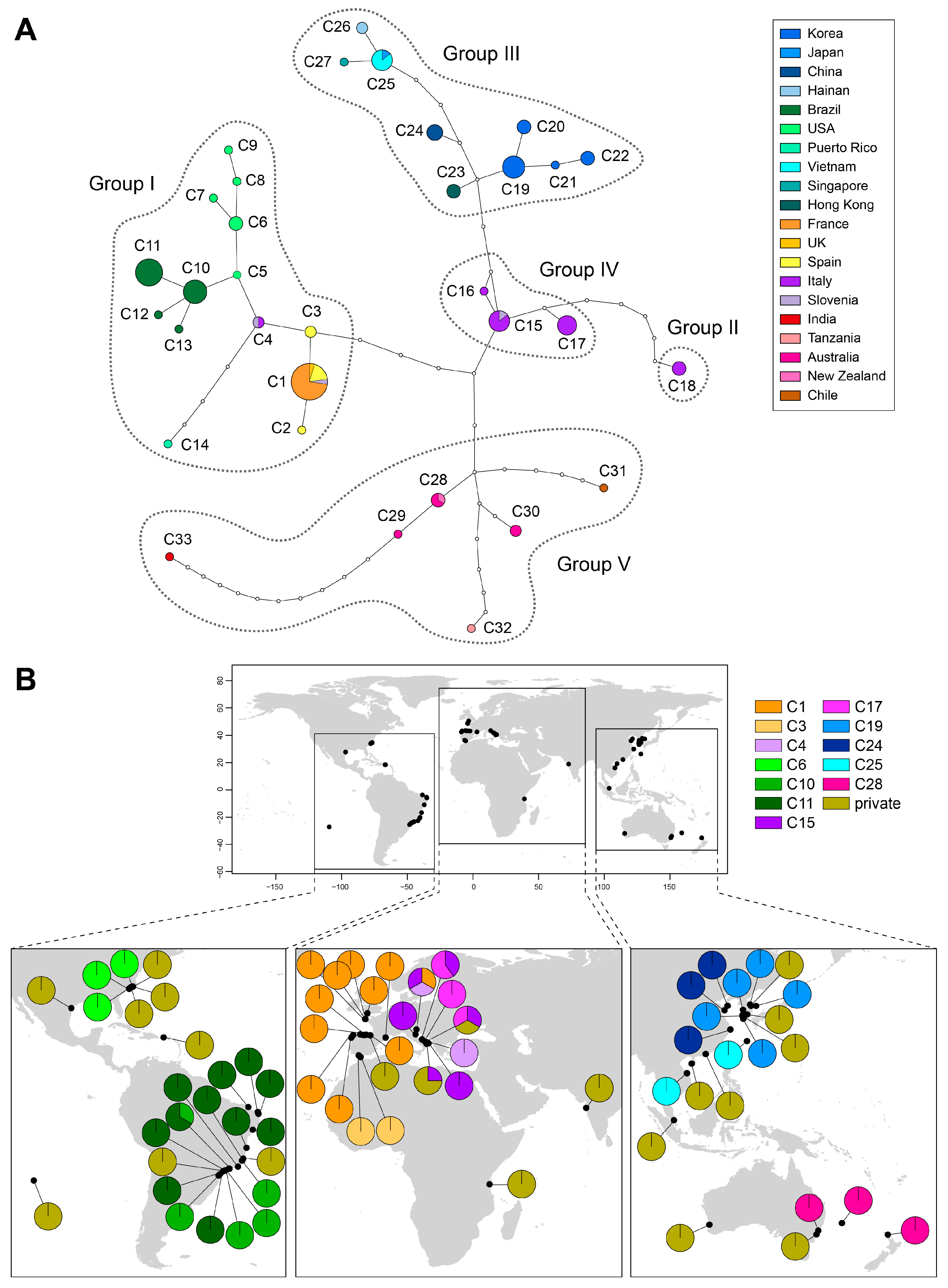
2.2. Genetic Diversity and Phylogeographic Structure Based on Mitochondrial COI-5P
2.3. Historical Demography Inferences Based on Mitochondrial COI-5P
3. Discussion
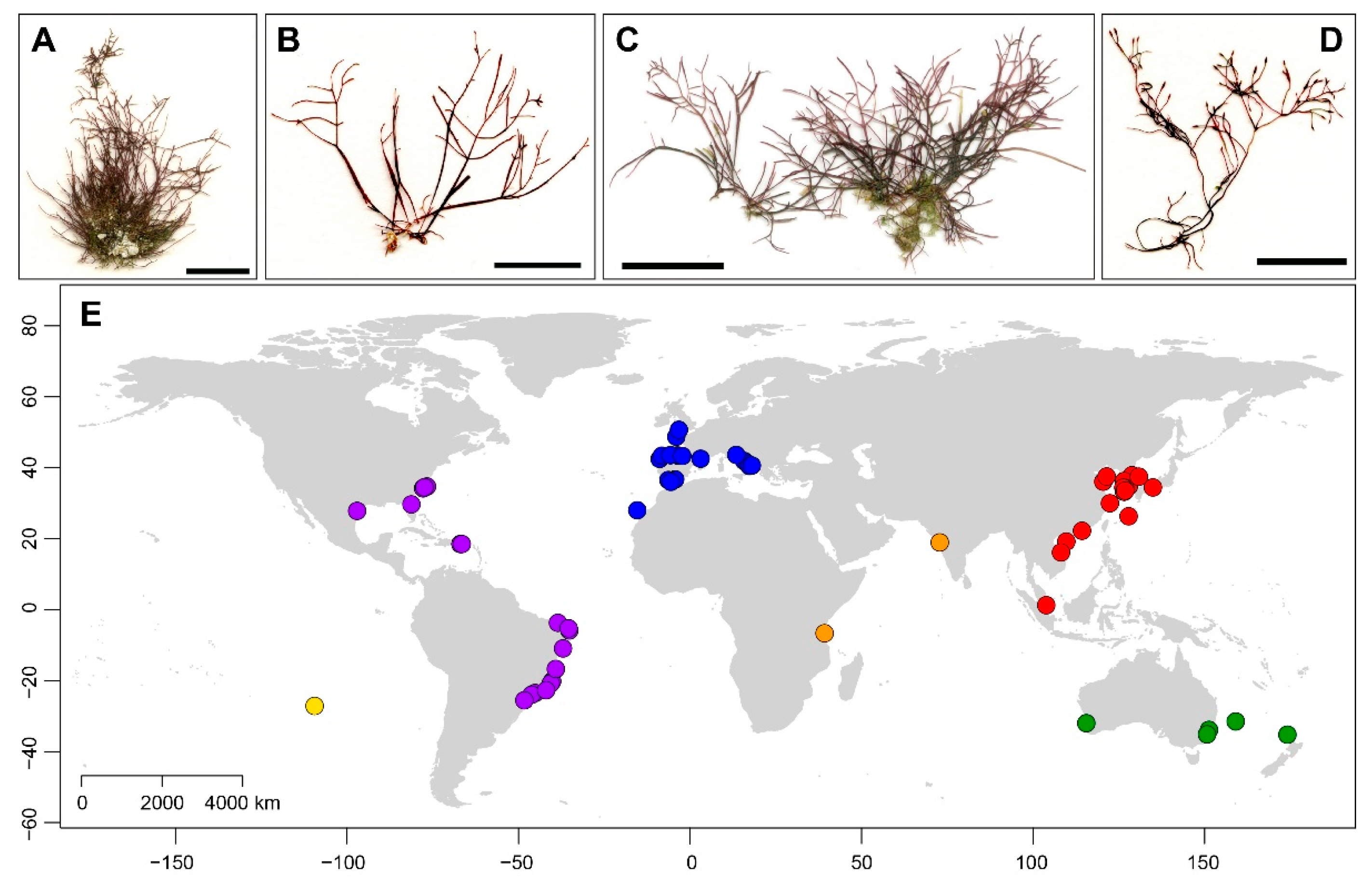
3.1. Genetic Diversity and Geographical Structure
3.2. Pleistocene Relicts for Cosmopolitan Distribution
3.3. Ecological Implication
3.4. Taxonomic Implication
Taxonomic Conclusion
4. Materials and Methods
4.1. Sampling and Herbarium Collections
4.2. DNA Extraction, PCR, and Sequencing
4.3. DNA Extraction and High Throughput Sequencing of Type Specimens of P. heteroplatos
4.4. Phylogenetic Analysis
4.5. Divergence Time Estimation
4.6. Phylogeographic Analysis of Mitochondrial COI-5P
4.7. Demographic History
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Van den Hoek, C. The possible significance of long-range dispersal for the biogeography of seaweeds. Helgoländer Meeresunters. 1987, 41, 261–272. [Google Scholar] [CrossRef]
- Wernberg, T.; Bennett, S.; Babcock, R.C.; de Bettignies, T.; Cure, K.; Depczynski, M.; Dufois, F.; Fromont, J.; Fulton, C.J.; Hovey, R.K.; et al. Climate-driven regime shift of a temperate marine ecosystem. Science 2016, 353, 169–172. [Google Scholar] [CrossRef]
- Pecl, G.T.; Araújo, M.B.; Bell, J.D.; Blanchard, J.; Bonebrake, T.C.; Chen, I.C.; Clark, T.D.; Colwell, R.K.; Danielsen, F.; Evengård, B.; et al. Biodiversity redistribution under climate change: Impacts on ecosystems and human well-being. Science 2017, 355, eaai9214. [Google Scholar] [CrossRef] [PubMed]
- Bertolini, C. Can secondary species maintain a primary role? Consistent inter-regional effects of understory algae on diversity. Mar. Biodiv. 2019, 49, 841–849. [Google Scholar] [CrossRef]
- Alfonso, B.; Hernández, J.C.; Sangil, C.; Martín, L.; Expósito, F.J.; Díaz, J.P.; Sansón, M. Fast climatic changes place an endemic Canary Island macroalga at extinction risk. Reg. Environ. Chang. 2021, 21, 113. [Google Scholar] [CrossRef]
- Womersley, H.B.S.; Guiry, M.D. Order Gelidiales Kyin 1923: 132. In The Marine Benthic Flora of Southern Australia. Rhodophyta–Part IIIA.; Womersley, H.B.S., Ed.; Australian Biological Resources Study: Canberra, Australia, 1994; pp. 118–142. [Google Scholar]
- Millar, A.J.K.; Freshwater, D.W. Morphology and molecular phylogeny of the marine algal order Gelidiales (Rhodophyta) from New South Wales, including Lord Howe and Norfolk Islands. Aust. Syst. Bot. 2005, 18, 215–263. [Google Scholar] [CrossRef]
- Kim, K.M.; Boo, S.M. Phylogenetic relationships and distribution of Gelidium crinale and G. pusillum (Gelidiales, Rhodophyta) using cox1 and rbcL sequences. Algae 2012, 27, 83–94. [Google Scholar] [CrossRef]
- Kapraun, D.F.; Zechman, F.W. Seasonality and vertical zonation of benthic marine algae on a North Carolina coastal jetty. Bull. Mar. Sci. 1982, 32, 702–715. [Google Scholar]
- Croce, M.E.; Parodi, E.R. The turf-forming alga Gelidium crinale (Florideophyceae, Rhodophyta) on Atlantic Patagonian coasts. Bot. Mar. 2013, 56, 131–141. [Google Scholar] [CrossRef]
- Díaz-Tapia, P.; Bárbara, I. Seaweeds from sand-covered rocks of the Atlantic Peninsula. Part 2. Palmariales, Ceramiales (excluding Rhodomelaceae), Gelidiales, Gigartinales, Plocamiales, Rhodymeniales and Scytothamnales. Cryptogamie Algol. 2014, 35, 157–199. [Google Scholar] [CrossRef]
- Nelson, W.A.; Farr, T.J. Field and morphological observations of Gelidium longipes (Gelidiales, Rhodophyta), a rare endemic red alga from northern New Zealand. N. Z. J. Bot. 2003, 41, 707–713. [Google Scholar] [CrossRef]
- Freshwater, D.W.; Tudor, K.; O’Shaughnessy, K.; Wysor, B. DNA barcoding in the red algal order Gelidiales: Comparison of COI with rbcL and verification of the “barcoding gap”. Cryptogamie Algol. 2010, 31, 435–449. [Google Scholar]
- Guiry, M.D.; Guiry, G.M.; AlgaeBase. World-Wide Electronic Publication, National University of Ireland, Galway. 2022. Available online: http://www.algaebase.org (accessed on 15 August 2022).
- Feldmann, J.; Hamel, G. Floridées de France VII. Gélidiales. Rev. Algol. 1936, 9, 85–140. [Google Scholar]
- Dixon, P.S.; Irvine, L.M. Seaweeds of the British Isles, Vol. 1. Rhodophyta Part 1. Introduction, Nemaliales, Gigartinales; British Museum (Natural History): London, UK, 1977. [Google Scholar]
- Iha, C.; Milstein, D.; Guimarães, S.M.P.B.; Freshwater, D.W.; Oliveira, M.C. DNA barcoding reveals high diversity in the Gelidiales of the Brazilian southeast coast. Bot. Mar. 2015, 58, 295–305. [Google Scholar] [CrossRef]
- Boo, G.H.; Kim, K.M.; Nelson, W.A.; Riosmena-Rodríguez, R.; Yoon, K.J.; Boo, S.M. Taxonomy and distribution of selected species of the agarophyte genus Gelidium (Gelidiales, Rhodophyta). J. Appl. Phycol. 2014, 26, 1243–1251. [Google Scholar] [CrossRef]
- Børgesen, F. Some Indian Rhodophyceae especially from the shores of Presidency of Bombay, IV. Kew Bull. 1934, 1, 1–30. [Google Scholar] [CrossRef]
- Santelices, B. A taxonomic review of the Hawaiian Gelidiales (Rhodophyta). Pac. Sci. 1977, 31, 61–84. [Google Scholar]
- Umamaheswara Rao, M.; Kaliaperumal, N. Genus Pterocladia (Rhodophyta) from India. Bull. Bot. Surv. India 1980, 22, 109–111. [Google Scholar]
- Kaliaperumal, N.; Umamaheswara Rao, M. Studies on the standing crop and phycocolloid of Gelidium pusillum and Pterocladia heteroplatos. Indian J. Bot. 1981, 4, 91–95. [Google Scholar]
- Freshwater, D.W.; Rueness, J. Phylogenetic relationships of some European Gelidium (Gelidiales, Rhodophyta) species, based on rbcL nucleotide sequence analysis. Phycologia 1994, 33, 187–194. [Google Scholar] [CrossRef]
- Sherwood, A.R.; Sauvage, T.; Kurihara, A.; Conklin, K.Y.; Presting, G.G. The Hawaiian Rhodophyta biodiversity survey (2006–2010): A summary of principal findings. BMC Plant Biol. 2010, 10, 258. [Google Scholar] [CrossRef] [PubMed]
- Boo, G.H.; Le Gall, L.; Miller, K.A.; Freshwater, D.W.; Wernberg, T.; Terada, R.; Yoon, K.J.; Boo, S.M. A novel phylogeny of the Gelidiales (Rhodophyta) based on five genes including nuclear CesA, with descriptions of Orthogonacladia gen. nov. and Orthogonacladiaceae fam. nov. Mol. Phylogenet. Evol. 2016, 101, 359–372. [Google Scholar] [CrossRef]
- Boo, G.H.; Zubia, M.; Hughey, J.R.; Sherwood, A.R.; Fujii, M.T.; Boo, S.M.; Miller, K.A. Complete mitochondrial genomes reveal population-level patterns in the widespread red alga Gelidiella fanii (Gelidiales, Rhodophyta). Front. Mar. Sci. 2020, 7, 583957. [Google Scholar] [CrossRef]
- Boo, G.H.; Qui, Y.-X.; Kim, J.Y.; Ang, P.O.; Bosch, S.; De Clerck, O.; He, P.; Higa, A.; Huang, B.; Kogame, K.; et al. Contrasting patterns of genetic structure and phylogeography in the marine agarophytes Gelidiophycus divaricatus and G. freshwateri (Gelidiales, Rhodophyta) from East Asia. J. Phycol. 2019, 55, 1319–1334. [Google Scholar] [CrossRef]
- Jamas, M.; Iha, C.; Oliveira, M.C.; Guimarães, S.M.P.B.; Fujii, M.T. Morphological and molecular studies on Gelidiaceae and Gelidiellaceae (Gelidiales, Rhodophyta) from Brazil with description of the new species Gelidium calidum. Phytotaxa 2017, 58, 195–218. [Google Scholar] [CrossRef]
- Shimada, S.; Horiguchi, T.; Masuda, M. Phylogenetic affinities of genera Acanthopeltis and Yatabella (Gelidiales, Rhodophyta) inferred from molecular analyses. Phycologia 1999, 38, 528–540. [Google Scholar] [CrossRef]
- Boo, G.H.; Le Gall, L.; Hwang, I.K.; Rousseau, F.; Yoon, H.S. Species diversity of Gelidium from southern Madagascar evaluated by an integrative taxonomic approach. Diversity 2022, 14, 826. [Google Scholar] [CrossRef]
- Díaz-Tapia, P.; Baldock, L.; Maggs, C.A. Discovery of Flabellia petiolata (Halimedaceae, Chlorophyta) in the southern British Isles: A relict population or a new introduction? Aquat. Bot. 2020, 160, 103160. [Google Scholar] [CrossRef]
- Petrocelli, A.; Cecere, E. A 20-year update on the state of seaweed resources in Italy. Bot. Mar. 2019, 62, 249–264. [Google Scholar] [CrossRef]
- Huisman, J.M.; Cowan, R.A.; De Clerck, O. Biogeography of Australian seaweeds. In Handbook of Australian Biogeography; Ebach, M.C., Ed.; CRC Press: Boca Raton, FL, USA, 2017; pp. 59–80. [Google Scholar]
- Bringloe, T.T.; Saunders, G.W. Trans-Arctic speciation of Florideophyceae (Rhodophyta) since the opening of the Bering Strait, with consideration of the “species pump” hypothesis. J. Biogeogr. 2019, 46, 694–705. [Google Scholar] [CrossRef]
- Dowsett, H.J.; Chandler, M.A.; Robinson, M.M. Surface temperatures of the Mid-Pliocene North Atlantic Ocean: Implications for future climate. Phil. Trans. R. Soc. A 2009, 367, 69–84. [Google Scholar] [CrossRef]
- Clark, P.U.; Dyke, A.S.; Shakun, J.D.; Carlson, A.E.; Clark, J.; Wohlfarth, B.; Mitrovika, J.X.; Hosteler, S.W.; Mccabe, A.M. The Last Glacial Maximum. Science 2009, 325, 710–714. [Google Scholar] [CrossRef]
- Provan, J.I.M.; Wattier, R.A.; Maggs, C.A. Phylogeographic analysis of the red seaweed Palmaria palmata reveals a Pleistocene marine glacial refugium in the English Channel. Mol. Ecol. 2005, 14, 793–803. [Google Scholar] [CrossRef] [PubMed]
- Montecinos, A.; Broitman, B.R.; Faugeron, S.; Haye, P.A.; Tellier, F.; Guillemin, M.L. Species replacement along a linear coastal habitat: Phylogeography and speciation in the red alga Mazzaella laminarioides along the south east Pacific. BMC Evol. Biol. 2012, 12, 97. [Google Scholar] [CrossRef]
- Maggs, C.A.; Castilho, R.; Foltz, D.; Henzler, C.; Taimour Jolly, M.; Kelly, J.; Olsen, J.; Pérez, K.E.; Stam, W.; Väinölä, R.; et al. Evaluating signatures of glacial refugia for north Atlantic benthic marine taxa. Ecology 2008, 89, S108–S122. [Google Scholar] [CrossRef] [PubMed]
- Yang, E.C.; Boo, S.M.; Bhattacharya, D.; Saunders, G.W.; Knoll, A.H.; Fredericq, S.; Graf, L.; Yoon, H.S. Divergence time estimates and the evolution of major lineages in the florideophyte red algae. Sci. Rep. 2016, 6, 21361. [Google Scholar] [CrossRef] [PubMed]
- Leliaert, F.; Payo, D.A.; Gurgel, C.F.D.; Schils, T.; Draisma, S.G.A.; Saunders, G.W.; Kamiya, M.; Sherwood, A.R.; Lin, S.-M.; Huisman, J.M.; et al. Patterns and drivers of species diversity in the Indo-Pacific red seaweed Portieria. J. Biogeogr. 2018, 45, 2299–2313. [Google Scholar] [CrossRef]
- Boo, G.H.; Leliaert, F.; Le Gall, L.; Coppejans, E.; De Clerck, O.; Nguyen, T.V.; Payri, C.E.; Miller, K.A.; Yoon, H.S. Ancient Tethyan vicariance and long-distance dispersal drive global diversification and cryptic speciation in the red seaweed Pterocladiella. Front. Plant Sci. 2022, 13, 849476. [Google Scholar] [CrossRef]
- Rius, M.; Ahyong, S.; Bieler, R.; Boudouresque, C.; Costello, M.J.; Downey, R.; Galil, B.S.; Gollasch, S.; Hutchings, P.; Kamburska, L.; et al. World Register of Introduced Marine Species (WRiMS). 2022. Available online: https://www.marinespecies.org/introduced (accessed on 15 August 2022). [CrossRef]
- Nelson, W.A.; D’Archino, N.F.; Neill, K.F.; Robinson, N.M. Introduced marine macroalgae: New perspectives on species recognition and distribution in New Zealand. Bot. Mar. 2021, 64, 379–393. [Google Scholar] [CrossRef]
- Turner, D. Fuci sive Plantarum Fucorum Generi a Botanicis Ascriptarum Icones Descriptiones et Historia; John and Arthur Arch: London, UK, 1819; Volume 4, pp. 1–153. [Google Scholar]
- Gaillon, B. Résumé méthodique des classifications des Thalassiophytes. Dict. Sci. Nat. 1828, 53, 350–406. [Google Scholar]
- Kützing, F.T. Phycologia Generalis Oder Anatomie, Physiologie und Systemkunde der Tange. Mit 80 farbig gedruckten Tafeln, Gezeichnet und Gravirt vom Verfasser; F.A. Brockhaus: Leipzig, Germany, 1843. [Google Scholar]
- Agardh, J.G. Species Genera et Ordines Algarum. Volume 3, Part 1—Epicrisis Systematiis Floridearum; Weigel: Leipzig, Germany, 1876. [Google Scholar]
- Winston, J.E. Dispersal in marine organisms without a pelagic larval phase. Integr. Comp. Biol. 2012, 52, 447–457. [Google Scholar] [CrossRef] [PubMed]
- Darling, J.A.; Carlton, J.T. A framework for understanding marine cosmopolitanism in the Anthropocene. Front. Mar. Sci. 2018, 5, 293. [Google Scholar] [CrossRef] [PubMed]
- Uwai, S.; Nelson, W.; Neill, K.; Wang, W.D.; Aguilar-Rosas, L.E.; Boo, S.M.; Kawai, H. Genetic diversity in Undaria pinnatifida (Laminariales, Phaeophyceae) deduced from mitochondrial genes—Origins and succession of introduced populations. Phycologia 2006, 45, 687–695. [Google Scholar] [CrossRef]
- Andreakis, N.; Costello, P.; Zanolla, M.; Saunders, G.W.; Mata, L. Endemic or introduced? Phylogeography of Asparagopsis (Florideophyceae) in Australasia reveals multiple introductions and a new mitochondrial lineage. J. Phycol. 2016, 52, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Edwards, P.; Kapraun, D.F. Benthic marine algal ecology in the Port Aransas, Texas area. Contrib. Mar. Sci. Univ. Texas 1973, 17, 15–52. [Google Scholar]
- Coutinho, R.; Seeliger, U. Seasonal occurrence and growth of benthic algae in the Patos Lagoon estuary, Brazil. Estuar. Coast. Shelf Sci. 1986, 23, 889–900. [Google Scholar] [CrossRef]
- Boulus, A.; Spaneir, E.; Friedlander, M. Effect of outdoor conditions on growth rate and chemical composition of Gelidium crinale in culture. J. Appl. Phycol. 2007, 19, 471–478. [Google Scholar] [CrossRef]
- Khaled, A.; Hessein, A.; Abdel-Halim, A.M.; Morsy, F.M. Distribution of heavy metals in seaweeds collected along Marsa-Matrouh beaches, Egyptian Mediterranean Sea. Egypt. J. Aquat. Res. 2014, 40, 363–371. [Google Scholar] [CrossRef]
- Bonometto, A.; Ponis, E.; Cacciatore, F.; Riccardi, E.; Pigozzi, S.; Parati, P.; Novello, M.; Ungaro, N.; Acquavita, A.; Manconi, P.; et al. A new multi-index method for the eutrophication assessment in transitional waters: Large-scale implementation in Italian lagoons. Environments 2022, 9, 41. [Google Scholar] [CrossRef]
- Kralj, M.; De Vittor, C.; Comici, C.; Relitti, F.; Auriemma, R.; Alabiso, G.; Del Negro, P. Recent evolution of the physical-chemical characteristics of a Site of National Interest-the Mar Piccolo of Taranto (Ionian Sea)-and changes over the last 20 years. Environ. Sci. Pollut. Res. Int. 2016, 23, 12675–12690. [Google Scholar] [CrossRef]
- Holloway-Adkins, K.G.; Hanisak, M.D. Macroalgal foraging preference of juvenile green turtles (Chelonia mydas) in a warm temperature/subtropical transition zone. Mar. Biol. 2017, 164, 161. [Google Scholar] [CrossRef]
- Whorff, J.S.; Whorff, L.L.; Sweet, M.H. Spatial variation in an algal turf community with respect to substratum slope and wave height. J. Mar. Biol. Ass. UK 1995, 75, 429444. [Google Scholar] [CrossRef]
- Thiers, W.R. Index Herbariorum: A global directory of public herbaria and associated staff. New York Botanical Garden’s Virtual Herbarium. 2022. Available online: http://sweetgum.nygb.org/ih/ (accessed on 15 August 2022).
- Cormaci, M.; Furnari, G.; Alongi, G. Benthic marine flora of the Mediterranean: Rhodophyta—Rhodymeniophycidae I. Acrosymphytales, Bonnemaisoniales, Gelidiales, Gigartinales, Gracilariales. Boll. Accad. Gioenia Nat. Sci. (Catania) 2020, 53, FP11–FP346. [Google Scholar] [CrossRef]
- Silva, P.C.; Basson, P.W.; Moe, R.L. Catalogue of the benthic marine algae of Indian Ocean. Univ. Calif. Publ. Bot. 1996, 79, 1–259. [Google Scholar]
- Okamura, K. Icones of Japanese Algae; Kazamashobo: Tokyo, Japan, 1923; Volume IV. [Google Scholar]
- Taylor, W.R. Marine algae from Haiti collected by H.H. Bartlett in 1941. Pap. Mich. Acad. Sci. Arts Lett. 1943, 28, 143–163. [Google Scholar]
- Lin, S.-M.; Fredericq, S.; Hommersand, M.H. Systematics of the Delesseriaceae (Ceramiales, Rhodophyta) based on large subunit rDNA and rbcL sequences, including the Phycodryoideae, subfam. nov. J. Phycol. 2001, 37, 881–889. [Google Scholar] [CrossRef]
- Gavio, B.; Fredericq, S. Grateloupia turuturu (Halymeniaceae, Rhodophyta) is the correct name of the non-native species in the Atlantic known as Grateloupia doryphora. Eur. J. Phycol. 2002, 37, 349–359. [Google Scholar] [CrossRef]
- Geraldino, P.J.L.; Yang, E.C.; Boo, S.M. Morphology and molecular phylogeny of Hypnea flexicaulis (Gigartinales, Rhodophyta) from Korea. Algae 2006, 21, 417–423. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Boo, G.H.; Hughey, J.R. Phylogenomics and multigene phylogenies decipher two new cryptic marine algae from California, Gelidium gabrielsonii and G. kathyanniae (Gelidiales, Rhodophyta). J. Phycol. 2019, 55, 160–172. [Google Scholar] [CrossRef]
- Perrone, C.; Bottalico, A.; Boo, G.H.; Boo, S.M.; Miller, K.A.; Freshwater, D.W. Gelidium adriaticum sp. nov. and Gelidium carolinianum sp. nov. (Gelidiales, Rhodophyta) from the Mediterranean Sea. Phycologia 2019, 58, 359–373. [Google Scholar] [CrossRef]
- Staats, M.; Cuence, A.; Richardson, J.E.; Ginkel, R.V.; Petersen, G.; Seberg, O.; Bakker, F.T. DNA damage in plant herbarium tissue. PLoS ONE 2011, 6, e28448. [Google Scholar] [CrossRef] [PubMed]
- Hughey, J.R.; Gabrielson, P.W. Comment on “Acquiring DNA sequence data from dried archival red algae (Florideophyceae) for the purpose of applying available names to contemporary genetic species: A critical assessment”. Botany 2012, 90, 1191–1194. [Google Scholar] [CrossRef]
- Boo, G.H.; Cai, Y.; Kim, J.Y.; Boo, S.M. Phylogeny and morphology of Parviphycus myriocladus (Børgesen) comb. nov. (Gelidiales, Rhodophyta) from Asian waters. Bot. Mar. 2015, 58, 475–483. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML Version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.; Darling, A.; Hohna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Posada, D.; Crandall, K.A. MODELTEST: Testing the model of DNA substitution. Bioinformatics 1998, 14, 817–818. [Google Scholar] [CrossRef]
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian Phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef] [PubMed]
- Gernhard, T. The conditioned reconstructed process. J. Theor. Biol. 2008, 253, 769–778. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Clement, M.; Posada, D.; Crandall, K.A. TCS: A computer program to estimate gene genealogies. Mol. Ecol. 2000, 9, 1657–1659. [Google Scholar] [CrossRef]
- Múrias dos Santos, A.; Cabezas, M.P.; Tavares, A.I.; Xavier, R.; Branco, M. tcsBU: A tool to extend TCS network layout and visualization. Bioinformatics 2016, 32, 627–628. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Pons, O.; Petit, R.J. Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics 1996, 144, 1237–1245. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Res. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Schneider, S.; Excoffier, L. Estimation of past demographic parameters from the distribution of pairwise differences when the mutation rates vary among sites: Application to human mitochondrial DNA. Genetics 1999, 152, 1079–1089. [Google Scholar] [CrossRef] [PubMed]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Fu, Y.-X. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 1997, 147, 915–925. [Google Scholar] [CrossRef] [PubMed]
- Heled, J.; Drummond, A.J. Bayesian inference of species trees from multilocus data. Mol. Biol. Evol. 2009, 27, 570–580. [Google Scholar] [CrossRef]


| Population | n | S | nh | π ± SD | Hd ± SD | Tajima’s D | Fu’s FS |
|---|---|---|---|---|---|---|---|
| Australia | 5 | 5 | 3 | 0.0057 ± 0.0014 | 0.800 ± 0.164 | 1.124 | 1.220 |
| Brazil | 23 | 3 | 4 | 0.0014 ± 0.0002 | 0.597 ± 0.063 | −0.368 | −0.694 |
| Chile | 1 | NA | 1 | NA | NA | NA | NA |
| China | 6 | 5 | 2 | 0.0054 ± 0.0017 | 0.533 ± 0.172 | 1.219 | 3.696 |
| France | 16 | 0 | 1 | 0.0000 ± 0.0000 | 0.000 ± 0.000 | NA | NA |
| Hong Kong | 3 | 0 | 1 | 0.0000 ± 0.0000 | 0.000 ± 0.000 | NA | NA |
| India | 1 | NA | 1 | NA | NA | NA | NA |
| Italy | 17 | 13 | 5 | 0.0073 ± 0.0016 | 0.757 ± 0.063 | −0.239 | 2.156 |
| Japan | 1 | NA | 1 | NA | NA | NA | NA |
| Korea | 15 | 3 | 4 | 0.0022 ± 0.0005 | 0.676 ± 0.101 | 0.587 | −0.116 |
| New Zealand | 1 | NA | 1 | NA | NA | NA | NA |
| Puerto Rico | 1 | NA | 1 | NA | NA | NA | NA |
| Singapore | 1 | NA | 1 | NA | NA | NA | NA |
| Slovenia | 3 | 7 | 3 | 0.0094 ± 0.0033 | 1.000 ± 0.272 | NA | 0.308 |
| Spain | 7 | 2 | 3 | 0.0015 ± 0.0005 | 0.667 ± 0.160 | −0.275 | −0.438 |
| Tanzania | 1 | NA | 1 | NA | NA | NA | NA |
| UK | 1 | NA | 1 | NA | NA | NA | NA |
| USA | 7 | 4 | 5 | 0.0027 ± 0.0007 | 0.857 ± 0. 137 | −0.876 | −2.477 * |
| Vietnam | 6 | 0 | 1 | 0.0000 ± 0.0000 | 0.000 ± 0.000 | NA | NA |
| Overall | 116 | 50 | 33 | 0.0145 ± 0.0006 | 0.933 ± 0.012 | −0.732 | −6.520 |
| Phylogenetic groups | |||||||
| Group I | 58 | 15 | 14 | 0.0058 ± 0.0003 | 0.796 ± 0.038 | −0.345 | −3.007 |
| Group II | 3 | 0 | 1 | 0.0000 ± 0.0000 | 0.000 ± 0.000 | NA | NA |
| Group III | 32 | 12 | 9 | 0.0071 ± 0.0005 | 0.869 ± 0.031 | 0.577 | 0.021 |
| Group IV | 14 | 3 | 3 | 0.0024 ± 0.0003 | 0.604 ± 0.076 | 0.826 | 1.252 |
| Group V | 9 | 23 | 6 | 0.0133 ± 0.0035 | 0.889 ± 0.091 | −1.100 | 0.607 |
| 6 realms | |||||||
| Eastern Atlantic | 44 | 15 | 8 | 0.0087 ± 0.0010 | 0.712 ± 0.061 | 0.762 | 2.346 |
| Western Atlantic | 31 | 12 | 10 | 0.0038 ± 0.0008 | 0.774 ± 0.055 | −1.231 | −3.397 * |
| Asia | 32 | 12 | 9 | 0.0071 ± 0.0005 | 0.869 ± 0.031 | 0.577 | 0.021 |
| Australasia | 6 | 5 | 3 | 0.0050 ± 0.0014 | 0.733 ± 0.155 | 0.708 | 1.420 |
| Western Indo-Pacific | 2 | 15 | 2 | 0.0303 ± 0.0152 | 1.000 ± 0.500 | NA | 2.708 |
| Eastern Indo-Pacific | 1 | NA | 1 | NA | NA | NA | NA |
| 1 | 2 | 3 | 4 | 5 | |
|---|---|---|---|---|---|
| 1. Group I | – | ||||
| 2. Group II | 0.7515 | – | |||
| 3. Group III | 0.7020 | 0.6966 | – | ||
| 4. Group IV | 0.6854 | 0.8600 | 0.5812 | – | |
| 5. Group V | 0.6815 | 0.5769 | 0.5140 | 0.5258 | – |
| Grouping | Source of Variation | d.f. | Sum of Squares | Variance Components | Percentage of Variation | F-Statistics |
|---|---|---|---|---|---|---|
| non-hierarchical | Among populations | 18 | 353.788 | 3.3152 | 82.49 | 0.8249 |
| Within populations | 97 | 68.241 | 0.7035 | 17.51 | ||
| 5 phylogroups | Among groups | 4 | 249.520 | 2.5789 | 54.71 | 0.5471 |
| Among populations within groups | 17 | 129.851 | 1.6808 | 35.66 | 0.7874 | |
| Within populations | 94 | 42.659 | 0.4538 | 9.63 | 0.9037 | |
| 4 realms * | Among groups | 3 | 208.992 | 1.9763 | 46.15 | 0.4616 |
| Among populations within groups | 12 | 115.196 | 1.6020 | 37.41 | 0.6949 | |
| Within populations | 97 | 68.242 | 0.7035 | 16.43 | 0.8357 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boo, G.H.; Bottalico, A.; Le Gall, L.; Yoon, H.S. Genetic Diversity and Phylogeography of a Turf-Forming Cosmopolitan Marine Alga, Gelidium crinale (Gelidiales, Rhodo-Phyta). Int. J. Mol. Sci. 2023, 24, 5263. https://doi.org/10.3390/ijms24065263
Boo GH, Bottalico A, Le Gall L, Yoon HS. Genetic Diversity and Phylogeography of a Turf-Forming Cosmopolitan Marine Alga, Gelidium crinale (Gelidiales, Rhodo-Phyta). International Journal of Molecular Sciences. 2023; 24(6):5263. https://doi.org/10.3390/ijms24065263
Chicago/Turabian StyleBoo, Ga Hun, Antonella Bottalico, Line Le Gall, and Hwan Su Yoon. 2023. "Genetic Diversity and Phylogeography of a Turf-Forming Cosmopolitan Marine Alga, Gelidium crinale (Gelidiales, Rhodo-Phyta)" International Journal of Molecular Sciences 24, no. 6: 5263. https://doi.org/10.3390/ijms24065263
APA StyleBoo, G. H., Bottalico, A., Le Gall, L., & Yoon, H. S. (2023). Genetic Diversity and Phylogeography of a Turf-Forming Cosmopolitan Marine Alga, Gelidium crinale (Gelidiales, Rhodo-Phyta). International Journal of Molecular Sciences, 24(6), 5263. https://doi.org/10.3390/ijms24065263






