Inter- vs. Intra-Molecular Hydrogen Bond in Complexes of Nitrophthalic Acids with Pyridine
Abstract
1. Introduction
2. Results and Discussion
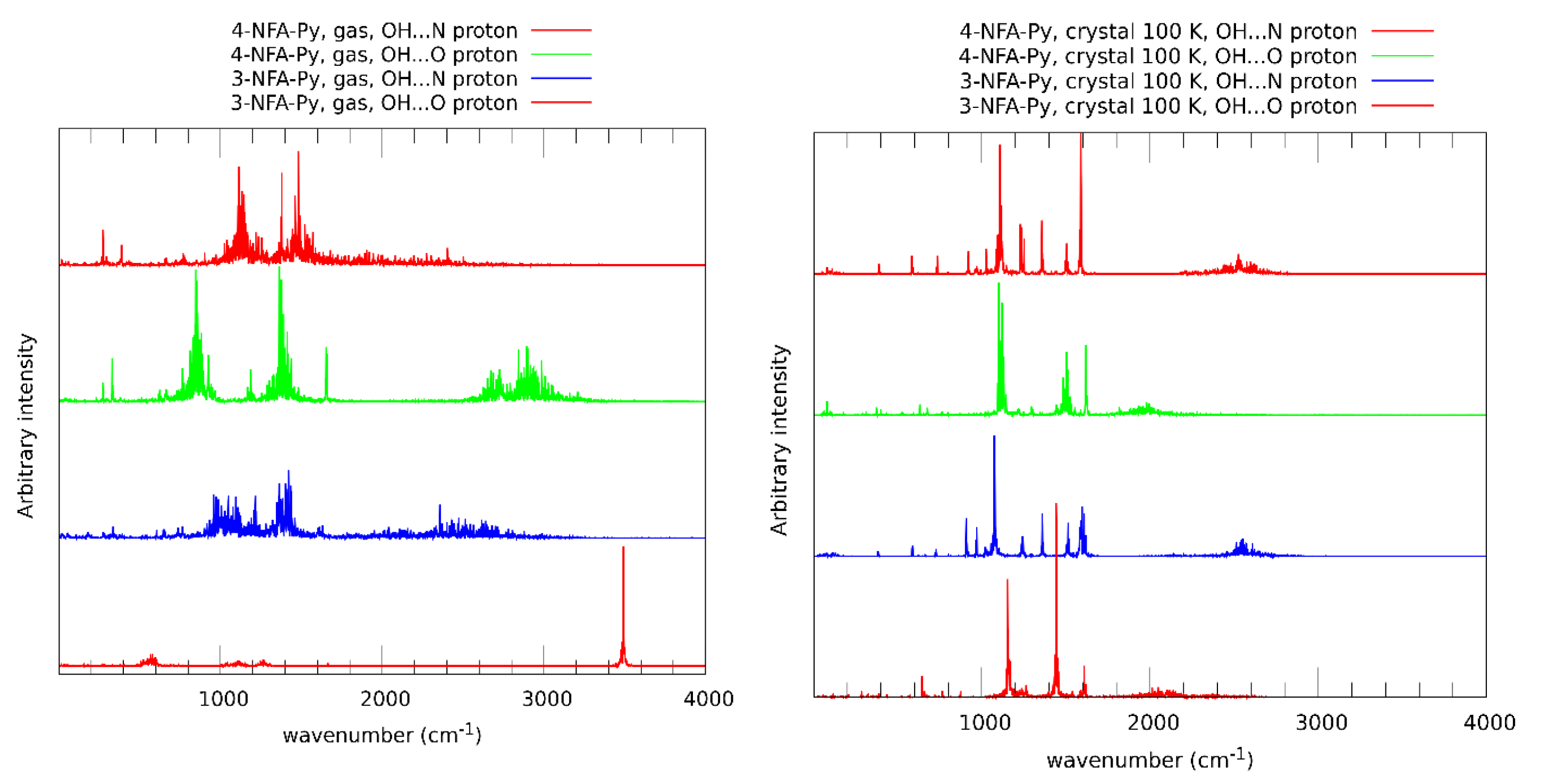
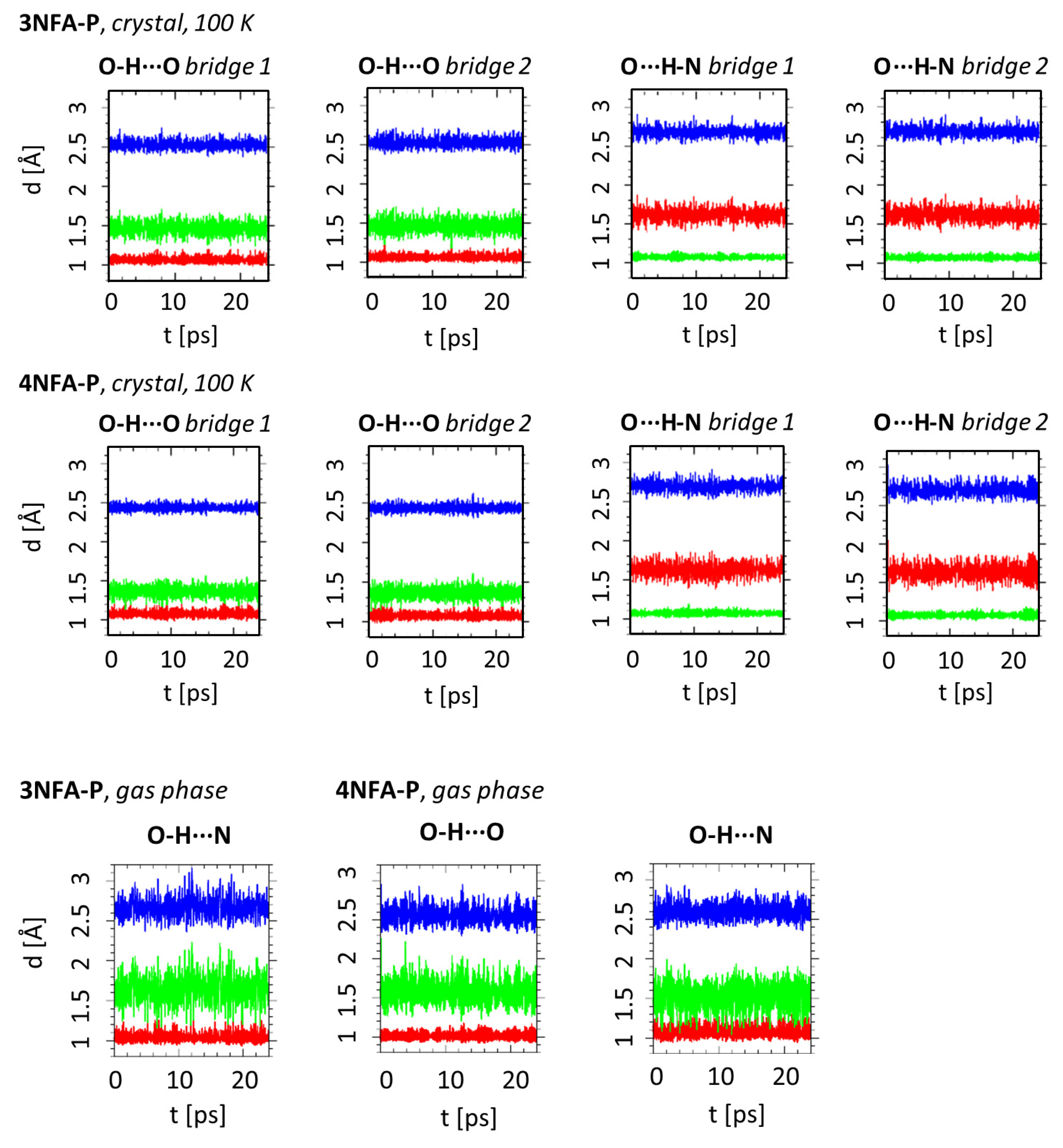
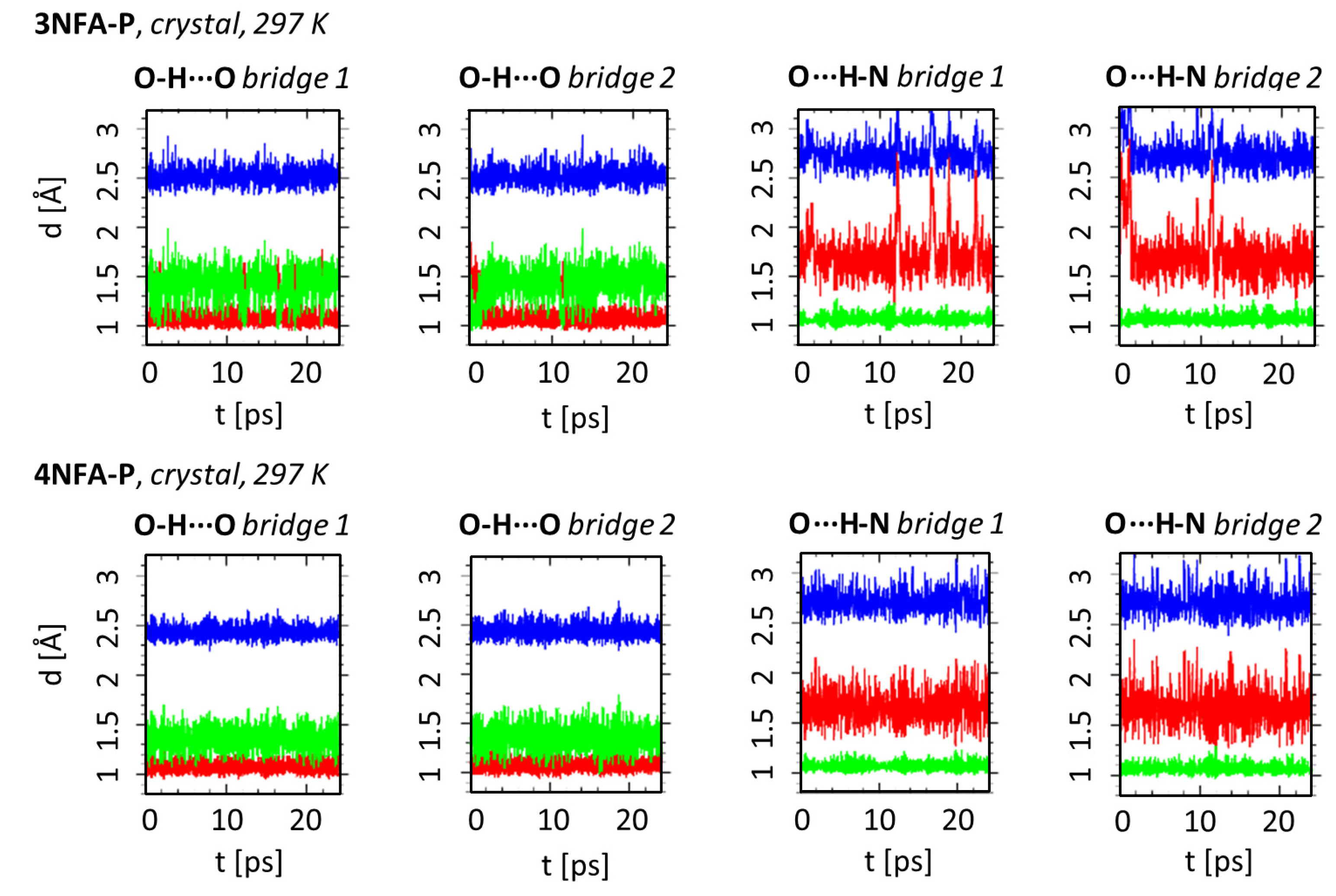
2.1. Strength of Hydrogen Bonds and Proton Dynamics in the Studied Complexes
2.2. Steric Analysis Based on X-ray Results of Studied Complexes
3. Materials and Methods
3.1. Compounds and Solvents
3.2. Single Crystal X-ray Structure Determination of Complexes
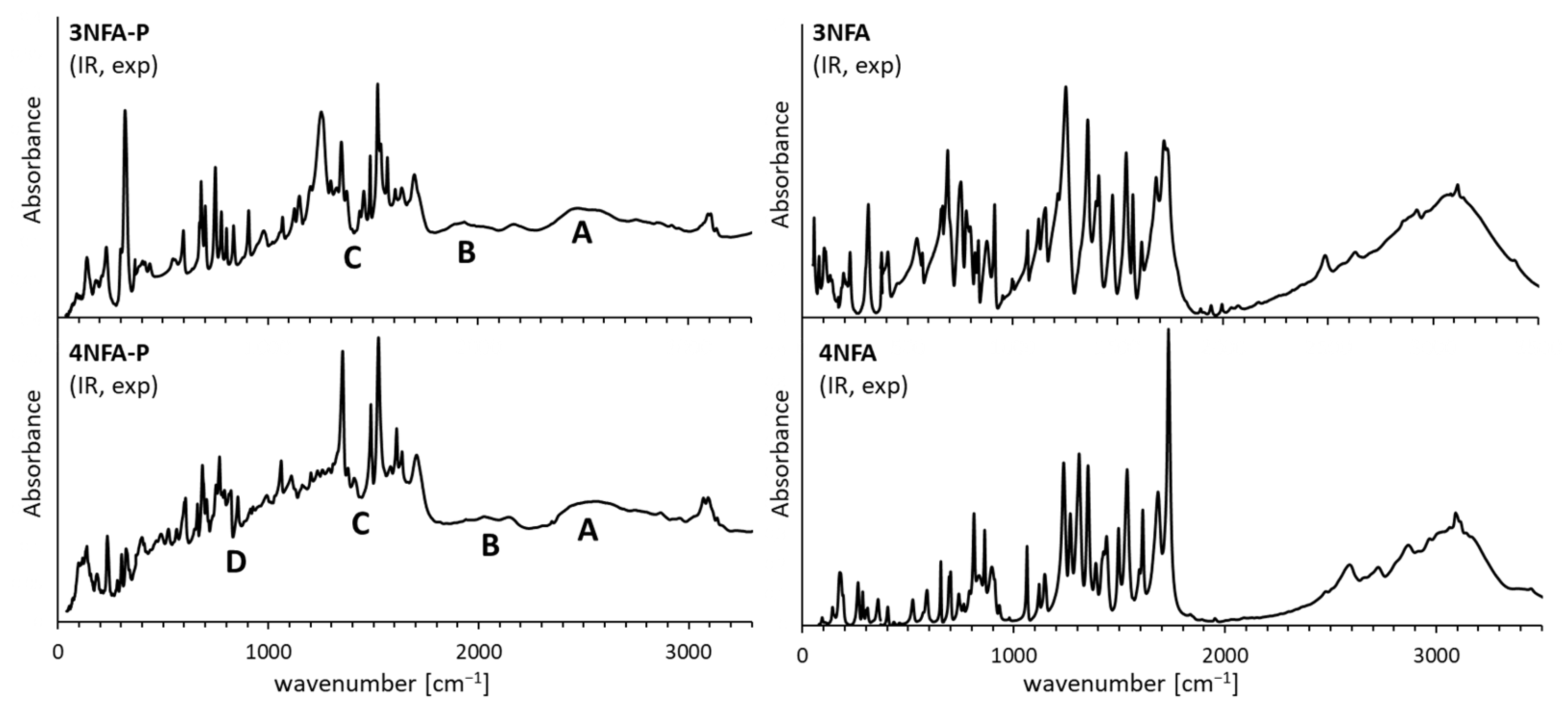
3.3. Raman and Infrared Measurements
3.4. Car-Parrinello Molecular Dynamics in the Gas and Crystalline Phases
3.5. DFT Calculations
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Crystal Data | 4NFA-P | 3NFA-P |
|---|---|---|
| Empirical formula | C13H10N2O6 | C13H10N2O6 |
| Formula weight | 290.23 | 290.23 |
| Temperature | 100(2) K | 100(2) K |
| Wavelength | 1.5418 Å | 1.5418 Å |
| Crystal system | Triclinic | Monoclinic |
| Space group | P-1(no.2) | P21/n(no.14) |
| Unit cell dimensions | a = 8.2455(3) Å, α = 87.866(3)° b = 8.5378(3) Å, β = 84.095(2)° c = 8.5726(3) Å, γ = 88.541(3)° | a = 7.5861(4) Å b = 15.0232(4) Å, β = 104.863(5)° c = 11.3482(5) Å |
| Volume | 599.74(4) Å3 | 1250.05(10) Å3 |
| Z | 2 | 4 |
| Density (calculated) | 1.607 Mg/m3 | 1.542 Mg/m3 |
| Absorption coefficient | 1.116 mm−1 | 1.070 mm−1 |
| F (000) | 300 | 600 |
| Crystal size | 0.19 × 0.18 × 0.07 mm3 | 0.20 × 0.20 × 0.16 mm3 |
| Theta range for data collection | 5.185 to 68.254° | 5.891 to 68.022° |
| Reflections collected | 3931 | 4776 |
| Independent reflections | 2155 [R(int) = 0.0673] | 2239 [R(int) = 0.0152] |
| Completeness to theta = 68.254° | 98.2% | 98.9% |
| Refinement method | Full-matrix least-squares on F2 | Full-matrix least-squares on F2 |
| Data/restraints/parameters | 2155/0/198 | 2239/0/198 |
| Goodness-of-fit on F2 | 1.033 | 1.019 |
| Final R indices [I > 2sigma(I)] | R1 = 0.0770, wR2 = 0.1902 | R1 = 0.0286, wR2 = 0.0737 |
| R indices (all data) | R1 = 0.1440, wR2 = 0.2342 | R1 = 0.0325, wR2 = 0.0767 |
| Extinction coefficient | n/a | n/a |
| Largest diff. peak and hole | 0.340 and −0.405 e. Å−3 | 0.225 and −0.256 e. Å−3 |
References
- Buemi, G.; Zuccarello, F. Importance of steric effect on the hydrogen bond strength of malondialdehyde and acetylacetone 3-substituted derivatives. An ab initio study. J. Theor. Chem. 1997, 2, 302–314. [Google Scholar]
- Pozharskii, A.F.; Ryabtsova, O.V.; Ozeryanskii, V.A.; Degtyarev, A.V.; Kazheva, O.N.; Alexandrov, G.G.; Dyachenko, O.A. Organometallic Synthesis, Molecular Structure, and Coloration of 2,7-Disubstituted 1,8-Bis(dimethylamino)naphthalenes. How Significant Is the Influence of “Buttressing Effect” on Their Basicity? J. Org. Chem. 2003, 68, 10109–10122. [Google Scholar] [CrossRef] [PubMed]
- Filatova, E.A.; Gulevskaya, A.V.; Pozharskii, A.F.; Ermolenko, E.A.; Ozeryanskii, V.A.; Misharev, A.D. Synthesis of 2-Aryl- and 2,7-Diaryl-1,8-bis(dimethylamino)naphthalenes. Overview of the “Buttressing effect” in 2,7-Disubstituted Proton Sponges. ChemistrySelect 2020, 5, 9932–9945. [Google Scholar] [CrossRef]
- Ozeryanskii, V.A.; Marchenko, A.V.; Pozharskii, A.F.; Filarowski, A.; Spiridonova, D.V. Combination of “buttressing” and “clothespin” effects for reaching the shortest NHN hydrogen bond in proton sponge cations. J. Org. Chem. 2021, 86, 3637–3647. [Google Scholar] [CrossRef] [PubMed]
- Hansen, P.E.; Ibsen, S.N.; Kristensen, T.; Bolvig, S. Deuterium and 18O isotope effects on 13C chemical shifts of sterically hindered and/or intra-molecularly hydrogen-bonded o-hydroxy acyl aromatics. Magn. Res. Chem. 1994, 32, 399–408. [Google Scholar] [CrossRef]
- Bolvig, S.; Wozniak, K.; Hansen, P.E. Steric compression effects of intramolecularly hydrogen bonded o-hydroxy acyl aromatics. An X-ray and 13C-NMR study. J. Mol. Struct. 2005, 749, 155–168. [Google Scholar]
- Hansen, P.E.; Spanget-Larsen, J. NMR and IR Investigations of Strong Intramolecular Hydrogen Bonds. Molecules 2017, 22, 552. [Google Scholar] [CrossRef]
- Kwocz, A.; Panek, J.J.; Jezierska, A.; Hetmańczyk, Ł.; Pawlukojć, A.; Kochel, A.; Lipkowski, P.; Filarowski, A. A molecular roundabout: Triple cycle-arranged hydrogen bonds in light of experiment and theory. New J. Chem. 2018, 42, 19467–19477. [Google Scholar] [CrossRef]
- Martyniak, A.; Panek, J.J.; Jezierska-Mazzarello, A.; Filarowski, A. Triple hydrogen bonding in a circular arrangement: Ab initio, DFT and first-principles MD studies of tris-hydroxyaryl enamines. J. Comp.-Aided Mol. Des. 2012, 9, 1045–1053. [Google Scholar] [CrossRef]
- Filarowski, A.; Koll, A.; Kochel, A.; Kalenik, J.; Hansen, P.E. The intramolecular hydrogen bond in ortho-hydroxy acetophenones. J. Mol. Struct. 2004, 700, 67–72. [Google Scholar] [CrossRef]
- Sobczyk, L.; Chudoba, D.M.; Tolstoy, T.M.; Filarowski, A. Some Brief Notes on Theoretical and Experimental Investigations of Intramolecular Hydrogen Bonding. Molecules 2016, 21, 1657. [Google Scholar] [CrossRef]
- Hussain, S.G.M.; Kumar, R.; Ali, M.M.N.; Kannappan, V. Steric effect in the formation of hydrogen bonded complexes of isopropylamine with alicyclic ethers by ultrasonic and DFT approach. J. Mol. Liq. 2020, 317, 2–5. [Google Scholar] [CrossRef]
- Liu, H.; Yang, X.; Cao, S.; Yu, F.; Long, S.; Chen, J.; Zhang, M.; Parkin, S.; Li, T.; Yang, Z. Steric Effect Determines the Formation of Lactam−Lactam Dimers or Amide C=O···NH (Lactam) Chain Motifs in N-Phenyl-2-Hydroxynicotinanilides. Cryst. Growth Des. 2020, 20, 4346–4357. [Google Scholar] [CrossRef]
- Huelsekopf, M.; Ludwig, R. Hydrogen Bonding in a sterically hindered alcohol. J. Mol. Liq. 2002, 98–99, 163–171. [Google Scholar] [CrossRef]
- Nowok, A.; Cieślik, W.; Dulski, M.; Jurkiewicz, K.; Grelska, J.; Alemán, J.; Musioł, R.; Szeremeta, A.Z.; Pawlus, S. Glass-forming Schiff bases: Peculiar self-organizing systems with bifurcated hydrogen bonds. J. Mol. Liq. 2022, 348, 118052. [Google Scholar] [CrossRef]
- Jezierska, A.; Tolstoy, P.M.; Panek, J.J.; Filarowski, A. Intramolecular Hydrogen Bonds in Selected Aromatic Compounds: Recent Developments. Catalysts 2019, 9, 909. [Google Scholar] [CrossRef]
- Majewska, P.; Pająk, J.; Rospenk, M.; Filarowski, A. Intra- versus intermolecular hydrogen bonding equilibrium in 2-hydroxy-N,N-diethylbenzamide. J. Phys. Org. Chem. 2009, 22, 130–137. [Google Scholar] [CrossRef]
- Kozlecki, T.; Tolstoy, P.M.; Kwocz, A.; Vovk, M.A.; Kochel, A.; Polowczyk, I.; Tretyakov, P.Y.; Filarowski, A. Conformational state of β-hydroxynaphthylamides: Barriers for the rotation of the amide group around CN bond and dynamics of the morpholine ring. Spectrochim. Acta A 2015, 149, 254–262. [Google Scholar] [CrossRef]
- Kawski, P.; Kochel, A.; Perevozkina, M.G.; Filarowski, A. The intramolecular hydrogen bond in 2-hydroxy-benzamides. J. Mol. Struct. 2006, 790, 65–73. [Google Scholar] [CrossRef]
- Szostak, M. (Ed.) Amide Bond Activation Molecules; MDPI: Basel, Switzerland; Beijing, China; Wuhan, China; Barcelona, Spain; Belgrade, Serbia, 2019. [Google Scholar]
- Ośmiałowski, B.; Kolehmainen, E.; Kauppinen, R.; Kowalska, M. Tuning the hydrogen-bonding strength in 2,6-bis(cycloalkylcarbonylamino)pyridine assemblies by variable flexibility. Association constants measured by hydrogen-bonded vs. non-hydrogen-bonded protons. Supramol. Chem. 2011, 23, 579–586. [Google Scholar] [CrossRef]
- Ośmiałowski, B. Conformational equilibrium and substituent effects in hydrogen-bonded complexes. Curr. Org. Chem. 2018, 22, 2182–2199. [Google Scholar] [CrossRef]
- Kwiatkowski, A.; Jędrzejewska, B.; Józefowicz, M.; Grela, I.; Ośmiałowski, B. The trans/cis photoisomerization in hydrogen bonded complexes with stability controlled by substituent effects: 3-(6-aminopyridin-3-yl) acrylate case study. RSC Adv. 2018, 42, 23698–23710. [Google Scholar] [CrossRef] [PubMed]
- Kolehmainen, E.; Ośmiałowski, B.; Nissinen, M.; Kauppinen, R.; Gawinecki, R. Substituent and temperature controlled tautomerism of 2-phenacylpyridine: The hydrogen bond as a configurational lock of (Z)-2-(2-hydroxy-2-phenylvinyl)pyridine. J. Chem. Soc. Perkin Trans. 2 2000, 11, 2185–2191. [Google Scholar] [CrossRef]
- Ośmiałowski, B.; Kolehmainen, E.; Dobosz, R.; Gawinecki, R.; Kauppinen, R.; Valkonen, A.; Koivukorpi, J.; Rissanen, K. Self-Organization of 2-Acylaminopyridines in the Solid State and in Solution. J. Phys. Chem. A 2010, 114, 10421–10426. [Google Scholar] [CrossRef]
- Ośmiałowski, B.; Kolehmainen, E.; Gawinecki, R.; Dobosz, R.; Kauppinen, R. Complexation of 2,6-Bis(acylamino)pyridines with Dipyridin-2-Ylamine and 4,4-Dimethylpiperidine-2,6-Dione. J. Phys. Chem. A 2010, 114, 12881–12887. [Google Scholar] [CrossRef]
- Glidewell, C.; Low, J.N.; Skakle, J.M.S.; Wardell, J.L. 3-Nitrophthalic acid: C(4) and R22(8) motifs of O-H…O hydrogen bonds generate sheets which are linked by C-H…O hydrogen bonds. Acta Cryst. 2003, C59, o144–o146. [Google Scholar]
- Smith, G.; Wermuth, U.D.; Young, D.J.; White, J.M. The 1:1 proton-transfer compounds of 4-(phenyldiazenyl)aniline (aniline yellow) with 3-nitrophthalic, 4-nitrophthalic and 5-nitroisophthalic acids. Acta Cryst. 2008, C64, o123–o127. [Google Scholar] [CrossRef]
- Smith, G.; Wermuth, U.D. Proton-transfer compounds of isonipecotamide with the aromatic dicarboxylic acids 4-nitrophthalic, 4,5-dichlorophthalic, 5-nitroisophthalic and terephthalic acid. Acta Cryst. 2011, C67, o259–o264. [Google Scholar] [CrossRef]
- Jin, S.; Wang, D.; Du, S.; Linhe, Q.; Fu, M.; Wu, S. Crystal and Molecular Structure of Two Proton Transfer Compounds from Quinolin-8-ol, 4-nitro-phthalic Acid, and 1,5-Naphthalenedisulfonic Acid. J. Chem. Crystallogr. 2014, 44, 435–441. [Google Scholar] [CrossRef]
- Saunders, L.K.; Nowell, H.; Hatcher, L.E.; Shepherd, H.J.; Teat, S.J.; Allan, D.R.; Raithby, P.R.; Wilson, C.C. Exploring short strong hydrogen bonds engineered in organic acid molecular crystals for temperature dependent proton migration behaviour using single crystal synchrotron X-ray diffraction (SCSXRD). CrystEngComm 2019, 21, 5249–5260. [Google Scholar] [CrossRef]
- Surov, A.O.; Vasilev, N.A.; Churakov, A.V.; Parashchuk, O.D.; Artobolevskii, S.V.; Alatortsev, O.A.; Makhrov, D.E.; Vener, M.V. Two Faces of Water in the Formation and Stabilization of Multicomponent Crystals of Zwitterionic Drug-Like Compounds. Symmetry 2021, 13, 425. [Google Scholar] [CrossRef]
- Jóźwiak, K.; Jezierska, A.; Panek, J.J.; Goremychkin, E.A.; Tolstoy, P.M.; Shenderovich, I.G.; Filarowski, A. Inter- vs. intra-molecular hydrogen bond patterns and proton dynamics in phthalic acid associates. Molecules 2020, 25, 4720. [Google Scholar] [CrossRef]
- Zhoujin, Y.; Yang, X.; Zhang, M.; Guo, J.; Parkin, S.; Li, T.; Yu, F.; Long, S. Synthon Polymorphism and π–π Stacking in N-Phenyl-2-hydroxynicotinanilides. Cryst. Growth Des. 2021, 21, 6155–6165. [Google Scholar] [CrossRef]
- Yadav, V.K. Steric and Stereoelectronic Effects in Organic Chemistry; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Kool, E.T. Hydrogen Bonding, Base Stacking, and Steric Effects in DNA Replication. Ann. Rev. Biophys. Biomol. Struct. 2001, 30, 1–22. [Google Scholar] [CrossRef]
- Poater, J.; Swart, M.; Fonseca Guerra, C.F.; Bickelhaupt, F.M. Selectivity in DNA replication. Interplay of steric shape, hydrogen bonds, p-stacking and solvent effects. Chem. Commun. 2011, 47, 7326–7328. [Google Scholar] [CrossRef]
- Hansen, T.; Sun, X.; Tiezza, M.D.; van Zeist, W.-J.; van Stralen, J.N.P.; Geerke, D.P.; Wolters, L.P.; Poater, J.; Hamlin, T.A.; Bickelhaupt, F.M. C-X Bond Activation by Palladium: Steric Shielding versus Steric Attraction. Chem. Eur. J. 2022, 28, e202201093. [Google Scholar]
- El-Hamdi, M.; Tiznado, W.; Poater, J.; Solà, M. An Analysis of the Isomerization Energies of 1,2-/1,3-Diazacyclobutadiene, Pyrazole/Imidazole, and Pyridazine/Pyrimidine with the Turn-Upside-Down Approach. J. Org. Chem. 2011, 76, 8913–8921. [Google Scholar] [CrossRef]
- Zundel, G. The Hydrogen Bond: Recent Developments in Theory and Experiments; Schuster, P., Zundel, G., Sandorfy, C., Eds.; North-Holland Publishing Company: Amsterdam, The Netherlands, 1976; Volume 2, pp. 683–766. [Google Scholar]
- Cleland, W.W.; Kreevoy, M.M. Low-barrier hydrogen-bonds and enzymatic catalysis. Science 1994, 264, 1887–1890. [Google Scholar] [CrossRef]
- Iogansen, A.V. Direct proportionality of the hydrogen bonding energy and the intensification of the stretching v(XH) vibration in infrared spectra. Spectr. Acta A 1999, 55, 1585–1612. [Google Scholar] [CrossRef]
- Sobczyk, L.; Lis, T.; Olejnik, Z.; Majerz, I. The OHN hydrogen bonding in the adduct of 2,4-dinitrobenzoic acid with pyridine. Low temperature X-ray diffraction and IR spectroscopic studies. J. Mol. Struct. 2000, 552, 233–241. [Google Scholar] [CrossRef]
- Steiner, T. The Hydrogen Bond in the Solid State. Angew. Chem. Int. Ed. 2002, 41, 48–76. [Google Scholar] [CrossRef]
- Jeffrey, G.A. An Introduction to Hydrogen Bonding; Oxford University Press: Oxford, UK, 1997. [Google Scholar]
- Latajka, Z.; Sobczyk, L. The Potential Energy Shape for the Proton Motion in Protonated Naphthalene Proton Sponges (DMAN-s) and Its Manifestations, in Practical Aspects of Computational Chemistry—Methods, Concepts and Applications; Leszczynski, J., Shukla, M.K., Eds.; Springer: Berlin/Heidelberg, Germany; Dordrecht, The Netherlands; London, UK; New York, NY, USA, 2010; pp. 371–386. [Google Scholar]
- Cai, W.; Katrusiak, A. Pressure effects on H-ordering in hydrogen bonds and interactions in benzoic acid. CrystEngComm 2012, 14, 4420–4424. [Google Scholar] [CrossRef]
- Wilson, C.C.; Shankland, N.; Florence, A.J. A single-crystal neutron diffraction study of the temperature dependence of hydrogen-atom disorder in benzoic acid dimers. J. Chem. Soc. Faraday Trans. 1996, 92, 5051–5057. [Google Scholar] [CrossRef]
- Rikagu Oxford Diffraction. CrysAlisPro; Agilent Technologies Inc.: Yarnton, Oxfordshire, UK, 2018. [Google Scholar]
- Sheldrick, G.M. Short History of SHELX. Acta Cryst. 2008, A64, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Sheldrick, G.M. Crystal Structure Refinement with SHELXL. Acta Cryst. 2015, C71, 3–8. [Google Scholar]
- Diamond—Crystal and Molecular Structure Visualization, Crystal Impact—Dr. H. Putz & Dr. K. Brandenburg GbR, Germany.
- CPMD 4.3, Copyright IBM Corp. (1990–2019) Copyright MPI für Festkoerperforschung Stuttgart (1997–2001). Available online: http://www.cpmd.org (accessed on 12 September 2022).
- Hohenberg, P.; Kohn, W. Inhomogeneous Electron Gas. Phys. Rev. 1964, 136, B864–B871. [Google Scholar] [CrossRef]
- Kohn, W.; Sham, L.J. Self-Consistent Equations Including Exchange and Correlation Effects. Phys. Rev. 1965, 140, A1133–A1138. [Google Scholar] [CrossRef]
- Hockney, R.W. The potential calculation and some applications. Meth. Comput. Phys. 1970, 9, 135–211. [Google Scholar]
- Perdew, J.P.; Ernzerhof, M.; Burke, K. Rationale for mixing exact exchange with density functional approximations. J. Chem. Phys. 1996, 105, 9982–9985. [Google Scholar] [CrossRef]
- Troullier, N.; Martins, J.L. Efficient pseudopotentials for plane-wave calculations. Phys. Rev. B 1991, 43, 1993–2006. [Google Scholar] [CrossRef]
- Nosé, S. A unified formulation of the constant temperature molecular dynamics methods. J. Chem. Phys. 1984, 81, 511–519. [Google Scholar] [CrossRef]
- Hoover, W.G. Canonical dynamics: Equilibrium phase-space distributions. Phys. Rev. A 1985, 31, 1695–1697. [Google Scholar] [CrossRef]
- Grimme, S. Semiempirical {GGA}-type density functional constructed with a long-range dispersion correction. J. Comput. Chem. 2006, 27, 1787–1799. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Mercury—Crystal Structure Visualisation. Available online: http://www.ccdc.cam.ac.uk/Solutions/CSDSystem/Pages/Mercury.aspx (accessed on 15 September 2022).
- Williams, T.; Kelley, C.; Lang, R.; Kotz, D.; Campbell, J.; Elber, G.; Woo, A. Gnuplot 4.4: An Interactive Plotting Program. 2010. Available online: http://gnuplot.sourceforge.net/ (accessed on 12 September 2022.).
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16, Revision C.01; Gaussian, Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Frisch, M.J.; Pople, J.A.; Binkley, J.S. Self-consistent molecular orbital methods 25. Supplementary functions for Gaussian basis sets. J. Chem. Phys. 1984, 80, 3265–3269. [Google Scholar] [CrossRef]
- Becke, A.D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993, 98, 5648–5652. [Google Scholar] [CrossRef]
- Lee, C.; Yang, W.; Parr, R.G. Development of the Colle-Salvetti Correlation-Energy Formula into a Functional of the Electron Density. Phys. Rev. 1993, 37, B785–B789. [Google Scholar] [CrossRef]
- Schaftenaar, G.; Noordik, J.H. Molden: A pre- and post-processing program for molecular and electronic structures. J. Comput.-Aided Mol. Des. 2000, 14, 123–134. [Google Scholar] [CrossRef]




| State, T | Type HB | D-H | H⋅⋅⋅A | D⋅⋅⋅A | D-H⋅⋅⋅A | |
|---|---|---|---|---|---|---|
| complex | X-ray measurements | |||||
| 3NFA-P | solid, 100 K | inter O(1)−H(1O)⋅⋅⋅O4i (i = 1/2 + x, 3/2 − y, 1/2 + z) | 0.96(2) | 1.57(2) | 2.526(1) | 176(2) |
| - | inter N(2)−H(2N)⋅⋅⋅O(4) | 0.93(2) | 1.74(2) | 2.659(1) | 173(2) | |
| 4NFA-P | - | intra O(1)−H(1O)⋅⋅⋅O(3) | 1.33(5) | 1.12(5) | 2.425(4) | 164(5) |
| - | inter N(2)−H(2N)⋅⋅⋅O(2) | 0.85(5) | 1.85(5) | 2.699(5) | 176(3) | |
| CPMD simulations | ||||||
| 3NFA-P | gas | inter O−H⋅⋅⋅N | 1.0504 | 1.6515 | 2.6725 | |
| solid, 297 K | inter O−H⋅⋅⋅O | 1.1138 | 1.4208 | 2.5248 | ||
| inter N−H⋅⋅⋅O | 1.7278 | 1.0679 | 2.7409 | |||
| solid, 100 K | inter O−H⋅⋅⋅O | 1.0666 | 1.4649 | 2.5277 | ||
| inter N−H⋅⋅⋅O | 1.6203 | 1.0716 | 2.6823 | |||
| 4NFA-P | gas | intra O−H⋅⋅⋅O | 1.0177 | 1.5933 | 2.5680 | |
| inter N−H⋅⋅⋅O | 1.0962 | 1.5244 | 2.6065 | |||
| solid, 297 K | intra O−H⋅⋅⋅O | 1.0882 | 1.3623 | 2.4392 | ||
| inter N−H⋅⋅⋅O | 1.6717 | 1.0717 | 2.7176 | |||
| solid, 100 K | intra O−H⋅⋅⋅O | 1.0776 | 1.3651 | 2.4352 | ||
| inter N−H⋅⋅⋅O | 1.6301 | 1.0737 | 2.6941 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jóźwiak, K.; Jezierska, A.; Panek, J.J.; Kochel, A.; Filarowski, A. Inter- vs. Intra-Molecular Hydrogen Bond in Complexes of Nitrophthalic Acids with Pyridine. Int. J. Mol. Sci. 2023, 24, 5248. https://doi.org/10.3390/ijms24065248
Jóźwiak K, Jezierska A, Panek JJ, Kochel A, Filarowski A. Inter- vs. Intra-Molecular Hydrogen Bond in Complexes of Nitrophthalic Acids with Pyridine. International Journal of Molecular Sciences. 2023; 24(6):5248. https://doi.org/10.3390/ijms24065248
Chicago/Turabian StyleJóźwiak, Kinga, Aneta Jezierska, Jarosław J. Panek, Andrzej Kochel, and Aleksander Filarowski. 2023. "Inter- vs. Intra-Molecular Hydrogen Bond in Complexes of Nitrophthalic Acids with Pyridine" International Journal of Molecular Sciences 24, no. 6: 5248. https://doi.org/10.3390/ijms24065248
APA StyleJóźwiak, K., Jezierska, A., Panek, J. J., Kochel, A., & Filarowski, A. (2023). Inter- vs. Intra-Molecular Hydrogen Bond in Complexes of Nitrophthalic Acids with Pyridine. International Journal of Molecular Sciences, 24(6), 5248. https://doi.org/10.3390/ijms24065248








