Function and Evolution of the Loop Extrusion Machinery in Animals
Abstract
1. The Origin and Evolution of the Loop Extrusion Machinery
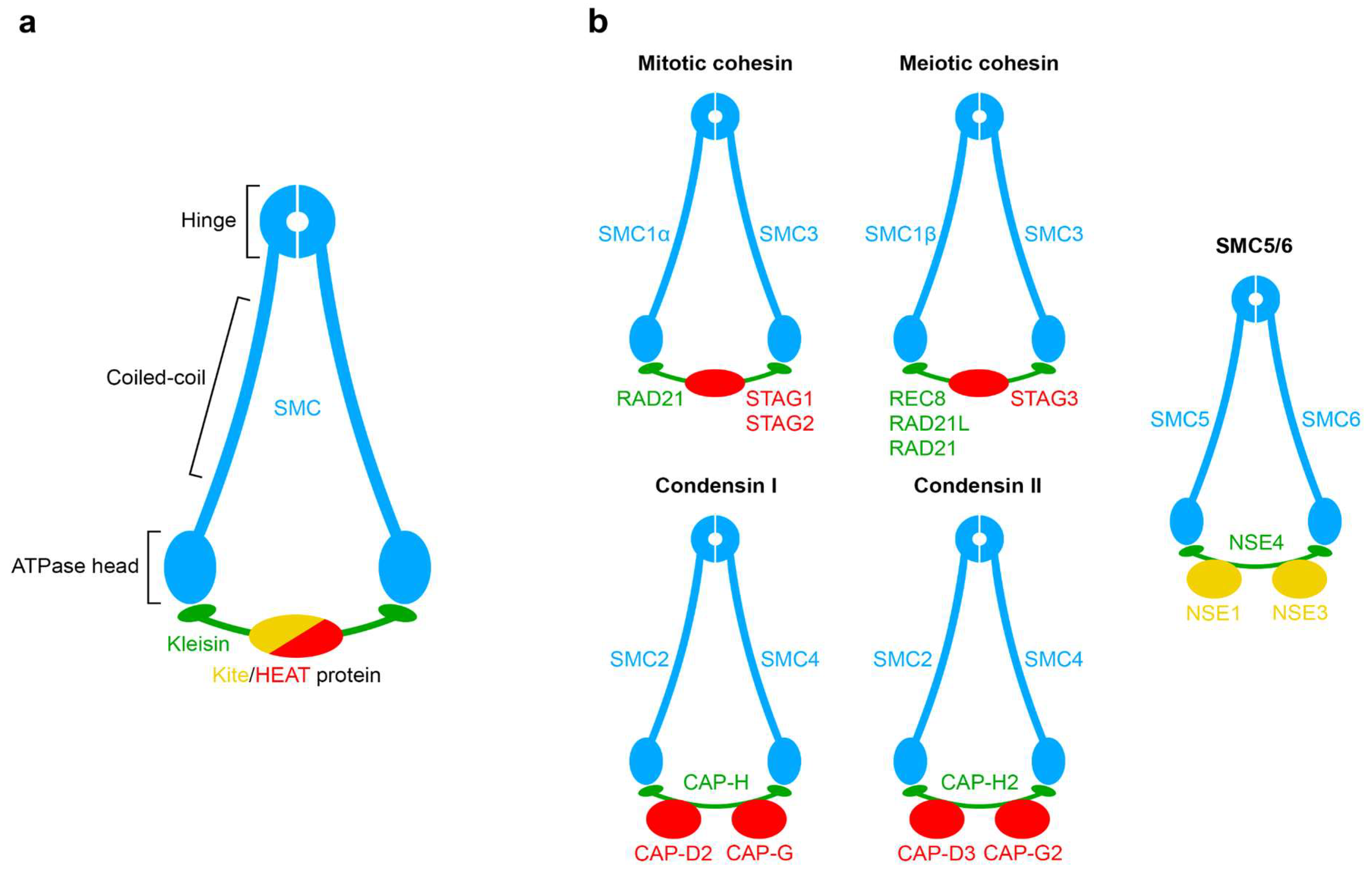
2. Structural Details of Interactions between SMC Complexes and DNA
3. The Mechanism of DNA Translocation by SMC Complexes
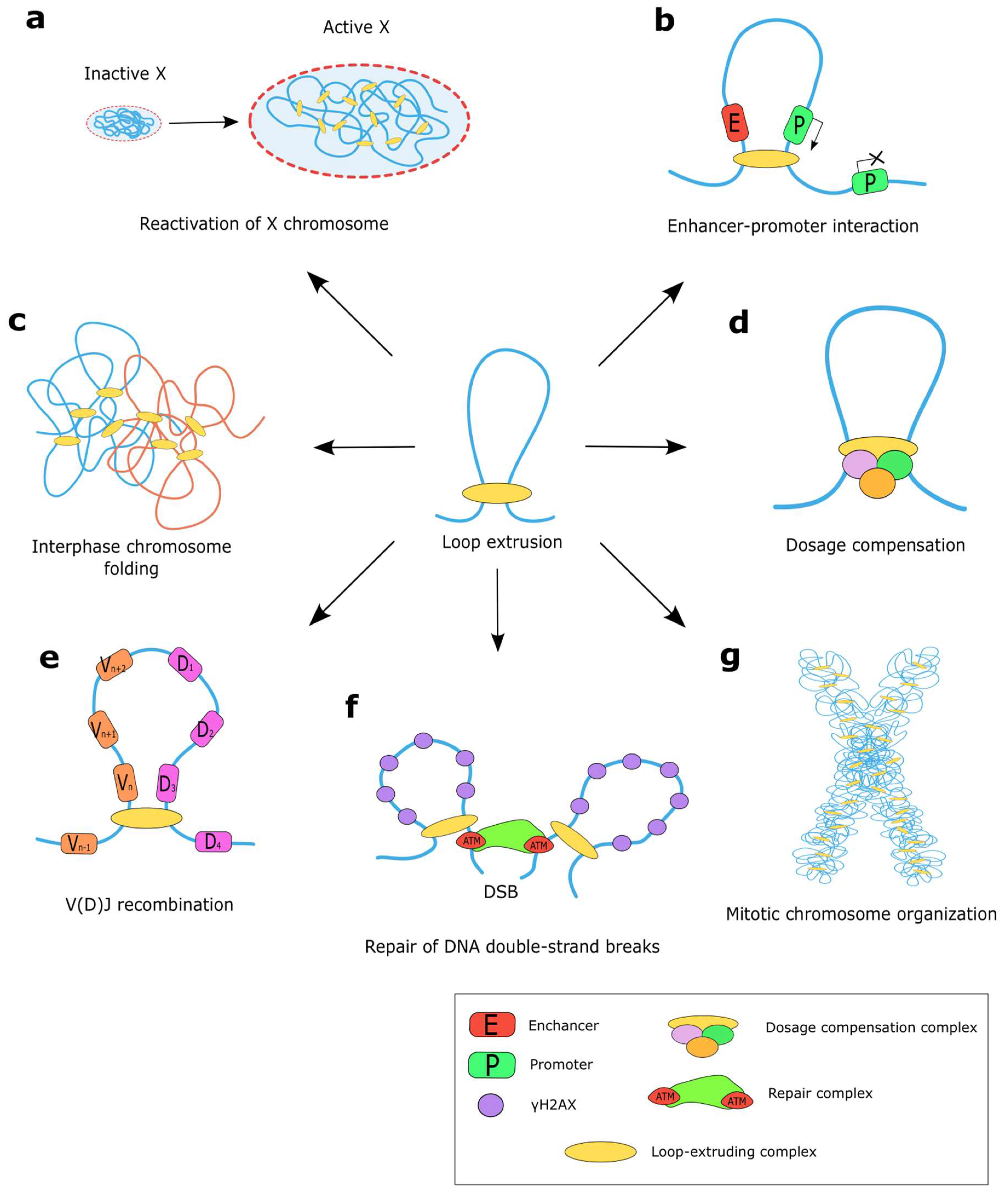
4. Functions of the Loop Extrusion Machinery
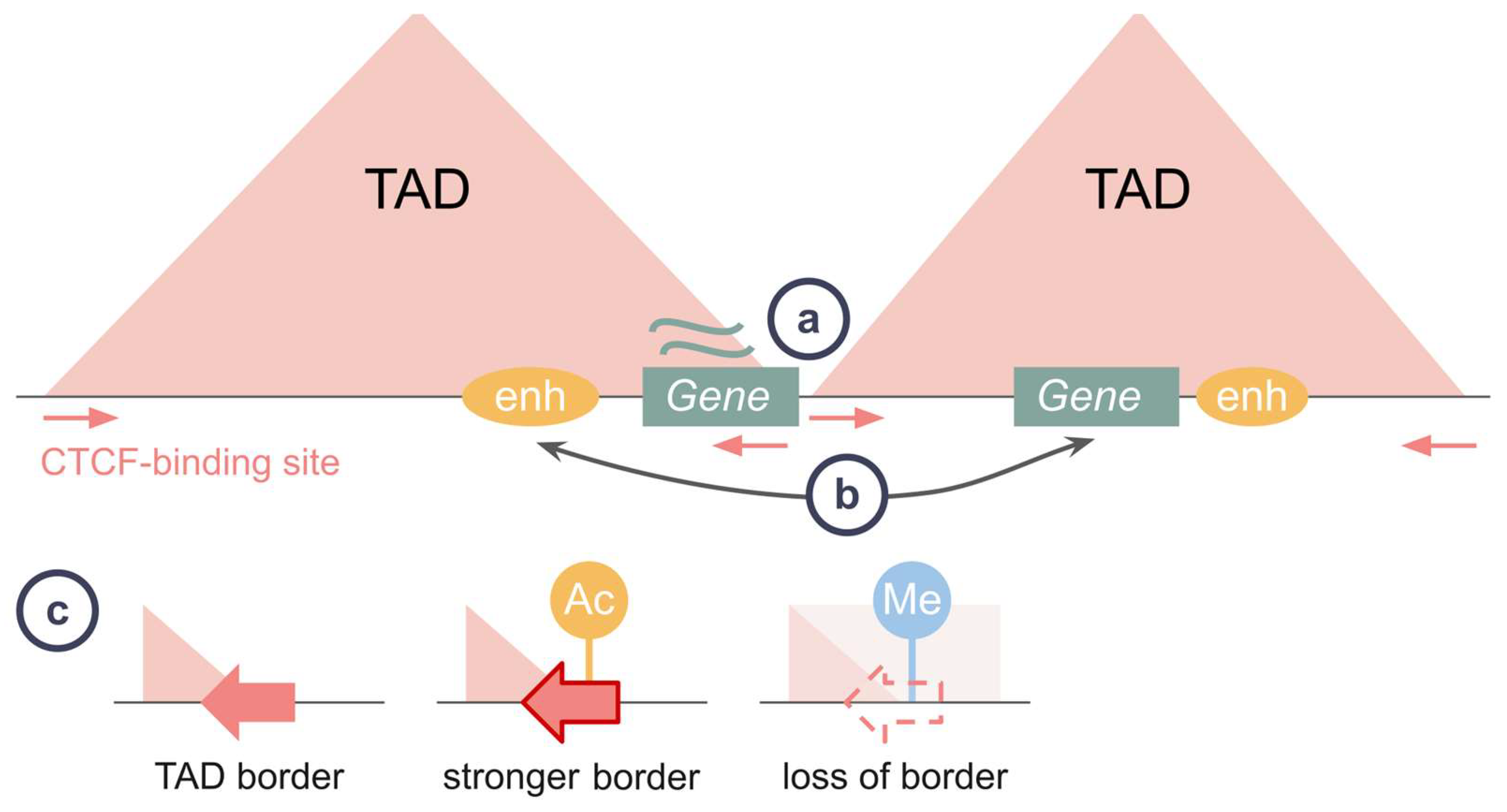
4.1. Loop Extrusion for Gene Regulation
4.2. Co-Evolution of CTCF and Cohesin Proteins in Bilateria
4.3. Loop Extrusion for DNA Repair
4.4. Loop Extrusion for Chromatin Topology
4.4.1. Formation of Mitotic Chromosomes by Loop Extrusion
4.4.2. Shaping Interphase Chromosomes by Loop Extrusion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cobbe, N.; Heck, M.M.S. The Evolution of SMC Proteins: Phylogenetic Analysis and Structural Implications. Mol. Biol. Evol. 2004, 21, 332–347. [Google Scholar] [CrossRef]
- Löwe, J.; Cordell, S.C.; van den Ent, F. Crystal Structure of the SMC Head Domain: An ABC ATPase with 900 Residues Antiparallel Coiled-Coil Inserted. J. Mol. Biol. 2001, 306, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Hirano, M.; Hirano, T. Hinge-Mediated Dimerization of SMC Protein Is Essential for Its Dynamic Interaction with DNA. EMBO J. 2002, 21, 5733–5744. [Google Scholar] [CrossRef] [PubMed]
- Kamada, K.; Miyata, M.; Hirano, T. Molecular Basis of SMC ATPase Activation: Role of Internal Structural Changes of the Regulatory Subcomplex ScpAB. Structure 2013, 21, 581–594. [Google Scholar] [CrossRef]
- Rybenkov, V.V.; Herrera, V.; Petrushenko, Z.M.; Zhao, H. MukBEF, a Chromosomal Organizer. J. Mol. Microbiol. Biotechnol. 2014, 24, 371–383. [Google Scholar] [CrossRef] [PubMed]
- Nolivos, S.; Sherratt, D. The Bacterial Chromosome: Architecture and Action of Bacterial SMC and SMC-like Complexes. FEMS Microbiol. Rev. 2014, 38, 380–392. [Google Scholar] [CrossRef]
- Gogou, C.; Japaridze, A.; Dekker, C. Mechanisms for Chromosome Segregation in Bacteria. Front. Microbiol. 2021, 12, 685687. [Google Scholar] [CrossRef]
- Yoshinaga, M.; Inagaki, Y. Ubiquity and Origins of Structural Maintenance of Chromosomes (SMC) Proteins in Eukaryotes. Genom. Biol. Evol. 2021, 13, evab256. [Google Scholar] [CrossRef]
- Schleiffer, A.; Kaitna, S.; Maurer-Stroh, S.; Glotzer, M.; Nasmyth, K.; Eisenhaber, F. Kleisins: A Superfamily of Bacterial and Eukaryotic SMC Protein Partners. Mol. Cell 2003, 11, 571–575. [Google Scholar] [CrossRef]
- Horsfield, J.A. Full Circle: A Brief History of Cohesin and the Regulation of Gene Expression. FEBS J. 2022. [CrossRef]
- Cheng, H.; Zhang, N.; Pati, D. Cohesin Subunit RAD21: From Biology to Disease. Gene 2020, 758, 144966. [Google Scholar] [CrossRef] [PubMed]
- Sakuno, T.; Hiraoka, Y. Rec8 Cohesin: A Structural Platform for Shaping the Meiotic Chromosomes. Genes 2022, 13, 200. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Hirano, T. RAD21L, a Novel Cohesin Subunit Implicated in Linking Homologous Chromosomes in Mammalian Meiosis. J. Cell Biol. 2011, 192, 263–276. [Google Scholar] [CrossRef]
- Gutiérrez-Caballero, C.; Herrán, Y.; Sánchez-Martín, M.; Suja, J.Á.; Barbero, J.L.; Llano, E.; Pendás, A.M. Identification and Molecular Characterization of the Mammalian α-Kleisin RAD21L. Cell Cycle 2011, 10, 1477–1487. [Google Scholar] [CrossRef] [PubMed]
- Hirano, T. Condensins: Universal Organizers of Chromosomes with Diverse Functions. Genes Dev. 2012, 26, 1659–1678. [Google Scholar] [CrossRef]
- Serrano, D.; Cordero, G.; Kawamura, R.; Sverzhinsky, A.; Sarker, M.; Roy, S.; Malo, C.; Pascal, J.M.; Marko, J.F.; D’Amours, D. The Smc5/6 Core Complex Is a Structure-Specific DNA Binding and Compacting Machine. Mol. Cell 2020, 80, 1025–1038.e5. [Google Scholar] [CrossRef]
- Palecek, J.J.; Gruber, S. Kite Proteins: A Superfamily of SMC/Kleisin Partners Conserved Across Bacteria, Archaea, and Eukaryotes. Structure 2015, 23, 2183–2190. [Google Scholar] [CrossRef]
- Wells, J.N.; Gligoris, T.G.; Nasmyth, K.A.; Marsh, J.A. Evolution of Condensin and Cohesin Complexes Driven by Replacement of Kite by Hawk Proteins. Curr. Biol. 2017, 27, R17–R18. [Google Scholar] [CrossRef]
- King, T.D.; Leonard, C.J.; Cooper, J.C.; Nguyen, S.; Joyce, E.F.; Phadnis, N. Recurrent Losses and Rapid Evolution of the Condensin II Complex in Insects. Mol. Biol. Evol. 2019, 36, 2195–2204. [Google Scholar] [CrossRef]
- Hoencamp, C.; Dudchenko, O.; Elbatsh, A.M.O.; Brahmachari, S.; Raaijmakers, J.A.; van Schaik, T.; Sedeño Cacciatore, Á.; Contessoto, V.G.; van Heesbeen, R.G.H.P.; van den Broek, B.; et al. 3D Genomics across the Tree of Life Reveals Condensin II as a Determinant of Architecture Type. Science 2021, 372, 984–989. [Google Scholar] [CrossRef]
- Lukyanchikova, V.; Nuriddinov, M.; Belokopytova, P.; Taskina, A.; Liang, J.; Reijnders, M.J.M.F.; Ruzzante, L.; Feron, R.; Waterhouse, R.M.; Wu, Y.; et al. Anopheles Mosquitoes Reveal New Principles of 3D Genome Organization in Insects. Nat. Commun. 2022, 13, 1960. [Google Scholar] [CrossRef]
- Ketharnathan, S.; Labudina, A.; Horsfield, J.A. Cohesin Components Stag1 and Stag2 Differentially Influence Haematopoietic Mesoderm Development in Zebrafish Embryos. Front. Cell Dev. Biol. 2020, 8, 617545. [Google Scholar] [CrossRef]
- Haering, C.H.; Schoffnegger, D.; Nishino, T.; Helmhart, W.; Nasmyth, K.; Löwe, J. Structure and Stability of Cohesin’s Smc1-Kleisin Interaction. Mol. Cell 2004, 15, 951–964. [Google Scholar] [CrossRef]
- Cryo-EM Structure of the Human Cohesin-NIPBL-DNA Complex|Science. Available online: https://www.science.org/doi/10.1126/science.abb0981?url_ver=Z39.88-2003&rfr_id=ori:rid:crossref.org&rfr_dat=cr_pub%20%200pubmed (accessed on 25 January 2023).
- Kikuchi, S.; Borek, D.M.; Otwinowski, Z.; Tomchick, D.R.; Yu, H. Crystal Structure of the Cohesin Loader Scc2 and Insight into Cohesinopathy. Proc. Natl. Acad. Sci. USA 2016, 113, 12444–12449. [Google Scholar] [CrossRef]
- van der Lelij, P.; Newman, J.A.; Lieb, S.; Jude, J.; Katis, V.; Hoffmann, T.; Hinterndorfer, M.; Bader, G.; Kraut, N.; Pearson, M.A.; et al. STAG1 Vulnerabilities for Exploiting Cohesin Synthetic Lethality in STAG2-Deficient Cancers. Life Sci. Alliance 2020, 3, e202000725. [Google Scholar] [CrossRef]
- Higashi, T.L.; Eickhoff, P.; Sousa, J.S.; Locke, J.; Nans, A.; Flynn, H.R.; Snijders, A.P.; Papageorgiou, G.; O’Reilly, N.; Chen, Z.A.; et al. A Structure-Based Mechanism for DNA Entry into the Cohesin Ring. Mol. Cell 2020, 79, 917–933.e9. [Google Scholar] [CrossRef]
- Davidson, I.F.; Bauer, B.; Goetz, D.; Tang, W.; Wutz, G.; Peters, J.-M. DNA Loop Extrusion by Human Cohesin. Science 2019, 366, 1338–1345. [Google Scholar] [CrossRef]
- Pradhan, B.; Barth, R.; Kim, E.; Davidson, I.F.; Bauer, B.; van Laar, T.; Yang, W.; Ryu, J.-K.; van der Torre, J.; Peters, J.-M.; et al. SMC Complexes Can Traverse Physical Roadblocks Bigger than Their Ring Size. Cell Rep. 2022, 41, 111491. [Google Scholar] [CrossRef]
- Banigan, E.J.; van den Berg, A.A.; Brandão, H.B.; Marko, J.F.; Mirny, L.A. Chromosome Organization by One-Sided and Two-Sided Loop Extrusion. eLife 2020, 9, e53558. [Google Scholar] [CrossRef]
- Ganji, M.; Shaltiel, I.A.; Bisht, S.; Kim, E.; Kalichava, A.; Haering, C.H.; Dekker, C. Real-Time Imaging of DNA Loop Extrusion by Condensin. Science 2018, 360, 102–105. [Google Scholar] [CrossRef]
- Kim, J.; Jimenez, D.S.; Ragipani, B.; Zhang, B.; Street, L.A.; Kramer, M.; Albritton, S.E.; Winterkorn, L.H.; Morao, A.K.; Ercan, S. Condensin DC Loads and Spreads from Recruitment Sites to Create Loop-Anchored TADs in C. Elegans. eLife 2022, 11, e68745. [Google Scholar] [CrossRef]
- Mirny, L.A.; Imakaev, M.; Abdennur, N. Two Major Mechanisms of Chromosome Organization. Curr. Opin. Cell Biol. 2019, 58, 142–152. [Google Scholar] [CrossRef]
- Kong, M.; Cutts, E.E.; Pan, D.; Beuron, F.; Kaliyappan, T.; Xue, C.; Morris, E.P.; Musacchio, A.; Vannini, A.; Greene, E.C. Human Condensin I and II Drive Extensive ATP-Dependent Compaction of Nucleosome-Bound DNA. Mol. Cell 2020, 79, 99–114.e9. [Google Scholar] [CrossRef] [PubMed]
- Higashi, T.L.; Pobegalov, G.; Tang, M.; Molodtsov, M.I.; Uhlmann, F. A Brownian Ratchet Model for DNA Loop Extrusion by the Cohesin Complex. eLife 2021, 10, e67530. [Google Scholar] [CrossRef] [PubMed]
- Nichols, M.H.; Corces, V.G. A Tethered-Inchworm Model of SMC DNA Translocation. Nat. Struct. Mol. Biol. 2018, 25, 906–910. [Google Scholar] [CrossRef]
- Vazquez Nunez, R.; Ruiz Avila, L.B.; Gruber, S. Transient DNA Occupancy of the SMC Interarm Space in Prokaryotic Condensin. Mol. Cell 2019, 75, 209–223.e6. [Google Scholar] [CrossRef]
- Marko, J.F.; De Los Rios, P.; Barducci, A.; Gruber, S. DNA-Segment-Capture Model for Loop Extrusion by Structural Maintenance of Chromosome (SMC) Protein Complexes. Nucleic Acids Res. 2019, 47, 6956–6972. [Google Scholar] [CrossRef]
- Ryu, J.-K.; Katan, A.J.; van der Sluis, E.O.; Wisse, T.; de Groot, R.; Haering, C.H.; Dekker, C. The Condensin Holocomplex Cycles Dynamically between Open and Collapsed States. Nat. Struct. Mol. Biol. 2020, 27, 1134–1141. [Google Scholar] [CrossRef]
- Bauer, B.W.; Davidson, I.F.; Canena, D.; Wutz, G.; Tang, W.; Litos, G.; Horn, S.; Hinterdorfer, P.; Peters, J.-M. Cohesin Mediates DNA Loop Extrusion by a “Swing and Clamp” Mechanism. Cell 2021, 184, 5448–5464.e22. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.H.; Abdullaev, Z.K.; Smith, A.D.; Ching, K.A.; Loukinov, D.I.; Green, R.D.; Zhang, M.Q.; Lobanenkov, V.V.; Ren, B. Analysis of the Vertebrate Insulator Protein CTCF-Binding Sites in the Human Genome. Cell 2007, 128, 1231–1245. [Google Scholar] [CrossRef]
- Nora, E.P.; Caccianini, L.; Fudenberg, G.; So, K.; Kameswaran, V.; Nagle, A.; Uebersohn, A.; Hajj, B.; Saux, A.L.; Coulon, A.; et al. Molecular Basis of CTCF Binding Polarity in Genome Folding. Nat. Commun. 2020, 11, 5612. [Google Scholar] [CrossRef]
- Li, Y.; Haarhuis, J.H.I.; Sedeño Cacciatore, Á.; Oldenkamp, R.; van Ruiten, M.S.; Willems, L.; Teunissen, H.; Muir, K.W.; de Wit, E.; Rowland, B.D.; et al. The Structural Basis for Cohesin–CTCF-Anchored Loops. Nature 2020, 578, 472–476. [Google Scholar] [CrossRef]
- Banigan, E.J.; Tang, W.; van den Berg, A.A.; Stocsits, R.R.; Wutz, G.; Brandão, H.B.; Busslinger, G.A.; Peters, J.-M.; Mirny, L.A. Transcription Shapes 3D Chromatin Organization by Interacting with Loop-Extruding Cohesin Complexes. bioRxiv 2022. [Google Scholar] [CrossRef]
- Dequeker, B.J.H.; Scherr, M.J.; Brandão, H.B.; Gassler, J.; Powell, S.; Gaspar, I.; Flyamer, I.M.; Lalic, A.; Tang, W.; Stocsits, R.; et al. MCM Complexes Are Barriers That Restrict Cohesin-Mediated Loop Extrusion. Nature 2022, 606, 197–203. [Google Scholar] [CrossRef]
- Fudenberg, G.; Imakaev, M.; Lu, C.; Goloborodko, A.; Abdennur, N.; Mirny, L.A. Formation of Chromosomal Domains by Loop Extrusion. Cell Rep. 2016, 15, 2038–2049. [Google Scholar] [CrossRef]
- Sanborn, A.L.; Rao, S.S.P.; Huang, S.-C.; Durand, N.C.; Huntley, M.H.; Jewett, A.I.; Bochkov, I.D.; Chinnappan, D.; Cutkosky, A.; Li, J.; et al. Chromatin Extrusion Explains Key Features of Loop and Domain Formation in Wild-Type and Engineered Genomes. Proc. Natl. Acad. Sci. USA 2015, 112, E6456–E6465. [Google Scholar] [CrossRef]
- Peters, J.-M. How DNA Loop Extrusion Mediated by Cohesin Enables V(D)J Recombination. Curr. Opin. Cell Biol. 2021, 70, 75–83. [Google Scholar] [CrossRef]
- Albritton, S.E.; Ercan, S. Caenorhabditis Elegans Dosage Compensation: Insights into Condensin-Mediated Gene Regulation. Trends Genet. 2018, 34, 41–53. [Google Scholar] [CrossRef]
- Generoso, S.F.; Neguembor, M.V.; Hershberg, E.A.; Sadreyev, R.I.; Kurimoto, K.; Yabuta, Y.; Ricci, R.; Audergon, P.; Bauer, M.; Saitou, M.; et al. Cohesin Controls X Chromosome Structure Remodeling and X-Reactivation during Mouse IPSC-Reprogramming. Proc. Natl. Acad. Sci. USA 2023, 120, e2213810120. [Google Scholar] [CrossRef]
- Arnould, C.; Rocher, V.; Finoux, A.-L.; Clouaire, T.; Li, K.; Zhou, F.; Caron, P.; Mangeot, P.E.; Ricci, E.P.; Mourad, R.; et al. Loop Extrusion as a Mechanism for Formation of DNA Damage Repair Foci. Nature 2021, 590, 660–665. [Google Scholar] [CrossRef]
- Batty, P.; Gerlich, D.W. Mitotic Chromosome Mechanics: How Cells Segregate Their Genome. Trends Cell Biol. 2019, 29, 717–726. [Google Scholar] [CrossRef]
- Rao, S.S.P.; Huntley, M.H.; Durand, N.C.; Stamenova, E.K.; Bochkov, I.D.; Robinson, J.T.; Sanborn, A.L.; Machol, I.; Omer, A.D.; Lander, E.S.; et al. A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell 2014, 159, 1665–1680. [Google Scholar] [CrossRef]
- Dixon, J.R.; Selvaraj, S.; Yue, F.; Kim, A.; Li, Y.; Shen, Y.; Hu, M.; Liu, J.S.; Ren, B. Topological Domains in Mammalian Genomes Identified by Analysis of Chromatin Interactions. Nature 2012, 485, 376–380. [Google Scholar] [CrossRef]
- Rowley, M.J.; Corces, V.G. The Three-Dimensional Genome: Principles and Roles of Long-Distance Interactions. Curr. Opin. Cell Biol. 2016, 40, 8–14. [Google Scholar] [CrossRef]
- Gabriele, M.; Brandão, H.B.; Grosse-Holz, S.; Jha, A.; Dailey, G.M.; Cattoglio, C.; Hsieh, T.-H.S.; Mirny, L.; Zechner, C.; Hansen, A.S. Dynamics of CTCF- and Cohesin-Mediated Chromatin Looping Revealed by Live-Cell Imaging. Science 2022, 376, 496–501. [Google Scholar] [CrossRef]
- Mach, P.; Kos, P.I.; Zhan, Y.; Cramard, J.; Gaudin, S.; Tünnermann, J.; Marchi, E.; Eglinger, J.; Zuin, J.; Kryzhanovska, M.; et al. Cohesin and CTCF Control the Dynamics of Chromosome Folding. Nat. Genet. 2022, 54, 1907–1918. [Google Scholar] [CrossRef]
- Yokoshi, M.; Segawa, K.; Fukaya, T. Visualizing the Role of Boundary Elements in Enhancer-Promoter Communication. Mol. Cell 2020, 78, 224–235.e5. [Google Scholar] [CrossRef]
- Krefting, J.; Andrade-Navarro, M.A.; Ibn-Salem, J. Evolutionary Stability of Topologically Associating Domains Is Associated with Conserved Gene Regulation. BMC Biol. 2018, 16, 87. [Google Scholar] [CrossRef]
- Huynh, L.; Hormozdiari, F. TAD Fusion Score: Discovery and Ranking the Contribution of Deletions to Genome Structure. Genom. Biol. 2019, 20, 60. [Google Scholar] [CrossRef]
- Real, F.M.; Haas, S.A.; Franchini, P.; Xiong, P.; Simakov, O.; Kuhl, H.; Schöpflin, R.; Heller, D.; Moeinzadeh, M.-H.; Heinrich, V.; et al. The Mole Genome Reveals Regulatory Rearrangements Associated with Adaptive Intersexuality. Science 2020, 370, 208–214. [Google Scholar] [CrossRef]
- Ibrahim, D.M.; Mundlos, S. The Role of 3D Chromatin Domains in Gene Regulation: A Multi-Facetted View on Genome Organization. Curr. Opin. Genet. Dev. 2020, 61, 1–8. [Google Scholar] [CrossRef]
- Glaser, J.; Mundlos, S. 3D or Not 3D: Shaping the Genome during Development. Cold Spring Harb. Perspect. Biol. 2021, 14, a040188. [Google Scholar] [CrossRef]
- Lupiáñez, D.G.; Kraft, K.; Heinrich, V.; Krawitz, P.; Brancati, F.; Klopocki, E.; Horn, D.; Kayserili, H.; Opitz, J.M.; Laxova, R.; et al. Disruptions of Topological Chromatin Domains Cause Pathogenic Rewiring of Gene-Enhancer Interactions. Cell 2015, 161, 1012–1025. [Google Scholar] [CrossRef]
- Hnisz, D.; Weintraub, A.S.; Day, D.S.; Valton, A.-L.; Bak, R.O.; Li, C.H.; Goldmann, J.; Lajoie, B.R.; Fan, Z.P.; Sigova, A.A.; et al. Activation of Proto-Oncogenes by Disruption of Chromosome Neighborhoods. Science 2016, 351, 1454–1458. [Google Scholar] [CrossRef]
- Flavahan, W.A.; Drier, Y.; Liau, B.B.; Gillespie, S.M.; Venteicher, A.S.; Stemmer-Rachamimov, A.O.; Suvà, M.L.; Bernstein, B.E. Insulator Dysfunction and Oncogene Activation in IDH Mutant Gliomas. Nature 2016, 529, 110–114. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, L.; Kucherlapati, M.; Chen, F.; Hadjipanayis, A.; Pantazi, A.; Bristow, C.A.; Lee, E.A.; Mahadeshwar, H.S.; Tang, J.; et al. A Pan-Cancer Compendium of Genes Deregulated by Somatic Genomic Rearrangement across More Than 1400 Cases. Cell Rep. 2018, 24, 515–527. [Google Scholar] [CrossRef]
- Ji, X.; Dadon, D.B.; Powell, B.E.; Fan, Z.P.; Borges-Rivera, D.; Shachar, S.; Weintraub, A.S.; Hnisz, D.; Pegoraro, G.; Lee, T.I.; et al. 3D Chromosome Regulatory Landscape of Human Pluripotent Cells. Cell Stem Cell 2016, 18, 262–275. [Google Scholar] [CrossRef]
- Gridina, M.; Fishman, V. Multilevel View on Chromatin Architecture Alterations in Cancer. Front. Genet. 2022, 13, 1059617. [Google Scholar] [CrossRef]
- Akdemir, K.C.; Le, V.T.; Chandran, S.; Li, Y.; Verhaak, R.G.; Beroukhim, R.; Campbell, P.J.; Chin, L.; Dixon, J.R.; Futreal, P.A. Disruption of Chromatin Folding Domains by Somatic Genomic Rearrangements in Human Cancer. Nat. Genet. 2020, 52, 294–305. [Google Scholar] [CrossRef]
- Xu, Z.; Lee, D.-S.; Chandran, S.; Le, V.T.; Bump, R.; Yasis, J.; Dallarda, S.; Marcotte, S.; Clock, B.; Haghani, N.; et al. Structural Variants Drive Context-Dependent Oncogene Activation in Cancer. Nature 2022, 612, 564–572. [Google Scholar] [CrossRef]
- Rajderkar, S.; Barozzi, I.; Zhu, Y.; Hu, R.; Zhang, Y.; Li, B.; Fukuda-Yuzawa, Y.; Kelman, G.; Akeza, A.; Blow, M.J.; et al. Topologically Associating Domain Boundaries Are Commonly Required for Normal Genome Function. bioRxiv 2021. [Google Scholar] [CrossRef]
- Despang, A.; Schöpflin, R.; Franke, M.; Ali, S.; Jerković, I.; Paliou, C.; Chan, W.-L.; Timmermann, B.; Wittler, L.; Vingron, M.; et al. Functional Dissection of the Sox9–Kcnj2 Locus Identifies Nonessential and Instructive Roles of TAD Architecture. Nat. Genet. 2019, 51, 1263–1271. [Google Scholar] [CrossRef]
- Rodríguez-Carballo, E.; Lopez-Delisle, L.; Willemin, A.; Beccari, L.; Gitto, S.; Mascrez, B.; Duboule, D. Chromatin Topology and the Timing of Enhancer Function at the HoxD Locus. Proc. Natl. Acad. Sci. USA 2020, 117, 31231–31241. [Google Scholar] [CrossRef]
- Rao, S.S.P.; Huang, S.-C.; St Hilaire, B.G.; Engreitz, J.M.; Perez, E.M.; Kieffer-Kwon, K.-R.; Sanborn, A.L.; Johnstone, S.E.; Bascom, G.D.; Bochkov, I.D.; et al. Cohesin Loss Eliminates All Loop Domains. Cell 2017, 171, 305–320.e24. [Google Scholar] [CrossRef]
- Phillips-Cremins, J.E.; Sauria, M.E.G.; Sanyal, A.; Gerasimova, T.I.; Lajoie, B.R.; Bell, J.S.K.; Ong, C.-T.; Hookway, T.A.; Guo, C.; Sun, Y.; et al. Architectural Protein Subclasses Shape 3D Organization of Genomes during Lineage Commitment. Cell 2013, 153, 1281–1295. [Google Scholar] [CrossRef]
- Cheutin, T.; Cavalli, G. The Multiscale Effects of Polycomb Mechanisms on 3D Chromatin Folding. Crit. Rev. Biochem. Mol. Biol. 2019, 54, 399–417. [Google Scholar] [CrossRef]
- Anania, C.; Acemel, R.D.; Jedamzick, J.; Bolondi, A.; Cova, G.; Brieske, N.; Kühn, R.; Wittler, L.; Real, F.M.; Lupiáñez, D.G. In Vivo Dissection of a Clustered-CTCF Domain Boundary Reveals Developmental Principles of Regulatory Insulation. Nat. Genet. 2022, 54, 1026–1036. [Google Scholar] [CrossRef]
- Barutcu, A.R.; Maass, P.G.; Lewandowski, J.P.; Weiner, C.L.; Rinn, J.L. A TAD Boundary Is Preserved upon Deletion of the CTCF-Rich Firre Locus. Nat. Commun. 2018, 9, 1444. [Google Scholar] [CrossRef]
- Williamson, I.; Kane, L.; Devenney, P.S.; Flyamer, I.M.; Anderson, E.; Kilanowski, F.; Hill, R.E.; Bickmore, W.A.; Lettice, L.A. Developmentally Regulated Shh Expression Is Robust to TAD Perturbations. Development 2019, 146, dev179523. [Google Scholar] [CrossRef]
- Narendra, V.; Rocha, P.P.; An, D.; Raviram, R.; Skok, J.A.; Mazzoni, E.O.; Reinberg, D. CTCF Establishes Discrete Functional Chromatin Domains at the Hox Clusters during Differentiation. Science 2015, 347, 1017–1021. [Google Scholar] [CrossRef]
- Rodríguez-Carballo, E.; Lopez-Delisle, L.; Zhan, Y.; Fabre, P.J.; Beccari, L.; El-Idrissi, I.; Huynh, T.H.N.; Ozadam, H.; Dekker, J.; Duboule, D. The HoxD Cluster Is a Dynamic and Resilient TAD Boundary Controlling the Segregation of Antagonistic Regulatory Landscapes. Genes Dev. 2017, 31, 2264–2281. [Google Scholar] [CrossRef]
- Zuin, J.; Roth, G.; Zhan, Y.; Cramard, J.; Redolfi, J.; Piskadlo, E.; Mach, P.; Kryzhanovska, M.; Tihanyi, G.; Kohler, H.; et al. Nonlinear Control of Transcription through Enhancer–Promoter Interactions. Nature 2022, 604, 571–577. [Google Scholar] [CrossRef]
- Bolt, C.C.; Lopez-Delisle, L.; Hintermann, A.; Mascrez, B.; Rauseo, A.; Andrey, G.; Duboule, D. Context-Dependent Enhancer Function Revealed by Targeted Inter-TAD Relocation. Nat. Commun. 2022, 13, 3488. [Google Scholar] [CrossRef]
- Islam, Z.; Saravanan, B.; Walavalkar, K.; Thakur, J.; Farooq, U.; Singh, A.K.; Sabarinathan, R.; Pandit, A.; Henikoff, S.; Notani, D. Active Enhancers Strengthen Insulation by RNA-Mediated CTCF Binding at TAD Boundaries. bioRxiv 2021. [Google Scholar] [CrossRef]
- Belokopytova, P.S.; Nuriddinov, M.A.; Mozheiko, E.A.; Fishman, D.; Fishman, V. Quantitative Prediction of Enhancer-Promoter Interactions. Genom. Res. 2020, 30, 72–84. [Google Scholar] [CrossRef]
- Chahar, S.; Zouari, Y.B.; Salari, H.; Molitor, A.M.; Kobi, D.; Maroquenne, M.; Erb, C.; Mossler, A.; Karasu, N.; Jost, D.; et al. Context-Dependent Transcriptional Remodeling of TADs during Differentiation. bioRxiv 2022. [Google Scholar] [CrossRef]
- Belokopytova, P.; Fishman, V. Predicting Genome Architecture: Challenges and Solutions. Front. Genet. 2020, 11, 617202. [Google Scholar] [CrossRef]
- International Nucleome Consortium. 3DGenBench: A Web-Server to Benchmark Computational Models for 3D Genomics. Nucleic Acids Res. 2022, 50, W4–W12. [Google Scholar] [CrossRef]
- Ringel, A.R.; Szabo, Q.; Chiariello, A.M.; Chudzik, K.; Schöpflin, R.; Rothe, P.; Mattei, A.L.; Zehnder, T.; Harnett, D.; Laupert, V.; et al. Repression and 3D-Restructuring Resolves Regulatory Conflicts in Evolutionarily Rearranged Genomes. Cell 2022, 185, 3689–3704.e21. [Google Scholar] [CrossRef]
- Schindler, M.; Osterwalder, M.; Harabula, I.; Wittler, L.; Tzika, A.C.; Dechmann, D.; Vingron, M.; Visel, A.; Haas, S.; Real, F.M. Co-Option of the Transcription Factor SALL1 in Mole Ovotestis Formation. bioRxiv 2022. [Google Scholar] [CrossRef]
- Kabirova, E.; Ryzhkova, A.; Lukyanchikova, V.; Khabarova, A.; Korablev, A.; Shnaider, T.; Nuriddinov, M.; Belokopytova, P.; Kontsevaya, G.; Serova, I.; et al. TAD Border Deletion at the Kit Locus Causes Tissue-Specific Ectopic Activation of a Neighboring Gene. bioRxiv 2022. [Google Scholar] [CrossRef]
- Martinez-Ara, M.; Comoglio, F.; van Arensbergen, J.; van Steensel, B. Systematic Analysis of Intrinsic Enhancer-Promoter Compatibility in the Mouse Genome. Mol. Cell 2022, 82, 2519–2531.e6. [Google Scholar] [CrossRef]
- van Arensbergen, J.; van Steensel, B.; Bussemaker, H.J. In Search of the Determinants of Enhancer–Promoter Interaction Specificity. Trends Cell Biol. 2014, 24, 695–702. [Google Scholar] [CrossRef]
- Bergman, D.T.; Jones, T.R.; Liu, V.; Ray, J.; Jagoda, E.; Siraj, L.; Kang, H.Y.; Nasser, J.; Kane, M.; Rios, A.; et al. Compatibility Rules of Human Enhancer and Promoter Sequences. Nature 2022, 607, 176–184. [Google Scholar] [CrossRef] [PubMed]
- Franke, M.; Ibrahim, D.M.; Andrey, G.; Schwarzer, W.; Heinrich, V.; Schöpflin, R.; Kraft, K.; Kempfer, R.; Jerković, I.; Chan, W.-L.; et al. Formation of New Chromatin Domains Determines Pathogenicity of Genomic Duplications. Nature 2016, 538, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Long, H.S.; Greenaway, S.; Powell, G.; Mallon, A.-M.; Lindgren, C.M.; Simon, M.M. Making Sense of the Linear Genome, Gene Function and TADs. Epigenet. Chromatin 2022, 15, 4. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Monahan, K.; Wu, H.; Gertz, J.; Varley, K.E.; Li, W.; Myers, R.M.; Maniatis, T.; Wu, Q. CTCF/Cohesin-Mediated DNA Looping Is Required for Protocadherin α Promoter Choice. Proc. Natl. Acad. Sci. USA 2012, 109, 21081–21086. [Google Scholar] [CrossRef]
- Matthews, N.E.; White, R. Chromatin Architecture in the Fly: Living without CTCF/Cohesin Loop Extrusion? BioEssays 2019, 41, 1900048. [Google Scholar] [CrossRef] [PubMed]
- Heger, P.; Marin, B.; Bartkuhn, M.; Schierenberg, E.; Wiehe, T. The Chromatin Insulator CTCF and the Emergence of Metazoan Diversity. Proc. Natl. Acad. Sci. USA 2012, 109, 17507–17512. [Google Scholar] [CrossRef]
- Filippova, G.N.; Fagerlie, S.; Klenova, E.M.; Myers, C.; Dehner, Y.; Goodwin, G.; Neiman, P.E.; Collins, S.J.; Lobanenkov, V.V. An Exceptionally Conserved Transcriptional Repressor, CTCF, Employs Different Combinations of Zinc Fingers to Bind Diverged Promoter Sequences of Avian and Mammalian c-Myc Oncogenes. Mol. Cell. Biol. 1996, 16, 2802–2813. [Google Scholar] [CrossRef]
- Bartkuhn, M.; Straub, T.; Herold, M.; Herrmann, M.; Rathke, C.; Saumweber, H.; Gilfillan, G.D.; Becker, P.B.; Renkawitz, R. Active Promoters and Insulators Are Marked by the Centrosomal Protein 190. EMBO J. 2009, 28, 877–888. [Google Scholar] [CrossRef] [PubMed]
- Kadota, M.; Hara, Y.; Tanaka, K.; Takagi, W.; Tanegashima, C.; Nishimura, O.; Kuraku, S. CTCF Binding Landscape in Jawless Fish with Reference to Hox Cluster Evolution. Sci. Rep. 2017, 7, 4957. [Google Scholar] [CrossRef]
- Kaushal, A.; Mohana, G.; Dorier, J.; Özdemir, I.; Omer, A.; Cousin, P.; Semenova, A.; Taschner, M.; Dergai, O.; Marzetta, F.; et al. CTCF Loss Has Limited Effects on Global Genome Architecture in Drosophila despite Critical Regulatory Functions. Nat. Commun. 2021, 12, 1011. [Google Scholar] [CrossRef] [PubMed]
- Klenova, E.M.; Chernukhin, I.V.; El-Kady, A.; Lee, R.E.; Pugacheva, E.M.; Loukinov, D.I.; Goodwin, G.H.; Delgado, D.; Filippova, G.N.; León, J.; et al. Functional Phosphorylation Sites in the C-Terminal Region of the Multivalent Multifunctional Transcriptional Factor CTCF. Mol. Cell. Biol. 2001, 21, 2221–2234. [Google Scholar] [CrossRef] [PubMed]
- Nora, E.P.; Goloborodko, A.; Valton, A.-L.; Gibcus, J.H.; Uebersohn, A.; Abdennur, N.; Dekker, J.; Mirny, L.A.; Bruneau, B.G. Targeted Degradation of CTCF Decouples Local Insulation of Chromosome Domains from Genomic Compartmentalization. Cell 2017, 169, 930–944.e22. [Google Scholar] [CrossRef]
- Li, D.; Ning, C.; Zhang, J.; Wang, Y.; Tang, Q.; Kui, H.; Wang, T.; He, M.; Jin, L.; Li, J.; et al. Dynamic Transcriptome and Chromatin Architecture in Granulosa Cells during Chicken Folliculogenesis. Nat. Commun. 2022, 13, 131. [Google Scholar] [CrossRef]
- Marlétaz, F.; de la Calle-Mustienes, E.; Acemel, R.D.; Nakamura, T.; Paliou, C.; Naranjo, S.; Martínez-García, P.M.; Cases, I.; Sleight, V.A.; Hirschberger, C.; et al. The Little Skate Genome and the Evolutionary Emergence of Wing-like Fin Appendages. bioRxiv 2022. [Google Scholar] [CrossRef]
- Fishman, V.; Battulin, N.; Nuriddinov, M.; Maslova, A.; Zlotina, A.; Strunov, A.; Chervyakova, D.; Korablev, A.; Serov, O.; Krasikova, A. 3D Organization of Chicken Genome Demonstrates Evolutionary Conservation of Topologically Associated Domains and Highlights Unique Architecture of Erythrocytes’ Chromatin. Nucleic Acids Res. 2019, 47, 648–665. [Google Scholar] [CrossRef]
- Heger, P.; Marin, B.; Schierenberg, E. Loss of the Insulator Protein CTCF during Nematode Evolution. BMC Mol. Biol. 2009, 10, 84. [Google Scholar] [CrossRef]
- Huang, Z.; Xu, L.; Cai, C.; Zhou, Y.; Liu, J.; Zhu, Z.; Kang, W.; Chen, D.; Pei, S.; Xue, T.; et al. Three Amphioxus Reference Genomes Reveal Gene and Chromosome Evolution of Chordates. bioRxiv 2022. [Google Scholar] [CrossRef] [PubMed]
- Acemel, R.D.; Lupiáñez, D.G. Evolution of 3D Chromatin Organization at Different Scales. Curr. Opin. Genet. Dev. 2023, 78, 102019. [Google Scholar] [CrossRef]
- Kim, J.-S.; Krasieva, T.; LaMorte, V.; Taylor, A.; Yokomori, K. Specific Recruitment of Human Cohesin to Laser-Induced DNA Damage. J. Biol. Chem. 2002, 277, 45149–45153. [Google Scholar] [CrossRef]
- Heale, J.T.; Ball, A.R.; Schmiesing, J.A.; Kim, J.-S.; Kong, X.; Zhou, S.; Hudson, D.F.; Earnshaw, W.C.; Yokomori, K. Condensin I Interacts with the PARP-1-XRCC1 Complex and Functions in DNA Single-Strand Break Repair. Mol. Cell 2006, 21, 837–848. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Stephens, J.; Ball, A.R.B., Jr.; Heale, J.T.; Newkirk, D.A.; Berns, M.W.; Yokomori, K. Condensin I Recruitment to Base Damage-Enriched DNA Lesions Is Modulated by PARP1. PLoS ONE 2011, 6, e23548. [Google Scholar] [CrossRef]
- Wood, J.L.; Liang, Y.; Li, K.; Chen, J. Microcephalin/MCPH1 Associates with the Condensin II Complex to Function in Homologous Recombination Repair. J. Biol. Chem. 2008, 283, 29586–29592. [Google Scholar] [CrossRef] [PubMed]
- Kaku, Y.; Barrington, C.; Barry, D.; Gerguri, T.; Fu, X.; Bates, P.; Khatri, B.; Uhlmann, F. Fission Yeast Condensin Contributes to Interphase Chromatin Organization and Prevents Transcription-Coupled DNA Damage. Genom. Biol. 2020, 21, 272. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Li, S.; Ser, Z.; Sanyal, T.; Choi, K.; Wan, B.; Kuang, H.; Sali, A.; Kentsis, A.; Patel, D.J.; et al. Integrative Analysis Reveals Unique Structural and Functional Features of the Smc5/6 Complex. Proc. Natl. Acad. Sci. USA 2021, 118, e2026844118. [Google Scholar] [CrossRef]
- Wu, N.; Yu, H. The Smc Complexes in DNA Damage Response. Cell Biosci. 2012, 2, 5. [Google Scholar] [CrossRef]
- Venegas, A.B.; Natsume, T.; Kanemaki, M.; Hickson, I.D. Inducible Degradation of the Human SMC5/6 Complex Reveals an Essential Role Only during Interphase. Cell Rep. 2020, 31, 107533. [Google Scholar] [CrossRef]
- Petermann, E.; Lan, L.; Zou, L. Sources, Resolution and Physiological Relevance of R-Loops and RNA–DNA Hybrids. Nat. Rev. Mol. Cell Biol. 2022, 23, 521–540. [Google Scholar] [CrossRef] [PubMed]
- Orthwein, A.; Noordermeer, S.M.; Wilson, M.D.; Landry, S.; Enchev, R.I.; Sherker, A.; Munro, M.; Pinder, J.; Salsman, J.; Dellaire, G.; et al. A Mechanism for the Suppression of Homologous Recombination in G1 Cells. Nature 2015, 528, 422–426. [Google Scholar] [CrossRef] [PubMed]
- Seeber, A.; Hegnauer, A.M.; Hustedt, N.; Deshpande, I.; Poli, J.; Eglinger, J.; Pasero, P.; Gut, H.; Shinohara, M.; Hopfner, K.-P.; et al. RPA Mediates Recruitment of MRX to Forks and Double-Strand Breaks to Hold Sister Chromatids Together. Mol. Cell 2016, 64, 951–966. [Google Scholar] [CrossRef] [PubMed]
- Scherzer, M.; Giordano, F.; Ferran, M.S.; Ström, L. Recruitment of Scc2/4 to Double-Strand Breaks Depends on ΓH2A and DNA End Resection. Life Sci. Alliance 2022, 5, e202101244. [Google Scholar] [CrossRef] [PubMed]
- Potts, P.R.; Porteus, M.H.; Yu, H. Human SMC5/6 Complex Promotes Sister Chromatid Homologous Recombination by Recruiting the SMC1/3 Cohesin Complex to Double-Strand Breaks. EMBO J. 2006, 25, 3377–3388. [Google Scholar] [CrossRef] [PubMed]
- Wassing, I.E.; Esashi, F. RAD51: Beyond the Break. Semin. Cell Dev. Biol. 2021, 113, 38–46. [Google Scholar] [CrossRef]
- Lee, J.; Terakawa, T.; Qi, Z.; Steinfeld, J.; Redding, S.; Kwon, Y.; Gaines, W.; Zhao, W.; Sung, P.; Greene, E. Base Triplet Stepping by the Rad51/RecA Family of Recombinases. Science 2015, 349, 977–981. [Google Scholar] [CrossRef]
- Bell, J.C.; Kowalczykowski, S.C. RecA: Regulation and Mechanism of a Molecular Search Engine. Trends Biochem. Sci. 2016, 41, 491–507. [Google Scholar] [CrossRef]
- Agmon, N.; Liefshitz, B.; Zimmer, C.; Fabre, E.; Kupiec, M. Effect of Nuclear Architecture on the Efficiency of Double-Strand Break Repair. Nat. Cell Biol. 2013, 15, 694–699. [Google Scholar] [CrossRef]
- Stinson, B.M.; Loparo, J.J. Repair of DNA Double-Strand Breaks by the Nonhomologous End Joining Pathway. Annu. Rev. Biochem. 2021, 90, 137–164. [Google Scholar] [CrossRef] [PubMed]
- Gelot, C.; Guirouilh-Barbat, J.; Le Guen, T.; Dardillac, E.; Chailleux, C.; Canitrot, Y.; Lopez, B.S. The Cohesin Complex Prevents the End Joining of Distant DNA Double-Strand Ends. Mol. Cell 2016, 61, 15–26. [Google Scholar] [CrossRef]
- Ström, L.; Lindroos, H.B.; Shirahige, K.; Sjögren, C. Postreplicative Recruitment of Cohesin to Double-Strand Breaks Is Required for DNA Repair. Mol. Cell 2004, 16, 1003–1015. [Google Scholar] [CrossRef]
- Litwin, I.; Pilarczyk, E.; Wysocki, R. The Emerging Role of Cohesin in the DNA Damage Response. Genes 2018, 9, 581. [Google Scholar] [CrossRef]
- Phipps, J.; Dubrana, K. DNA Repair in Space and Time: Safeguarding the Genome with the Cohesin Complex. Genes 2022, 13, 198. [Google Scholar] [CrossRef]
- Soutoglou, E.; Misteli, T. Mobility and Immobility of Chromatin in Transcription and Genome Stability. Curr. Opin. Genet. Dev. 2007, 17, 435–442. [Google Scholar] [CrossRef]
- Piazza, A.; Bordelet, H.; Dumont, A.; Thierry, A.; Savocco, J.; Girard, F.; Koszul, R. Cohesin Regulates Homology Search during Recombinational DNA Repair. Nat. Cell Biol. 2021, 23, 1176–1186. [Google Scholar] [CrossRef]
- Unal, E.; Arbel-Eden, A.; Sattler, U.; Shroff, R.; Lichten, M.; Haber, J.E.; Koshland, D. DNA Damage Response Pathway Uses Histone Modification to Assemble a Double-Strand Break-Specific Cohesin Domain. Mol. Cell 2004, 16, 991–1002. [Google Scholar] [CrossRef]
- Kong, X.; Ball, A.R.; Pham, H.X.; Zeng, W.; Chen, H.-Y.; Schmiesing, J.A.; Kim, J.-S.; Berns, M.; Yokomori, K. Distinct Functions of Human Cohesin-SA1 and Cohesin-SA2 in Double-Strand Break Repair. Mol. Cell. Biol. 2014, 34, 685–698. [Google Scholar] [CrossRef]
- Countryman, P.; Fan, Y.; Gorthi, A.; Pan, H.; Strickland, J.; Kaur, P.; Wang, X.; Lin, J.; Lei, X.; White, C.; et al. Cohesin SA2 Is a Sequence-Independent DNA-Binding Protein That Recognizes DNA Replication and Repair Intermediates. J. Biol. Chem. 2018, 293, 1054–1069. [Google Scholar] [CrossRef]
- Ström, L.; Karlsson, C.; Lindroos, H.; Wedahl, S.; Katou, Y.; Shirahige, K.; Sjögren, C. Postreplicative Formation of Cohesion Is Required for Repair and Induced by a Single DNA Break. Science 2007, 317, 242–245. [Google Scholar] [CrossRef]
- Unal, E.; Heidinger-Pauli, J.M.; Koshland, D. DNA Double-Strand Breaks Trigger Genome-Wide Sister-Chromatid Cohesion through Eco1 (Ctf7). Science 2007, 317, 245–248. [Google Scholar] [CrossRef]
- McLellan, J.L.; O’Neil, N.J.; Barrett, I.; Ferree, E.; Pel, D.M.V.; Ushey, K.; Sipahimalani, P.; Bryan, J.; Rose, A.M.; Hieter, P. Synthetic Lethality of Cohesins with PARPs and Replication Fork Mediators. PLoS Genet. 2012, 8, e1002574. [Google Scholar] [CrossRef]
- Heidinger-Pauli, J.M.; Unal, E.; Koshland, D. Distinct Targets of the Eco1 Acetyltransferase Modulate Cohesion in S Phase and in Response to DNA Damage. Mol. Cell 2009, 34, 311–321. [Google Scholar] [CrossRef]
- Hou, W.; Li, Y.; Zhang, J.; Xia, Y.; Wang, X.; Chen, H.; Lou, H. Cohesin in DNA Damage Response and Double-Strand Break Repair. Crit. Rev. Biochem. Mol. Biol. 2022, 57, 333–350. [Google Scholar] [CrossRef]
- Iacovoni, J.S.; Caron, P.; Lassadi, I.; Nicolas, E.; Massip, L.; Trouche, D.; Legube, G. High-Resolution Profiling of GammaH2AX around DNA Double Strand Breaks in the Mammalian Genome. EMBO J. 2010, 29, 1446–1457. [Google Scholar] [CrossRef]
- Liu, Y.; Zou, R.S.; He, S.; Nihongaki, Y.; Li, X.; Razavi, S.; Wu, B.; Ha, T. Very Fast CRISPR on Demand. Science 2020, 368, 1265–1269. [Google Scholar] [CrossRef]
- de Wit, E.; Nora, E.P. New Insights into Genome Folding by Loop Extrusion from Inducible Degron Technologies. Nat. Rev. Genet. 2023, 24, 73–85. [Google Scholar] [CrossRef]
- Rogakou, E.P.; Boon, C.; Redon, C.; Bonner, W.M. Megabase Chromatin Domains Involved in DNA Double-Strand Breaks in Vivo. J. Cell Biol. 1999, 146, 905–916. [Google Scholar] [CrossRef]
- Stucki, M.; Clapperton, J.A.; Mohammad, D.; Yaffe, M.B.; Smerdon, S.J.; Jackson, S.P. MDC1 Directly Binds Phosphorylated Histone H2AX to Regulate Cellular Responses to DNA Double-Strand Breaks. Cell 2005, 123, 1213–1226. [Google Scholar] [CrossRef]
- Caron, P.; Aymard, F.; Iacovoni, J.S.; Briois, S.; Canitrot, Y.; Bugler, B.; Massip, L.; Losada, A.; Legube, G. Cohesin Protects Genes against ΓH2AX Induced by DNA Double-Strand Breaks. PLoS Genet. 2012, 8, e1002460. [Google Scholar] [CrossRef]
- Natale, F.; Rapp, A.; Yu, W.; Maiser, A.; Harz, H.; Scholl, A.; Grulich, S.; Anton, T.; Hörl, D.; Chen, W.; et al. Identification of the Elementary Structural Units of the DNA Damage Response. Nat. Commun. 2017, 8, 15760. [Google Scholar] [CrossRef]
- Collins, P.; Purman, C.; Porter, S.; Nganga, V.; Saini, A.; Hayer, K.; Gurewitz, G.; Sleckman, B.; Bednarski, J.; Bassing, C.; et al. DNA Double-Strand Breaks Induce H2Ax Phosphorylation Domains in a Contact-Dependent Manner. Nat. Commun. 2020, 11, 3158. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Shi, Z.; Zhang, H.; Finkelstein, I.J.; Yu, H. Human Cohesin Compacts DNA by Loop Extrusion. Science 2019, 366, 1345–1349. [Google Scholar] [CrossRef]
- Gobbini, E.; Casari, E.; Colombo, C.V.; Bonetti, D.; Longhese, M.P. The 9-1-1 Complex Controls Mre11 Nuclease and Checkpoint Activation during Short-Range Resection of DNA Double-Strand Breaks. Cell Rep. 2020, 33, 108287. [Google Scholar] [CrossRef] [PubMed]
- Libri, A.; Marton, T.; Deriano, L. The (Lack of) DNA Double-Strand Break Repair Pathway Choice During V(D)J Recombination. Front. Genet. 2022, 12, 823943. [Google Scholar] [CrossRef]
- Ishiguro, K.-I. The Cohesin Complex in Mammalian Meiosis. Genes Cells 2019, 24, 6–30. [Google Scholar] [CrossRef] [PubMed]
- Gothe, H.J.; Bouwman, B.A.M.; Gusmao, E.G.; Piccinno, R.; Petrosino, G.; Sayols, S.; Drechsel, O.; Minneker, V.; Josipovic, N.; Mizi, A.; et al. Spatial Chromosome Folding and Active Transcription Drive DNA Fragility and Formation of Oncogenic MLL Translocations. Mol. Cell 2019, 75, 267–283.e12. [Google Scholar] [CrossRef]
- Dyson, S.; Segura, J.; Martínez-García, B.; Valdés, A.; Roca, J. Condensin Minimizes Topoisomerase II-Mediated Entanglements of DNA in Vivo. EMBO J. 2021, 40, e105393. [Google Scholar] [CrossRef]
- McAleenan, A.; Clemente-Blanco, A.; Cordon-Preciado, V.; Sen, N.; Esteras, M.; Jarmuz, A.; Aragón, L. Post-Replicative Repair Involves Separase-Dependent Removal of the Kleisin Subunit of Cohesin. Nature 2013, 493, 250–254. [Google Scholar] [CrossRef]
- Carré-Simon, À.; Fabre, E. 3D Genome Organization: Causes and Consequences for DNA Damage and Repair. Genes 2021, 13, 7. [Google Scholar] [CrossRef]
- Gibcus, J.H.; Samejima, K.; Goloborodko, A.; Samejima, I.; Naumova, N.; Nuebler, J.; Kanemaki, M.T.; Xie, L.; Paulson, J.R.; Earnshaw, W.C.; et al. A Pathway for Mitotic Chromosome Formation. Science 2018, 359, eaao6135. [Google Scholar] [CrossRef]
- Larson, A.G.; Elnatan, D.; Keenen, M.M.; Trnka, M.J.; Johnston, J.B.; Burlingame, A.L.; Agard, D.A.; Redding, S.; Narlikar, G.J. Liquid Droplet Formation by HP1α Suggests a Role for Phase Separation in Heterochromatin. Nature 2017, 547, 236–240. [Google Scholar] [CrossRef] [PubMed]
- Flyamer, I.M.; Gassler, J.; Imakaev, M.; Brandão, H.B.; Ulianov, S.V.; Abdennur, N.; Razin, S.V.; Mirny, L.A.; Tachibana-Konwalski, K. Single-Nucleus Hi-C Reveals Unique Chromatin Reorganization at Oocyte-to-Zygote Transition. Nature 2017, 544, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Hartl, T.A.; Smith, H.F.; Bosco, G. Chromosome Alignment and Transvection Are Antagonized by Condensin II. Science 2008, 322, 1384–1387. [Google Scholar] [CrossRef]
- AlHaj Abed, J.; Erceg, J.; Goloborodko, A.; Nguyen, S.C.; McCole, R.B.; Saylor, W.; Fudenberg, G.; Lajoie, B.R.; Dekker, J.; Mirny, L.A.; et al. Highly Structured Homolog Pairing Reflects Functional Organization of the Drosophila Genome. Nat. Commun. 2019, 10, 4485. [Google Scholar] [CrossRef]
- Rowley, M.J.; Lyu, X.; Rana, V.; Ando-Kuri, M.; Karns, R.; Bosco, G.; Corces, V.G. Condensin II Counteracts Cohesin and RNA Polymerase II in the Establishment of 3D Chromatin Organization. Cell Rep. 2019, 26, 2890–2903.e3. [Google Scholar] [CrossRef]
- Houlard, M.; Cutts, E.E.; Shamim, M.S.; Godwin, J.; Weisz, D.; Presser Aiden, A.; Lieberman Aiden, E.; Schermelleh, L.; denini, A.; Nasmyth, K. MCPH1 Inhibits Condensin II during Interphase by Regulating Its SMC2-Kleisin Interface. eLife 2021, 10, e73348. [Google Scholar] [CrossRef]
- Brahmachari, S.; Marko, J.F. Chromosome Disentanglement Driven via Optimal Compaction of Loop-Extruded Brush Structures. Proc. Natl. Acad. Sci. USA 2019, 116, 24956–24965. [Google Scholar] [CrossRef] [PubMed]
- Brahmachari, S.; Contessoto, V.G.; Di Pierro, M.; Onuchic, J.N. Shaping the Genome via Lengthwise Compaction, Phase Separation, and Lamina Adhesion. Nucleic Acids Res. 2022, 50, 4258–4271. [Google Scholar] [CrossRef] [PubMed]
- Cremer, M.; Brandstetter, K.; Maiser, A.; Rao, S.S.P.; Schmid, V.J.; Guirao-Ortiz, M.; Mitra, N.; Mamberti, S.; Klein, K.N.; Gilbert, D.M.; et al. Cohesin Depleted Cells Rebuild Functional Nuclear Compartments after Endomitosis. Nat. Commun. 2020, 11, 6146. [Google Scholar] [CrossRef] [PubMed]
- Gridina, M.; Mozheiko, E.; Valeev, E.; Nazarenko, L.P.; Lopatkina, M.E.; Markova, Z.G.; Yablonskaya, M.I.; Voinova, V.Y.; Shilova, N.V.; Lebedev, I.N.; et al. A Cookbook for DNase Hi-C. Epigenet. Chromatin 2021, 14, 15. [Google Scholar] [CrossRef]
- Hong, P.; Jiang, H.; Xu, W.; Lin, D.; Xu, Q.; Cao, G.; Li, G. The DLO Hi-C Tool for Digestion-Ligation-Only Hi-C Chromosome Conformation Capture Data Analysis. Genes 2020, 11, 289. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Cao, Y.; Cui, K.; Tang, Q.; Zhao, K. Hi-TrAC Reveals Division of Labor of Transcription Factors in Organizing Chromatin Loops. Nat. Commun. 2022, 13, 6679. [Google Scholar] [CrossRef] [PubMed]
- Lafontaine, D.L.; Yang, L.; Dekker, J.; Gibcus, J.H. Hi-C 3.0: Improved Protocol for Genome-Wide Chromosome Conformation Capture. Curr. Protoc. 2021, 1, e198. [Google Scholar] [CrossRef] [PubMed]
- Fiorillo, L.; Musella, F.; Conte, M.; Kempfer, R.; Chiariello, A.M.; Bianco, S.; Kukalev, A.; Irastorza-Azcarate, I.; Esposito, A.; Abraham, A.; et al. Comparison of the Hi-C, GAM and SPRITE Methods Using Polymer Models of Chromatin. Nat. Methods 2021, 18, 482–490. [Google Scholar] [CrossRef]
- Yunusova, A.; Smirnov, A.; Shnaider, T.; Lukyanchikova, V.; Afonnikova, S.; Battulin, N. Evaluation of the OsTIR1 and AtAFB2 AID Systems for Genome Architectural Protein Degradation in Mammalian Cells. Front. Mol. Biosci. 2021, 8, 757394. [Google Scholar] [CrossRef] [PubMed]
- Gridina, M.; Taskina, A.; Lagunov, T.; Nurislamov, A.; Kulikova, T.; Krasikova, A.; Fishman, V. Comparison and Critical Assessment of Single-Cell Hi-C Protocols. Heliyon 2022, 8, e11023. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kabirova, E.; Nurislamov, A.; Shadskiy, A.; Smirnov, A.; Popov, A.; Salnikov, P.; Battulin, N.; Fishman, V. Function and Evolution of the Loop Extrusion Machinery in Animals. Int. J. Mol. Sci. 2023, 24, 5017. https://doi.org/10.3390/ijms24055017
Kabirova E, Nurislamov A, Shadskiy A, Smirnov A, Popov A, Salnikov P, Battulin N, Fishman V. Function and Evolution of the Loop Extrusion Machinery in Animals. International Journal of Molecular Sciences. 2023; 24(5):5017. https://doi.org/10.3390/ijms24055017
Chicago/Turabian StyleKabirova, Evelyn, Artem Nurislamov, Artem Shadskiy, Alexander Smirnov, Andrey Popov, Pavel Salnikov, Nariman Battulin, and Veniamin Fishman. 2023. "Function and Evolution of the Loop Extrusion Machinery in Animals" International Journal of Molecular Sciences 24, no. 5: 5017. https://doi.org/10.3390/ijms24055017
APA StyleKabirova, E., Nurislamov, A., Shadskiy, A., Smirnov, A., Popov, A., Salnikov, P., Battulin, N., & Fishman, V. (2023). Function and Evolution of the Loop Extrusion Machinery in Animals. International Journal of Molecular Sciences, 24(5), 5017. https://doi.org/10.3390/ijms24055017








