Rapid Generation of Barley Homozygous Transgenic Lines Based on Microspore Culture: HvPR1 Overexpression as an Example
Abstract
1. Introduction
2. Results
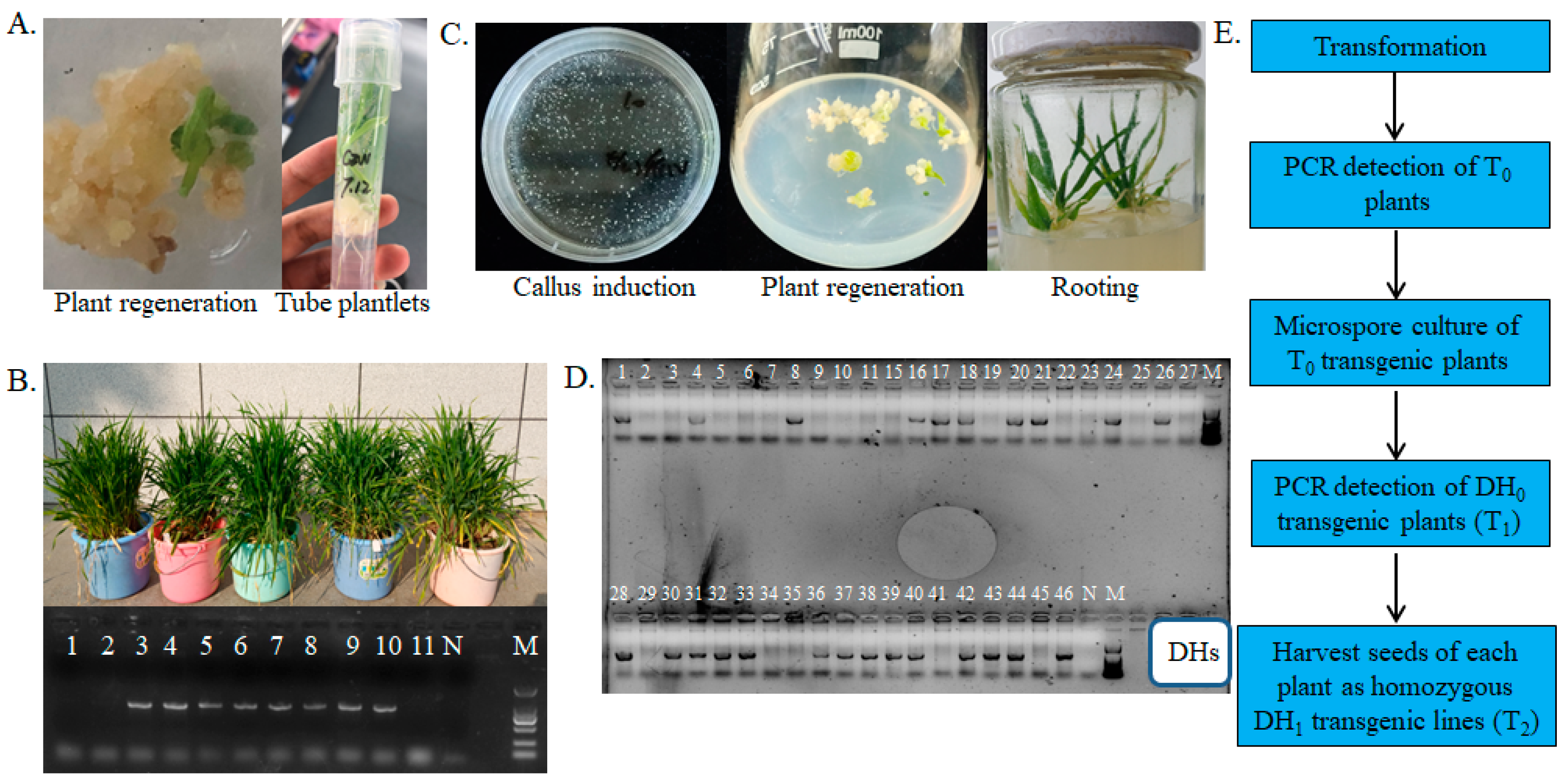
2.1. Plant Transformation and Microspore Culture
2.2. PCR Detection and qPCR Validation of Transgenic Plants
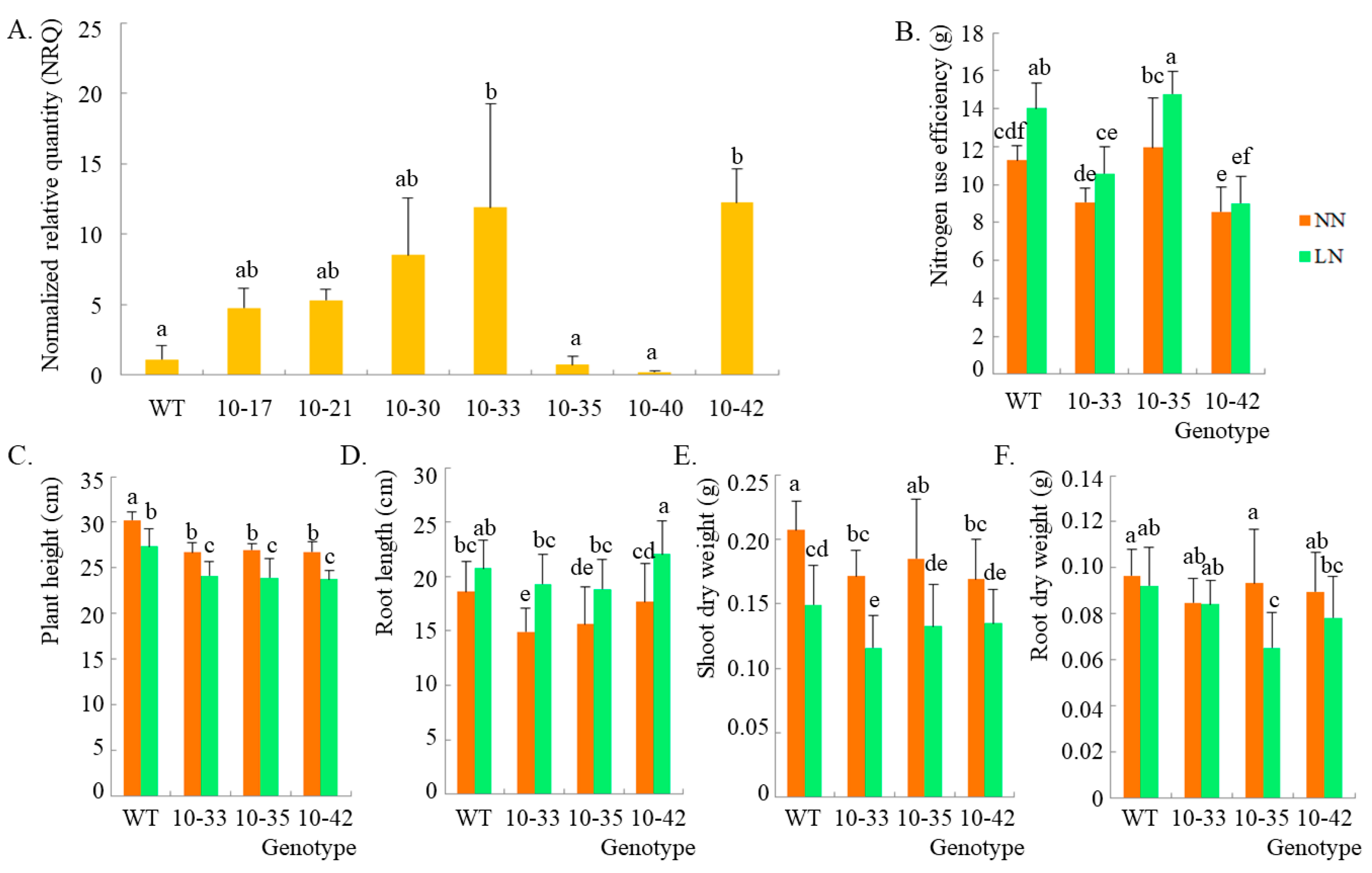
2.3. Phenotypic Identification by Using HvPR1 Overexpression Plants under Low-Nitrogen Treatment
3. Discussion
4. Materials and Methods
4.1. Gene Cloning and Overexpression Vector Construction for the HvPR1 Gene
4.2. Barley Transformation of HvPR1 Gene Mediated with Agrobacterium
4.3. Microspore Culture of Transgenic Barley Plants
4.4. Gene Expression Analysis of HvPR1 by qPCR
4.5. Plant Growth and Low-Nitrogen Treatment
4.6. Biomass and Total Nitrogen Measurement
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, J.; Li, X. Genetic breeding technology and new variety breeding. In Theories and Practices of China Agriculture and Research System (Barley); China Agriculture Press: Beijing, China, 2021. (In Chinese) [Google Scholar]
- Lu, R.; Wang, Y.; Sun, Y.; Shan, L.; Chen, P.; Huang, J. Improvement of isolated microspore culture of barley (Hordeum vulgare L.): The effect of floret co-culture. Plant Cell Tissue Organ Cult. 2008, 93, 21–27. [Google Scholar] [CrossRef]
- Lu, R.; Chen, Z.; Gao, R.; He, T.; Wang, Y.; Xu, H.; Guo, G.; Li, Y.; Liu, C.; Huang, J. Genotypes-independent optimization of nitrogen supply for isolated microspore cultures in barley. Biomed. Res. Int. 2016, 2016, 8. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Guo, G.; Fang, C.; Huang, S.; Chen, J.; Lu, R.; Huang, J.; Fan, X.; Liu, C. Rapid generation of barley mutant lines with high nitrogen uptake efficiency by microspore mutagenesis and field screening. Front. Plant Sci. 2018, 9, 450. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Li, Y.; Gao, R.; Xu, R.; Guo, G.; Lu, R.; Halford, N.G.; Chen, Z.; Liu, C. Rapid Generation and Analysis of a Barley Doubled Haploid Line with Higher Nitrogen Use Efficiency Than Parental Lines by F1 Microspore Embryogenesis. Plants 2021, 10, 1588. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Okamoto, M.; Beatty, P.H.; Rothstein, S.J.; Good, A.G. The genetics of nitrogen use efficiency in crop plants. Annu. Rev. Genet. 2015, 49, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Ray, D.K.; Ramankutty, N.; Mueller, N.D.; West, P.C.; Foley, J.A. Recent patterns of crop yield growth and stagnation. Nat. Commun. 2012, 3, 1293. [Google Scholar] [CrossRef]
- Zhang, Q.; He, K.; Huo, H. Policy: Cleaning China’s air. Nature 2012, 484, 161–162. [Google Scholar] [CrossRef]
- Chen, Z.; Li, Y.; Liu, C.; Wang, Y.; He, T.; Guo, G.; Fang, C.; Gao, R.; Xu, H.; Zhou, L.; et al. Reference gene selection for quantitative RT-PCR normalisation in barley under low-nitrogen stress, based on RNA-seq data. J. Cereal Sci. 2018, 82, 213–215. [Google Scholar] [CrossRef]
- Jähne, A.; Becker, D.; Brettschneider, R.; Lӧrz, H. Regeneration of transgenic, microspore-derived, fertile barley. Theor. Appl. Genet. 1994, 89, 525–533. [Google Scholar] [CrossRef]
- Shim, Y.S.; Kasha, K.J. Barley microspore transformation protocol by biolistic gun. In Doubled Haploid Production in Crop Plants; Maluszynski, M., Kasha, K.J., Forster, B.P., Szarejko, I., Eds.; Springer: Dordrecht, The Netherlands, 2003. [Google Scholar]
- Obert, B.; Middlefell-Williams, J.; Millam, S. Genetic transformation of barley microspores using anther bombardment. Biotechnol. Lett. 2008, 30, 945–949. [Google Scholar] [CrossRef]
- Han, Y.; Broughton, S.; Liu, L.; Zhang, X.Q.; Zeng, J.; He, X.; Li, C. Highly efficient and genotype-independent barley gene editing based on anther culture. Plant Commun. 2020, 2, 100082. [Google Scholar] [CrossRef]
- Ohnoutková, L.; Vlčko, T. Homozygous transgenic barley (Hordeum vulgare L.) plants by anther culture. Plants 2020, 9, 918. [Google Scholar] [CrossRef]
- Esteves, P.; Belzile, F.J. Isolated microspore culture in barley. Methods Mol. Biol. 2019, 1900, 53–71. [Google Scholar]
- Liu, C.; Guo, G.; He, T.; Lu, R.; Huang, J. Establishment of a highly efficient regeneration system of isolated microspores of barley and hulless barley and its utilization in genetic breeding. Barley Cereal Sci. 2018, 35, 1–6, (In Chinese with English abstract). [Google Scholar]
- Ghorbel, M.; Zribi, I.; Missaoui, K.; Drira-Fakhfekh, M.; Azzouzi, B.; Brini, F. Differential regulation of the durum wheat pathogenesis-related protein (PR1) by calmodulin TdCaM1.3 protein. Mol. Biol. Rep. 2021, 48, 347–362. [Google Scholar] [CrossRef]
- Wang, L.; Lu, H.; Zhan, J.; Shang, Q.; Wang, L.; Yin, W.; Sa, W.; Liang, J. Pathogenesis-related protein-4 (PR-4) gene family in Qingke (Hordeum vulgare L. var. nudum): Genome-wide identification, structural analysis and expression profile under stresses. Mol. Biol. Rep. 2022, 49, 9397–9408. [Google Scholar] [CrossRef]
- Sidorenko, L.V.; Lee, T.F.; Woosley, A.; Moskal, W.A.; Bevan, S.A.; Merlo, P.A.O.; Walsh, T.A.; Wang, X.; Weaver, S.; Glancy, T.P.; et al. GC-rich coding sequences reduce transposon-like, small RNA-mediated transgene silencing. Nat. Plants 2017, 3, 875–884. [Google Scholar] [CrossRef]
- Chen, Z.; Jiang, Q.; Jiang, P.; Zhang, W.; Huang, J.; Liu, C.; Halford, N.G.; Lu, R. Novel low-nitrogen stress-responsive long non-coding RNAs (lncRNA) in barley landrace B968 (Liuzhutouzidamai) at seedling stage. BMC Plant Biol. 2020, 20, 142. [Google Scholar] [CrossRef]
- He, J.; Zhang, C.; Dai, H.; Liu, H.; Zhang, X.; Yang, J.; Chen, X.; Zhu, Y.; Wang, D.; Qi, X.; et al. A LysM receptor heteromer mediates perception of arbuscular mycorrhizal symbiotic signal in rice. Mol. Plant 2019, 12, 1561–1576. [Google Scholar] [CrossRef]
- Shen, Q.F.; Fu, L.B.; Su, T.T.; Ye, L.Z.; Huang, L.; Kuang, L.H.; Wu, L.Y.; Wu, D.Z.; Chen, Z.H.; Zhang, G.P. Calmodulin HvCaM1 negatively regulates salt tolerance via modulation of HvHKT1s and HvCAMTA4. Plant Physiol. 2020, 183, 1650–1662. [Google Scholar] [CrossRef]
- Chen, Z.; Liu, C.; Wang, Y.; He, T.; Gao, R.; Xu, H.; Guo, G.; Li, Y.; Zhou, L.; Lu, R.; et al. Expression analysis of nitrogen metabolism-related genes reveals differences in adaptation to low-nitrogen stress between two different barley cultivars at seedling stage. Int. J. Genom. 2018, 2018, 8152860. [Google Scholar] [CrossRef] [PubMed]
- Chardon, F.; Barthélémy, J.; Daniel-Vedele, F.; Masclaux-Daubresse, C. Natural variation of nitrate uptake and nitrogen use efficiency in Arabidopsis thaliana cultivated with limiting and ample nitrogen supply. J. Exp. Bot. 2010, 61, 2293–2302. [Google Scholar] [CrossRef] [PubMed]
- Decouard, B.; Bailly, M.; Rigault, M.; Marmagne, A.; Arkoun, M.; Soulay, F.; Caïus, J.; Paysant-Le, R.C.; Louahlia, S.; Jacquard, C.; et al. Genotypic variation of nitrogen use efficiency and amino acid metabolism in barley. Front. Plant Sci. 2022, 12, 807798. [Google Scholar] [CrossRef] [PubMed]
| Description | Primer Name | Sequence (5′-3′) | Origin |
|---|---|---|---|
| HvPR1 cloning and overexpression vector construction | HvPR1-F | TTATACACCGAACCGAGAATG | This study |
| HvPR1-R | GCATCACGGTTAGTATGGTTT | ||
| HvPR1-BamH I-F | CGGGATCCATGCAGACGCCCAAGCTAG | ||
| HvPR1-EcoR I-R | GGAATTCTTAGTATGGTTTCTGTCCAATGATA | ||
| HvPR1-FLAG-EcoR I-R | GGAATTCGTATGGTTTCTGTCCAATGATATTC | ||
| Transgenic detection | Hyg-F | TGAAAAAGCCTGAACTCACCG | Ertao Wang’s lab |
| Hyg-R | TATTTCTTTGCCCTCGGACG | ||
| qPCR | HvPR1-q-F | ATCAACGACTGCAAGCTCCA | This study |
| HvPR1-q-R | GTCCTTCTTCTCGCTCACCC | ||
| HvTUBB6-F | TCCCGAACAATGTCAAGTCA | [9] | |
| HvTUBB6-R | GTGGAGTTGCCAATGAAGGT | ||
| HvGAPDH-F | AAGCATGAAGATACAGGGAGTGTG | ||
| HvGAPDH-R | AAATTTATTCTCGGAAGAGGTTGTACA |
| Code of Transgenic T0 Lines | Number of Spikes for Anther Separation | Tubes of Separated Anthers | Petri Dishes within Microspore Mixtures | Petri Dishes with Inductive Calli | Number of Regenerated Plants |
|---|---|---|---|---|---|
| T0-3 | 44 | 4 | Contaminated during callus induction | ||
| T0-4 | 41 | 5 | 3 smaller Petri dishes | No calli | |
| T0-5 | 29 | 3 | 1 bigger Petri dish and 2 smaller Petri dishes | Less than 15 mg and 5 mg callus per dish of the bigger Petri dish and the 2 smaller Petri dishes, respectively | No plants |
| T0-6 | 47 | 5 | 5 bigger Petri dishes | Less than 15 mg callus per dish of the 5 bigger Petri dishes | 1 plant |
| T0-7 | 15 | 2 | Contaminated during callus induction | ||
| T0-8 | 42 | 4 | 1 bigger Petri dish and 3 smaller Petri dishes | Less than 15 mg and 5 mg callus per dish of the bigger Petri dish and the 3 smaller Petri dish, respectively | No plants |
| T0-9 | 25 | 3 | 1 bigger Petri dish | Less than 15 mg callus per dish of the bigger Petri dish | Contaminated during differentiation |
| T0-10 | 24 | 4 | 4 bigger Petri dishes | Approximately 90 mg callus per dish of 3 of the 4 bigger Petri dishes | 46 plants |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Z.; Jiang, Q.; Guo, G.; Shen, Q.; Yang, J.; Wang, E.; Zhang, G.; Lu, R.; Liu, C. Rapid Generation of Barley Homozygous Transgenic Lines Based on Microspore Culture: HvPR1 Overexpression as an Example. Int. J. Mol. Sci. 2023, 24, 4945. https://doi.org/10.3390/ijms24054945
Chen Z, Jiang Q, Guo G, Shen Q, Yang J, Wang E, Zhang G, Lu R, Liu C. Rapid Generation of Barley Homozygous Transgenic Lines Based on Microspore Culture: HvPR1 Overexpression as an Example. International Journal of Molecular Sciences. 2023; 24(5):4945. https://doi.org/10.3390/ijms24054945
Chicago/Turabian StyleChen, Zhiwei, Qi Jiang, Guimei Guo, Qiufang Shen, Jun Yang, Ertao Wang, Guoping Zhang, Ruiju Lu, and Chenghong Liu. 2023. "Rapid Generation of Barley Homozygous Transgenic Lines Based on Microspore Culture: HvPR1 Overexpression as an Example" International Journal of Molecular Sciences 24, no. 5: 4945. https://doi.org/10.3390/ijms24054945
APA StyleChen, Z., Jiang, Q., Guo, G., Shen, Q., Yang, J., Wang, E., Zhang, G., Lu, R., & Liu, C. (2023). Rapid Generation of Barley Homozygous Transgenic Lines Based on Microspore Culture: HvPR1 Overexpression as an Example. International Journal of Molecular Sciences, 24(5), 4945. https://doi.org/10.3390/ijms24054945










